Acinetobacter baumannii Antibiotic Resistance Mechanisms
Abstract
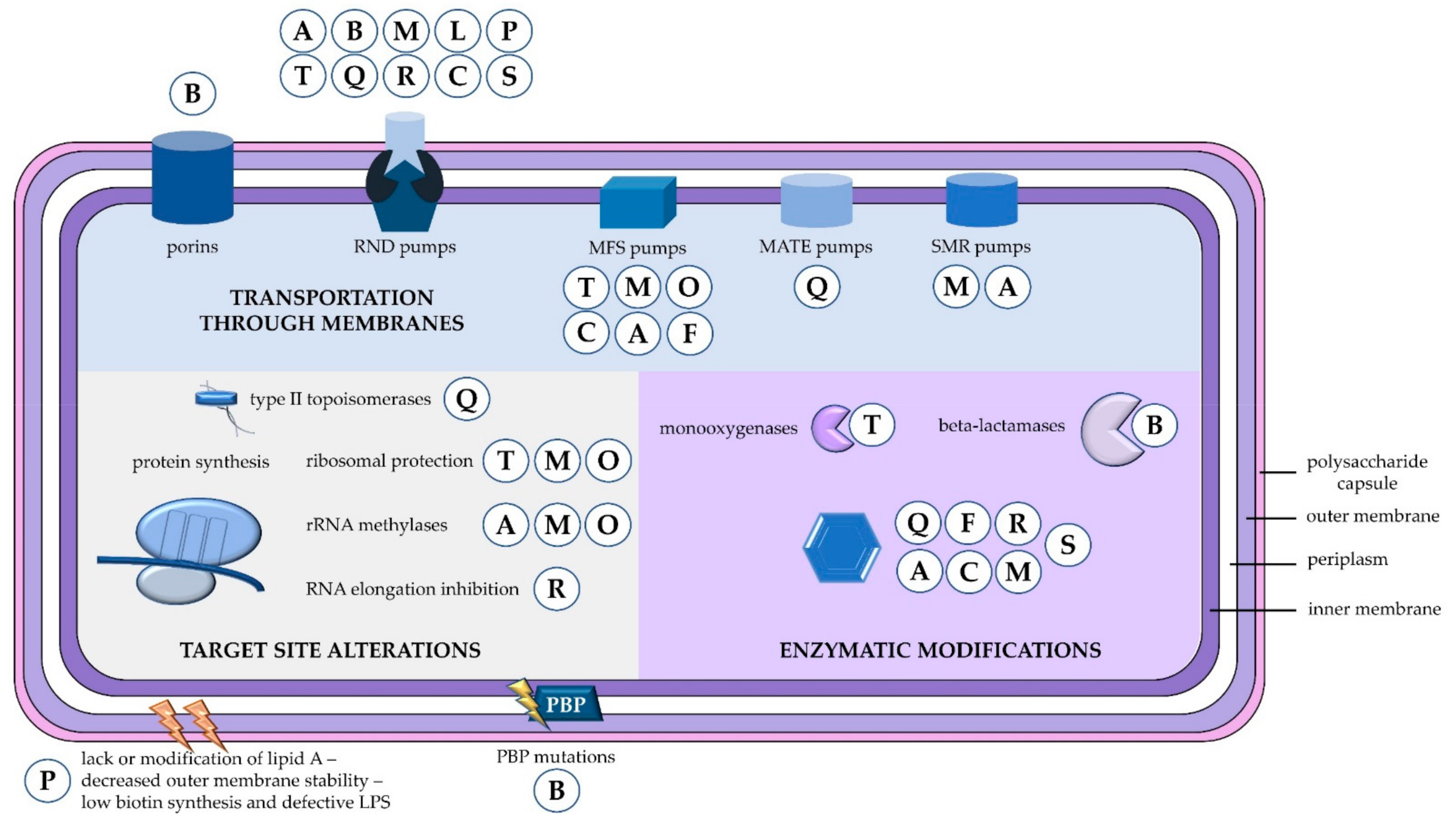
:1. Introduction
2. Resistance to Beta-Lactams
2.1. Beta-Lactamases
2.1.1. Class A
2.1.2. Class B
2.1.3. Class C
2.1.4. Class D
2.2. Outer Membrane Proteins
2.3. Efflux Pumps
2.4. Penicillin-Binding Proteins
3. Resistance to Aminoglycosides
4. Resistance to Tetracyclines
5. Resistance to Fluoroquinolones
6. Resistance to Macrolides—Lincosamides—Streptogramin Antibiotics
7. Resistance to Polymyxins
8. Resistance to Others
8.1. Resistance to Amphenicols—Oxazolidinones
8.2. Resistance to Glycopeptide and Lipopetide Antibiotics
8.3. Resistance to Rifamycins
8.4. Resistance to Fosfomycin
8.5. Resistance to Diaminopyrimidines—Sulfonamides
9. SARS-CoV-2 and Resistant A. baumannii Coinfections
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [Green Version]
- Vrancianu, C.O.; Gheorghe, I.; Czobor, I.B.; Chifiriuc, M.C. Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of acinetobacter baumannii. Microorganisms 2020, 8, 935. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [Green Version]
- Levy-Blitchtein, S.; Roca, I.; Plasencia-Rebata, S.; Vicente-Taboada, W.; Velásquez-Pomar, J.; Muñoz, L.; Moreno-Morales, J.; Pons, M.J.; Del Valle-Mendoza, J.; Vila, J. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru article. Emerg. Microbes Infect. 2018, 7, 119. [Google Scholar] [CrossRef] [Green Version]
- Nasr, P. Genetics, epidemiology, and clinical manifestations of multidrugresistant Acinetobacter baumannii. J. Hosp. Infect. 2020, 104, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Murray, G.L.; Peleg, A.Y. Acinetobacter baumannii: Evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015, 36, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Kyriakidis, I.; Palabougiouki, M.; Vasileiou, E.; Tragiannidis, A.; Stamou, M.; Moudiou, T.; Vyzantiadis, T.; Gombakis, N.; Hatzilianou, M. Candidemia complicating biliary atresia in an infant with hemoglobinopathy. Turk. Pediatri. Ars. 2019, 54, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Mohd Sazlly Lim, S.; Zainal Abidin, A.; Liew, S.M.; Roberts, J.A.; Sime, F.B. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J. Infect. 2019, 79, 593–600. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2019; ECDC: Stockholm, Sweden, 2020. [Google Scholar]
- Moghnieh, R.A.; Kanafani, Z.A.; Tabaja, H.Z.; Sharara, S.L.; Awad, L.S.; Kanj, S.S. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect. Dis. 2018, 18, e379–e394. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis 2019, 69, S521–S528. [Google Scholar] [CrossRef] [Green Version]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Garnacho-Montero, J.; Timsit, J.F. Managing Acinetobacter baumannii infections. Curr. Opin. Infect. Dis. 2019, 32, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: An approach based on the mechanisms of resistance to carbapenems. Infection 2020, 48, 835–851. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Pakharukova, N.; Tuittila, M.; Paavilainen, S.; Malmi, H.; Parilova, O.; Teneberg, S.; Knight, S.D.; Zavialov, A.V. Structural basis for Acinetobacter baumannii biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 5558–5563. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Su, P.W.; Moi, S.H.; Chuang, L.Y. Biofilm formation in Acinetobacter baumannii: Genotype-phenotype correlation. Molecules 2019, 24, 1849. [Google Scholar] [CrossRef] [Green Version]
- Krzyściak, P.; Chmielarczyk, A.; Pobiega, M.; Romaniszyn, D.; Wójkowska-Mach, J. Acinetobacter baumannii isolated from hospital-acquired infection: Biofilm production and drug susceptibility. APMIS 2017, 125, 1017–1026. [Google Scholar] [CrossRef]
- Green, D.W. The bacterial cell wall as a source of antibacterial targets. Expert. Opin. Ther. Targets 2002, 6, 1–19. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Corvec, S.; Rapoport, M.; Mugnier, P.; Petroni, A.; Pasteran, F.; Faccone, D.; Galas, M.; Drugeon, H.; Cattoir, V.; et al. Identification of the Novel Narrow-Spectrum β-Lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 2007, 51, 2179–2184. [Google Scholar] [CrossRef] [Green Version]
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended spectrum beta-lactamases: Definition, classification and epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Smiline, A.S.G.; Vijayashree, J.P.; Paramasivam, A. Molecular characterization of plasmid-encoded blaTEM, blaSHV and blaCTX-M among extended spectrum β-lactamases [ESBLs] producing Acinetobacter baumannii. Br. J. Biomed. Sci. 2018, 75, 200–202. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Nordmann, P.; Potron, A.; Lecuyer, H.; Zahar, J.R.; Poirel, L. Carbapenem-hydrolyzing GES-type extended-spectrum β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 349–354. [Google Scholar] [CrossRef] [Green Version]
- Moubareck, C.; Brémont, S.; Conroy, M.C.; Courvalin, P.; Lambert, T. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 3579–3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnin, R.A.; Potron, A.; Poirel, L.; Lecuyer, H.; Neri, R.; Nordmann, P. PER-7, an extended-spectrum β-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 2424–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirel, L.; Menuteau, O.; Agoli, N.; Cattoen, C.; Nordmann, P. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 2003, 41, 3542–3547. [Google Scholar] [CrossRef] [Green Version]
- Walther-Rasmussen, J.; Høiby, N. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum β-lactamases. Can. J. Microbiol. 2004, 50, 137–165. [Google Scholar] [CrossRef]
- Martinez, T.; Martinez, I.; Vazquez, G.J.; Aquino, E.E.; Robledo, I.E. Genetic environment of the KPC gene in Acinetobacter baumannii ST2 clone from Puerto Rico and genomic insights into its drug resistance. J. Med. Microbiol. 2016, 65, 784–792. [Google Scholar] [CrossRef]
- Cornaglia, G.; Giamarellou, H.; Rossolini, G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011, 11, 381–393. [Google Scholar] [CrossRef]
- Shivaprasad, A.; Antony, B.; Shenoy, P. Comparative evaluation of four phenotypic tests for detection of Metallo-β-lactamase and Carbapenemase production in Acinetobacter baumannii. J. Clin. Diagn. Res. 2014, 8, 5–8. [Google Scholar] [CrossRef]
- Ikonomidis, A.; Ntokou, E.; Maniatis, A.N.; Tsakris, A.; Pournaras, S. Hidden VIM-1 metallo-β-lactamase phenotypes among Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 2008, 46, 346–349. [Google Scholar] [CrossRef] [Green Version]
- Moulana, Z.; Babazadeh, A.; Eslamdost, Z.; Shokri, M.; Ebrahimpour, S. Phenotypic and genotypic detection of metallo-beta-lactamases in Carbapenem resistant Acinetobacter baumannii. Casp. J. Intern. Med. 2020, 11, 171–176. [Google Scholar] [CrossRef]
- Chatterjee, S.; Datta, S.; Roy, S.; Ramanan, L.; Saha, A.; Viswanathan, R.; Som, T.; Basu, S. Carbapenem resistance in Acinetobacter baumannii and other Acinetobacter spp. causing neonatal sepsis: Focus on NDM-1 and its linkage to ISAba125. Front. Microbiol. 2016, 7, 1126. [Google Scholar] [CrossRef] [Green Version]
- Amin, M.; Navidifar, T.; Shooshtari, F.S.; Goodarzi, H. Association of the genes encoding metallo-β-lactamase with the presence of integrons among multidrug-resistant clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 2019, 12, 1171–1180. [Google Scholar] [CrossRef] [Green Version]
- El-Ageery, S.M.; Al-Hazmi, S.S. Microbiological and molecular detection of VIM-1 metallo beta lactamase-producing Acinetobacter baumannii. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 965–970. [Google Scholar] [PubMed]
- López, C.; Ayala, J.A.; Bonomo, R.A.; González, L.J.; Vila, A.J. Protein determinants of dissemination and host specificity of metallo-β-lactamases. Nat. Commun. 2019, 10, 3617. [Google Scholar] [CrossRef]
- Héritier, C.; Poirel, L.; Nordmann, P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 2006, 12, 123–130. [Google Scholar] [CrossRef] [Green Version]
- Hamidian, M.; Hall, R.M. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J. Antimicrob. Chemother. 2013, 68, 2682–2683. [Google Scholar] [CrossRef] [Green Version]
- Lopes, B.S.; Amyes, S.G.B. Role of ISAba1 and ISAba125 in governing the expression of bla ADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J. Med. Microbiol. 2012, 61, 1103–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamidian, M.; Hancock, D.P.; Hall, R.M. Horizontal transfer of an ISAba125-activated ampC gene between Acinetobacter baumannii strains leading to cephalosporin resistance. J. Antimicrob. Chemother. 2013, 68, 244–245. [Google Scholar] [CrossRef] [Green Version]
- Hamidian, M.; Hall, R.M. Tn6168, a transposon carrying an ISAba1-activated ampC gene and conferring cephalosporin resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2014, 69, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Couchoud, C.; Bertrand, X.; Valot, B.; Hocquet, D. Deciphering the role of insertion sequences in the evolution of bacterial epidemic pathogens with panisa software. Microb. Genom. 2020, 6, e000356. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, J.M.; Poirel, L.; Nordmann, P. Genetic and functional variability of AmpC-type β-lactamases from Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4930–4933. [Google Scholar] [CrossRef] [Green Version]
- Tian, G.B.; Adams-Haduch, J.M.; Taracila, M.; Bonomo, R.A.; Wang, H.N.; Doi, Y. Extended-spectrum AmpC cephalosporinase in Acinetobacter baumannii: ADC-56 confers resistance to cefepime. Antimicrob. Agents Chemother. 2011, 55, 4922–4925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingti, B.; Upadhyay, S.; Hazarika, M.; Khyriem, A.B.; Paul, D.; Bhattacharya, P.; Joshi, S.R.; Bora, D.; Dhar, D.; Bhattacharjee, A. Distribution of carbapenem resistant Acinetobacter baumannii with blaADC-30 and induction of ADC-30 in response to beta-lactam antibiotics. Res. Microbiol. 2020, 171, 128–133. [Google Scholar] [CrossRef]
- Kuo, S.C.; Lee, Y.T.; Lauderdale, T.L.Y.; Huang, W.C.; Chuang, M.F.; Chen, C.P.; Su, S.C.; Lee, K.R.; Chen, T.L. Contribution of Acinetobacter-derived cephalosporinase-30 to sulbactam resistance in Acinetobacter baumannii. Front. Microbiol. 2015, 6, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, J.H.; Yang, J.T.; Chern, J.; Chen, T.L.; Wu, W.L.; Liao, J.H.; Tsai, S.F.; Liang, S.Y.; Chou, C.C.; Wu, S.H. Comparative phosphoproteomics reveals the role of AmpC β-lactamase phosphorylation in the clinical imipenem-resistant strain Acinetobacter baumannii SK17. Mol. Cell Proteom. 2016, 15, 12–25. [Google Scholar] [CrossRef] [Green Version]
- Antunes, N.T.; Fisher, J.F. Acquired class D β-Lactamases. Antibiotics 2014, 3, 398–434. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nordmann, P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 2010, 54, 24–38. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.H.-Y.; Chan, B.K.-W.; Chan, E.W.-C.; Chen, S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ß-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019, 10, 2809. [Google Scholar] [CrossRef] [Green Version]
- Ewers, C.; Klotz, P.; Leidner, U.; Stamm, I.; Prenger-Berninghoff, E.; Göttig, S.; Semmler, T.; Scheufen, S. OXA-23 and ISAba1–OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int. J. Antimicrob. Agents 2017, 49, 37–44. [Google Scholar] [CrossRef]
- Morakchi, H.; Loucif, L.; Gacemi-Kirane, D.; Rolain, J.M. Molecular characterisation of carbapenemases in urban pigeon droppings in France and Algeria. J. Glob. Antimicrob. Resist. 2017, 9, 103–110. [Google Scholar] [CrossRef]
- Sugawara, E.; Nikaido, H. OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J. Bacteriol. 2012, 194, 4089–4096. [Google Scholar] [CrossRef] [Green Version]
- Jyothisri, K.; Deepak, V.; Rajeswari, M.R. Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii. FEBS Lett. 1999, 443, 57–60. [Google Scholar] [CrossRef] [Green Version]
- Smani, Y.; Fàbrega, A.; Roca, I.; Sańchez-Encinales, V.; Vila, J.; Pachón, J. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 1806–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, R.; Moussa, S.H.; Durand-Réville, T.F.; Tommasi, R.; Miller, A. Acinetobacter baumannii OmpA Is a Selective Antibiotic Permeant Porin. ACS Infect. Dis. 2018, 4, 373–381. [Google Scholar] [CrossRef]
- Kwon, H.I.; Kim, S.; Oh, M.H.; Na, S.H.; Kim, Y.J.; Jeon, Y.H.; Lee, J.C. Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 2017, 72, 3012–3015. [Google Scholar] [CrossRef]
- Zhong, X.; Wu, X.; Schweppe, D.K.; Chavez, J.D.; Mathay, M.; Eng, J.K.; Keller, A.; Bruce, J.E. In Vivo Cross-Linking MS Reveals Conservation in OmpA Linkage to Different Classes of β-Lactamase Enzymes. J Am. Soc. Mass Spectrom. 2020, 31, 190–195. [Google Scholar] [CrossRef]
- Limansky, A.S.; Mussi, M.A.; Viale, A.M. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 2002, 40, 4776–4778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mussi, M.A.; Limansky, A.S.; Viale, A.M. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: Natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 2005, 49, 1432–1440. [Google Scholar] [CrossRef] [Green Version]
- Mussi, M.A.; Relling, V.M.; Limansky, A.S.; Viale, A.M. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for l-ornithine uptake. FEBS Lett. 2007, 581, 5573–5578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mussi, M.A.; Limansky, A.S.; Relling, V.; Ravasi, P.; Arakaki, A.; Actis, L.A.; Viale, A.M. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J. Bacteriolnal. Bacteriol. 2011, 193, 4736–4748. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.W.; Cheong, H.J.; Kim, W.J.; Kim, M.J.; Song, K.J.; Song, J.W.; Kim, H.S.; Roh, K.H. Loss of the 29-kilodalton outer membrane protein in the presence of OXA-51-like enzymes in Acinetobacter baumannii is associated with decreased imipenem susceptibility. Microb. Drug Resist. 2009, 15, 151–158. [Google Scholar] [CrossRef]
- Khorsi, K.; Messai, Y.; Ammari, H.; Hamidi, M.; Bakour, R. ISAba36 inserted into the outer membrane protein gene carO and associated with the carbapenemase gene blaOXA-24-like in Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2018, 15, 107–108. [Google Scholar] [CrossRef]
- Zhu, L.J.; Chen, X.Y.; Hou, P.F. Mutation of CarO participates in drug resistance in imipenem-resistant Acinetobacter baumannii. J. Clin. Lab. Anal. 2019, 33, e22976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahn, M.; D’Agostino, T.; Eren, E.; Baslé, A.; Ceccarelli, M.; Van Berg, B.D. Small-molecule transport by CarO, an abundant eight-stranded β-barrel outer membrane protein from Acinetobacter baumannii. J. Mol. Biol. 2015, 427, 2329–2339. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, E.L.; Scheidegger, E.; Freitas, F.S.; Cipriano, R.; Vicente, A.C.P. Carbapenem-resistant Acinetobacter baumannii from Brazil: Role of carO alleles expression and blaOXA-23 gene. BMC Microbiol. 2013, 13, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catel-Ferreira, M.; Coadou, G.; Molle, V.; Mugnier, P.; Nordmann, P.; Siroy, A.; Jouenne, T.; Dé, E. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J. Antimicrob. Chemother. 2011, 66, 2053–2056. [Google Scholar] [CrossRef] [Green Version]
- Siroy, A.; Molle, V.; Lemaître-Guillier, C.; Vallenet, D.; Pestel-Caron, M.; Cozzone, A.J.; Jouenne, T.; Dé, E. Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 4876–4883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, V.B.; Vaidyanathan, V.; Rajamohan, G. AbuO, a tolc-like outer membrane protein of Acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance. Antimicrob. Agents Chemother. 2015, 59, 1236–1245. [Google Scholar] [CrossRef] [Green Version]
- Clark, R.B. Imipenem resistance among Acinetobacter baumannii: Association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 1996, 38, 245–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomás, M.D.M.; Beceiro, A.; Pérez, A.; Velasco, D.; Moure, R.; Villanueva, R.; Martínez-Beltrán, J.; Bou, G. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 5172–5175. [Google Scholar] [CrossRef] [Green Version]
- Zahn, M.; Bhamidimarri, S.P.; Baslé, A.; Winterhalter, M.; Van Den Berg, B. Structural Insights into Outer Membrane Permeability of Acinetobacter baumannii. Structure 2016, 24, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Catel-Ferreira, M.; Nehmé, R.; Molle, V.; Aranda, J.; Bouffartigues, E.; Chevalier, S.; Bou, G.; Jouenne, T.; Dé, E. Deciphering the function of the outer membrane protein OprD homologue of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2012, 56, 3826–3832. [Google Scholar] [CrossRef] [Green Version]
- Cecchini, T.; Yoon, E.J.; Charretier, Y.; Bardet, C.; Beaulieu, C.; Lacoux, X.; Docquier, J.D.; Lemoine, J.; Courvalin, P.; Grillot-Courvalin, C.; et al. Deciphering multifactorial resistance phenotypes in Acinetobacter baumannii by genomics and targeted label-free proteomics. Mol. Cellul Proteom. 2018, 17, 442–456. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Sun, L.; Xu, G.; Xia, T. Differential susceptibility to carbapenems due to the AdeABC efflux pump among nosocomial outbreak isolates of Acinetobacter baumannii in a Chinese hospital. Diagn. Microbiol. Infect. Dis. 2008, 62, 326–332. [Google Scholar] [CrossRef]
- Hawkey, J.; Ascher, D.B.; Judd, L.M.; Wick, R.R.; Kostoulias, X.; Cleland, H.; Spelman, D.W.; Padiglione, A.; Peleg, A.Y.; Holt, K.E. Evolution of carbapenem resistance in Acinetobacter baumannii during a prolonged infection. Microb. Genom. 2018, 4, e000165. [Google Scholar] [CrossRef] [Green Version]
- Magnet, S.; Courvalin, P.; Lambert, T. Resistance-Nodulation-Cell Division-Type Efflux Pump Involved in Aminoglycoside Resistance in Acinetobacter baumannii Strain BM4454. Antimicrob. Agents Chemother. 2001, 45, 3375–3380. [Google Scholar] [CrossRef] [Green Version]
- Vila, J.; Martí, S.; Sánchez-Céspedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef] [Green Version]
- Marchand, I.; Marchand, I.; Damier-piolle, L.; Damier-piolle, L.; Courvalin, P.; Courvalin, P.; Lambert, T.; Lambert, T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 2004, 48, 3298–3304. [Google Scholar] [CrossRef] [Green Version]
- Leus, I.V.; Weeks, J.W.; Bonifay, V.; Smith, L.; Richardson, S.; Zgurskaya, H.I. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 2018, 200, e00049-18. [Google Scholar] [CrossRef] [Green Version]
- Hou, P.F.; Chen, X.Y.; Yan, G.F.; Wang, Y.P.; Ying, C.M. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy 2012, 58, 152–158. [Google Scholar] [CrossRef]
- Yoon, E.J.; Chabane, Y.N.; Goussard, S.; Snesrud, E.; Courvalin, P.; Dé, E.; Grillot-Courvalin, C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 2015, 6, e00309-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [Green Version]
- Zapun, A.; Contreras-Martel, C.; Vernet, T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayô, R.; Rodríguez, M.C.; Espinal, P.; Fernández-Cuenca, F.; Ocampo-Sosa, A.A.; Pascual, Á.; Ayala, J.A.; Vila, J.; Martínez-Martínez, L. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 5907–5913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehrlein, M.; Leying, H.; Cullmann, W.; Wendt, S.; Opferkuch, W. Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy 1991, 37, 405–412. [Google Scholar] [CrossRef]
- Yun, S.H.; Choi, C.W.; Kwon, S.O.; Park, G.W.; Cho, K.; Kwon, K.H.; Kim, J.Y.; Yoo, J.S.; Lee, J.C.; Choi, J.S.; et al. Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J. Proteome Res. 2011, 10, 459–469. [Google Scholar] [CrossRef]
- Fernández-Cuenca, F.; Martínez-Martínez, L.; Conejo, M.C.; Ayala, J.A.; Perea, E.J.; Pascual, A. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2003, 51, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vashist, J.; Tiwari, V.; Das, R.; Kapil, A.; Rajeswari, M.R. Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant Acinetobacter baumannii. Indian J. Med. Res. 2011, 133, 332–338. [Google Scholar] [PubMed]
- Siroy, A.; Cosette, P.; Seyer, D.; Lemaître-Guillier, C.; Vallenet, D.; Van Dorsselaer, A.; Boyer-Mariotte, S.; Jouenne, T.; Dé, E. Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J. Proteome Res. 2006, 5, 3385–3398. [Google Scholar] [CrossRef] [PubMed]
- Pathogen Detection Microbial Browser for Identification of Genetic and Genomic Elements (MicroBIGG-E)—Pathogen Detection—NCBI. Available online: https://www.ncbi.nlm.nih.gov/pathogens/microbigge/# (accessed on 5 March 2021).
- Vázquez-López, R.; Solano-Gálvez, S.G.; Vignon-Whaley, J.J.J.; Vaamonde, J.A.A.; Alonzo, L.A.P.; Reséndiz, A.R.; Álvarez, M.M.; López, E.N.V.; Franyuti-Kelly, G.; Álvarez-Hernández, D.A.; et al. Acinetobacter baumannii resistance: A real challenge for clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. MedChemComm 2016, 7, 11–27. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat 2010, 13, 151–171. [Google Scholar] [CrossRef] [Green Version]
- Aínsa, J.A.; Martin, C.; Gicquel, B.; Gomez-Lus, R. Characterization of the chromosomal aminoglycoside 2’-N- acetyltransferase gene from Mycobacterium fortuitum. Antimicrob. Agents Chemother. 1996, 40, 2350–2355. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, J.; Yu, P.; Ge, P.; Jiang, Y.; Xu, R.; Chen, R.; Liu, X. Identification of antibiotic resistance genes in the multidrug-resistant Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int. J. Mol. Med. 2017, 39, 364–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheikhalizadeh, V.; Hasani, A.; Rezaee, M.A.; Rahmati-Yamchi, M.; Hasani, A.; Ghotaslou, R.; Goli, H.R. Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J. Infect. Chemother. 2017, 23, 74–79. [Google Scholar] [CrossRef]
- Jana, S.; Deb, J.K. Molecular understanding of aminoglycoside action and resistance. Appl. Microbiol. Biotechnol. 2006, 70, 140–150. [Google Scholar] [CrossRef]
- De Silva, P.M.; Kumar, A. Signal transduction proteins in Acinetobacter baumannii: Role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019, 10, 49. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Bilya, S.R.; Xu, W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef]
- Xu, A.; Zhu, H.; Gao, B.; Weng, H.; Ding, Z.; Li, M.; Weng, X.; He, G. Diagnosis of severe community-acquired pneumonia caused by Acinetobacter baumannii through next-generation sequencing: A case report. BMC Infect. Dis. 2020, 20, 45. [Google Scholar] [CrossRef]
- Chukwudi, C.U. rRNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 2016, 60, 4433–4441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warburton, P.J.; Amodeo, N.; Roberts, A.P. Mosaic tetracycline resistance genes encoding ribosomal protection proteins. J. Antimicrob. Chemother. 2016, 71, 3333–3339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, X.; Pan, C.; Zhu, L.; Liu, Z.; Xu, Q.; Wang, H.; Yu, Y. Complete genome sequence of Acinetobacter baumannii A1296 (ST1469) with a small plasmid harbouring the tet(39) tetracycline resistance gene. J. Glob. Antimicrob. Resist. 2017, 11, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Rumbo, C.; Gato, E.; López, M.; Ruiz De Alegría, C.; Fernández-Cuenca, F.; Martínez-Martínez, L.; Vila, J.; Pachón, J.; Cisneros, J.M.; Rodríguez-Baño, J.; et al. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 5247–5257. [Google Scholar] [CrossRef] [Green Version]
- Nikolakopoulou, T.L.; Giannoutsou, E.P.; Karabatsou, A.A.; Karagouni, A.D. Prevalence of tetracycline resistance genes in Greek seawater habitats. J. Microbiol. 2008, 46, 633–640. [Google Scholar] [CrossRef]
- Lin, L.; Wang, S.F.; Yang, T.Y.; Hung, W.C.; Chan, M.Y.; Tseng, S.P. Antimicrobial resistance and genetic diversity in ceftazidime non-susceptible bacterial pathogens from ready-to-eat street foods in three Taiwanese cities. Sci. Rep. 2017, 7, 15515. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Kehrenberg, C.; Ulep, C.; Schwarz, S.; Roberts, M.C. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 2003, 47, 883–888. [Google Scholar] [CrossRef] [Green Version]
- Yuhan, Y.; Ziyun, Y.; Yongbo, Z.; Fuqiang, L.; Qinghua, Z. Over expression of AdeABC and AcrAB-TolC efflux systems confers tigecycline resistance in clinical isolates of Acinetobacter baumannii and Klebsiella pneumoniae. Rev. Soc. Bras. Med. Trop. 2016, 49, 165–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huys, G.; Cnockaert, M.; Vaneechoutte, M.; Woodford, N.; Nemec, A.; Dijkshoorn, L.; Swings, J. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res. Microbiol. 2005, 156, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Foong, W.E.; Wilhelm, J.; Tam, H.K.; Pos, K.M. Tigecycline efflux in Acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. J. Antimicrob. Chemother. 2020, 75, 1135–1139. [Google Scholar] [CrossRef]
- Dönhöfer, A.; Franckenberg, S.; Wickles, S.; Berninghausen, O.; Beckmann, R.; Wilson, D.N. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 16900–16905. [Google Scholar] [CrossRef] [Green Version]
- Ribera, A.; Ruiz, J.; Vila, J. Presence of the tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2003, 47, 2310–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Hassan, K.A.; Tetu, S.G.; Naidu, V.; Pokhrel, A.; Cain, A.K.; Paulsen, I.T. The Transcriptomic Signature of Tigecycline in Acinetobacter baumannii. Front. Microbiol. 2020, 11, 565438. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of Plasmid-Mediated High-Level Tigecycline Resistance Genes in Animals and Humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Lv, Y.; Cui, L.; Li, Y.; Li, T.; Song, H.; Hao, Y.; Shen, J.; Wang, Y.; et al. Novel Plasmid-Mediated tet(X5) Gene Conferring Resistance to Tigecycline, Eravacycline, and Omadacycline in a Clinical Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 2020, 64, e01326-19. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Su, X.Z.; Chen, J.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 2005, 49, 4362–4364. [Google Scholar] [CrossRef] [Green Version]
- Perez, F.; Hujer, A.M.; Hujer, K.M.; Decker, B.K.; Rather, P.N.; Bonomo, R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 3471–3484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaki, M.E.S.; Abou ElKheir, N.; Mofreh, M. Molecular Study of Quinolone Resistance Determining Regions of gyrA Gene and parC Genes in Clinical Isolates of Acintobacter baumannii Resistant to Fluoroquinolone. Open Microbiol. J. 2018, 12, 116–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.H.; Deng, Y.T.; Zeng, Z.L.; Gao, J.H.; Chen, L.; Arakawa, Y.; Chen, Z.L. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob. Agents Chemother. 2008, 52, 2992–2993. [Google Scholar] [CrossRef] [Green Version]
- Lari, A.R.; Ardebili, A.; Hashemi, A. Ader-ades mutations & overexpression of the AdeABC efflux system in ciprofloxacin-resistant Acinetobacter baumannii clinical isolates. Indian J. Med. Res. 2018, 147, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Coyne, S.; Courvalin, P.; Périchon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardebili, A.; Lari, A.R.; Beheshti, M.; Lari, E.R. Association between mutations in gyrA and parC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran. J. Basic Med. Sci 2015, 18, 623–626. [Google Scholar] [CrossRef]
- D’Souza, R.; Pinto, N.A.; Le Phuong, N.; Higgins, P.G.; Vu, T.N.; Byun, J.H.; Cho, Y.L.; Choi, J.R.; Yong, D. Phenotypic and genotypic characterization of Acinetobacter spp. panel strains: A cornerstone to facilitate antimicrobial development. Front. Microbiol. 2019, 10, 559. [Google Scholar] [CrossRef] [Green Version]
- Robicsek, A.; Jacoby, G.A.; Hooper, D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006, 6, 629–640. [Google Scholar] [CrossRef]
- Xiong, X.; Bromley, E.H.C.; Oelschlaeger, P.; Woolfson, D.N.; Spencer, J. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: Conserved surface loops direct the activity of a Qnr protein from a Gram-negative bacterium. Nucleic Acids Res. 2011, 39, 3917–3927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Chi, H.P.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol 2017, 7, 55. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Morinaga, Y.; Yanagihara, K.; Kaku, N.; Harada, Y.; Uno, N.; Nakamura, S.; Imamura, Y.; Hasegawa, H.; Miyazaki, T.; et al. Azithromycin inhibits MUC5AC induction via multidrug-resistant Acinetobacter baumannii in human airway epithelial cells. Pulm. Pharmacol. Ther. 2014, 28, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yang, S.; Jia, M.; Zhao, L.; Hou, C.; You, X.; Zhao, J.; Chen, A. Comparative study between macrolide regulatory proteins MphR(A) and MphR(E) in ligand identification and DNA binding based on the rapid in vitro detection system. Anal. Bioanal. Chem. 2016, 408, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Chopjitt, P.; Kerdsin, A.; Takeuchi, D.; Hatrongjit, R.; Boueroy, P.; Akeda, Y.; Tomono, K.; Hamada, S. Whole genome analysis of extensively drug-resistant Acinetobacter baumannii clinical isolates in Thailand. Infect. Disord Drug Targets 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Rajamohan, G.; Srinivasan, V.B.; Gebreyes, W.A. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2010, 65, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Lean, S.S.; Yeo, C.C.; Suhaili, Z.; Thong, K.L. Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: Insights into the mechanisms of resistance of an ST195 clone from Malaysia. Int. J. Antimicrob. Agents 2015, 45, 178–182. [Google Scholar] [CrossRef]
- Schroeder, M.R.; Lohsen, S.; Chancey, S.T.; Stephens, D.S. High-Level Macrolide Resistance Due to the Mega Element [mef(E)/mel] in Streptococcus pneumoniae. Front. Microbiol. 2019, 10, 868. [Google Scholar] [CrossRef]
- Munita, J.; Arias, C. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Okada, U.; Yamashita, E.; Neuberger, A.; Morimoto, M.; Van Veen, H.W.; Murakami, S. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 2017, 8, 1336. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G.; Gebreyes, W.A. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 5312–5316. [Google Scholar] [CrossRef] [Green Version]
- Richmond, G.E.; Evans, L.P.; Anderson, M.J.; Wand, M.E.; Bonney, L.C.; Ivens, A.; Chua, K.L.; Webber, M.A.; Mark Sutton, J.; Peterson, M.L.; et al. The Acinetobacter baumannii two-component system aders regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 2016, 7, e00430-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclercq, R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez-Cordova, J.; Kumar, A. Activity of the efflux pump inhibitor phenylalanine-arginine β-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int. J. Antimicrob. Agents 2011, 37, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Damier-Piolle, L.; Magnet, S.; Brémont, S.; Lambert, T.; Courvalin, P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008, 52, 557–562. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.J.; Balloy, V.; Fiette, L.; Chignard, M.; Courvalin, P.; Grillot-Courvalin, C. Contribution of the ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. mBio 2016, 7, e00697-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; D’Souza, R.; Yong, D.; Lee, K. Prediction of putative resistance islands in a carbapenem-resistant Acinetobacter baumannii global clone 2 clinical isolate. Ann. Lab. Med. 2016, 36, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Huang, Y.; Yu, L.; Liu, Y.; Niu, W.; Zou, D.; Liu, H.; Zheng, J.; Yin, X.; Yuan, J.; et al. Isolation and Whole-genome Sequence Analysis of the Imipenem Heteroresistant Acinetobacter baumannii Clinical Isolate HRAB-85. Int. J. Infect. Dis 2017, 62, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Durán, P.A.; Fonseca-Coronado, S.; Lozano-Trenado, L.M.; Araujo-Betanzos, S.; Rugerio-Trujillo, D.A.; Vaughan, G.; Saldaña-Rivera, E. Nosocomial transmission of extensively drug resistant Acinetobacter baumannii strains in a tertiary level hospital. PLoS ONE 2020, 15, e0231829. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. Adv. Exp. Med. Biol. 2019, 1145, 55–71. [Google Scholar] [CrossRef]
- Bojkovic, J.; Richie, D.L.; Six, D.A.; Rath, C.M.; Sawyer, W.S.; Hu, Q.; Dean, C.R. Characterization of an Acinetobacter baumannii lptD deletion strain: Permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J. Bacteriol. 2016, 198, 731–741. [Google Scholar] [CrossRef] [Green Version]
- Luke, N.R.; Sauberan, S.L.; Russo, T.A.; Beanan, J.M.; Olson, R.; Loehfelm, T.W.; Cox, A.D.; St. Michael, F.; Vinogradov, E.V.; Campagnari, A.A. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun. 2010, 78, 2017–2023. [Google Scholar] [CrossRef] [Green Version]
- The Comprehensive Antibiotic Resistance Database (CARD). Available online: https://card.mcmaster.ca/ontology/35989 (accessed on 5 March 2021).
- Sun, B.; Liu, H.; Jiang, Y.; Shao, L.; Yang, S.; Chen, D. New Mutations Involved in Colistin Resistance in Acinetobacter baumannii. mSphere 2020, 5, e00895-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.Y.; Gregg, K.A.; Napier, B.A.; Ernst, R.K.; Weiss, D.S. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015, 59, 7911–7914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerson, S.; Betts, J.W.; Lucaßen, K.; Nodari, C.S.; Wille, J.; Josten, M.; Göttig, S.; Nowak, J.; Stefanik, D.; Roca, I.; et al. Investigation of Novel pmrB and eptA Mutations in Isogenic Acinetobacter baumannii Isolates Associated with Colistin Resistance and Increased Virulence In Vivo. Antimicrob. Agents Chemother. 2019, 63, e01586-18. [Google Scholar] [CrossRef] [Green Version]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Karpíšková, R. Detection of colistin-resistant Acinetobacter baumannii with the mcr-4 gene. Klin. Mikrobiol. Infekc. Lek. 2019, 25, 4–6. [Google Scholar] [PubMed]
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020, 75, 60–64. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: First report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef] [Green Version]
- Hood, M.I.; Becker, K.W.; Roux, C.M.; Dunman, P.M.; Skaar, E.P. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 2013, 81, 542–551. [Google Scholar] [CrossRef] [Green Version]
- Nhu, N.T.K.; Riordan, D.W.; Nhu, T.D.H.; Thanh, D.P.; Thwaites, G.; Lan, N.P.H.; Wren, B.W.; Baker, S.; Stabler, R.A. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 2016, 6, 28291. [Google Scholar] [CrossRef]
- Paul, D.; Mallick, S.; Das, S.; Saha, S.; Ghosh, A.K.; Mandal, S.M. Colistin Induced Assortment of Antimicrobial Resistance in a Clinical Isolate of Acinetobacter baumannii SD01. Infect. Disord. Drug Targets 2019, 20, 501–505. [Google Scholar] [CrossRef]
- Traglia, G.; Chiem, K.; Quinn, B.; Fernandez, J.S.; Montaña, S.; Almuzara, M.; Mussi, M.A.; Tolmasky, M.E.; Iriarte, A.; Centrón, D.; et al. Genome sequence analysis of an extensively drug-resistant Acinetobacter baumannii indigo-pigmented strain depicts evidence of increase genome plasticity. Sci. Rep. 2018, 8, 16961. [Google Scholar] [CrossRef] [PubMed]
- Foong, W.E.; Tam, H.K.; Crames, J.J.; Averhoff, B.; Pos, K.M. The chloramphenicol/H+ antiporter CraA of Acinetobacter baumannii AYE reveals a broad substrate specificity. J. Antimicrob. Chemother. 2019, 74, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Karalewitz, A.P.A.; Millera, S.I. Multidrug-resistant Acinetobacter baumannii chloramphenicol resistance requires an inner membrane permease. Antimicrob. Agents Chemother. 2018, 62, e00513-18. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Cui, C.Y.; Wu, X.T.; Fang, L.X.; He, Q.; He, B.; Long, T.F.; Liao, X.P.; Chen, L.; Liu, Y.H.; et al. Spread of tet(X5) and tet(X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet. Microbiol. 2021, 253, 108954. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Quan, J.; Yang, Y.; Ji, J.; Liu, L.; Fu, Y.; Hua, X.; Chen, Y.; Pi, B.; Jiang, Y.; et al. Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1371–1375. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Q.; Wang, R.; Zhang, Y.; Wang, X.; Wang, H. Global regulator SoxR is a negative regulator of efflux pump gene expression and affects antibiotic resistance and fitness in Acinetobacter baumannii. Med. Baltim. 2017, 96, e7188. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, Z.; He, F.; Ruan, Z.; Jiang, Y.; Hua, X.; Yu, Y. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS ONE 2018, 13, e0192288. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.L.; Guo, Y.Z.; Wu, Y.M.; Gong, W.T.; Sun, J.; Huang, Z. In vivo bactericidal effect of colistin–linezolid combination in a murine model of MDR and XDR Acinetobacter baumannii pneumonia. Sci. Rep. 2020, 10, 17518. [Google Scholar] [CrossRef]
- Gu, B.; Kelesidis, T.; Tsiodras, S.; Hindler, J.; Humphries, R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013, 68, 4–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wareth, G.; Linde, J.; Hammer, P.; Nguyen, N.H.; Nguyen, T.N.M.; Splettstoesser, W.D.; Makarewicz, O.; Neubauer, H.; Sprague, L.D.; Pletz, M.W. Phenotypic and WGS-derived antimicrobial resistance profiles of clinical and non-clinical Acinetobacter baumannii isolates from Germany and Vietnam. Int. J. Antimicrob. Agents 2020, 56, 106127. [Google Scholar] [CrossRef]
- Ducas-Mowchun, K.; De Silva, P.M.; Crisostomo, L.; Fernando, D.M.; Chao, T.C.; Pelka, P.; Schweizer, H.P.; Kumar, A. Next generation of Tn7-based single-copy insertion elements for use in multi- and pan-drug-resistant strains of Acinetobacter baumannii. Appl. Environ. Microbiol. 2019, 85, e00066-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luna, B.M.; Ulhaq, A.; Yan, J.; Pantapalangkoor, P.; Nielsen, T.B.; Davies, B.W.; Actis, L.A.; Spellberg, B. Selectable Markers for Use in Genetic Manipulation of Extensively Drug-Resistant (XDR) Acinetobacter baumannii HUMC1. mSphere 2017, 2, e00140-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claeys, K.C.; Fiorvento, A.D.; Rybak, M.J. A Review of Novel Combinations of Colistin and Lipopeptide or Glycopeptide Antibiotics for the Treatment of Multidrug-Resistant Acinetobacter baumannii. Infect. Ther. 2014, 3, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Rolain, J.M.; Diene, S.M.; Kempf, M.; Gimenez, G.; Robert, C.; Raoult, D. Real-time sequencing to decipher the molecular mechanism of resistance of a clinical pan-drug-resistant Acinetobacter baumannii isolate from Marseille, France. Antimicrob. Agents Chemother. 2013, 57, 592–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannouli, M.; Di Popolo, A.; Durante-Mangoni, E.; Bernardo, M.; Cuccurullo, S.; Amato, G.; Tripodi, M.F.; Triassi, M.; Utili, R.; Zarrilli, R. Molecular epidemiology and mechanisms of rifampicin resistance in Acinetobacter baumannii isolates from Italy. Int. J. Antimicrob. Agents 2012, 39, 58–63. [Google Scholar] [CrossRef]
- Luna, B.; Trebosc, V.; Lee, B.; Bakowski, M.; Ulhaq, A.; Yan, J.; Lu, P.; Cheng, J.; Nielsen, T.; Lim, J.; et al. A nutrient-limited screen unmasks rifabutin hyperactivity for extensively drug-resistant Acinetobacter baumannii. Nat. Microbiol. 2020, 5, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Mustapha, M.M.; Tomich, A.D.; Callaghan, J.D.; McElheny, C.L.; Mettus, R.T.; Shanks, R.M.Q.; Sluis-Cremer, N.; Doi, Y. Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio 2017, 8, e00749-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Sharma, R.; Bhattacharyya, T.; Bhando, T.; Pathania, R. Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J. Antimicrob. Chemother. 2017, 72, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Sonkar, A.; Shukla, H.; Shukla, R.; Kalita, J.; Pandey, T.; Tripathi, T. UDP-N-Acetylglucosamine enolpyruvyl transferase (MurA) of Acinetobacter baumannii (AbMurA): Structural and functional properties. Int. J. Biol. Macromol. 2017, 97, 106–114. [Google Scholar] [CrossRef]
- Sánchez-Osuna, M.; Cortés, P.; Llagostera, M.; Barbé, J.; Erill, I. Exploration into the origins and mobilization of di-hydrofolate reductase genes and the emergence of clinical resistance to trimethoprim. Microb. Genom. 2020, 6, mgen000440. [Google Scholar] [CrossRef] [PubMed]
- Ghaffoori Kanaan, M.H.; Al-Shadeedi, S.M.J.; Al-Massody, A.J.; Ghasemian, A. Drug resistance and virulence traits of Acinetobacter baumannii from Turkey and chicken raw meat. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101451. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Long, Q.; Qian, K.; Fu, T.; Zhang, Z.; Liao, P.; Xie, J. Resistance and integron characterization of Acinetobacter baumannii in a teaching hospital in Chongqing, China. New Microbes New Infect. 2015, 8, 103–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, B.K.; Wang, M.; Chi, H.P.; Kim, E.C.; Jacoby, G.A.; Hooper, D.C. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 3582–3584. [Google Scholar] [CrossRef] [Green Version]
- Osei Sekyere, J.; Reta, M.A. Genomic and Resistance Epidemiology of Gram-Negative Bacteria in Africa: A Systematic Review and Phylogenomic Analyses from a One Health Perspective. mSystems 2020, 5, e00897-20. [Google Scholar] [CrossRef] [PubMed]
- Girija, A.S.S.; Vijayashree Priyadharsini, J.; Paramasivam, A. Plasmid-encoded resistance to trimethoprim/sulfamethoxazole mediated by dfrA1, dfrA5, sul1 and sul2 among Acinetobacter baumannii isolated from urine samples of patients with severe urinary tract infection. J. Glob. Antimicrob. Resist. 2019, 17, 145–146. [Google Scholar] [CrossRef]
- Duployez, C.; Guern, R.L.; Milliere, L.; Caplan, M.; Loïez, C.; Ledoux, G.; Jaillette, E.; Favory, R.; Mathieu, D.; Wallet, F. An outbreak can hide another. Jpn. J. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Ripa, M.; Galli, L.; Poli, A.; Oltolini, C.; Spagnuolo, V.; Mastrangelo, A.; Muccini, C.; Monti, G.; De Luca, G.; Landoni, G.; et al. Secondary infections in patients hospitalized with COVID-19: Incidence and predictive factors. Clin. Microbiol. Infect. 2021, 27, 451–457. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yang, B.; Li, Q.; Wen, L.; Zhang, R. Clinical Features of 69 Cases With Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020, 71, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contou, D.; Claudinon, A.; Pajot, O.; Micaëlo, M.; Longuet Flandre, P.; Dubert, M.; Cally, R.; Logre, E.; Fraissé, M.; Mentec, H.; et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care 2020, 10, 119. [Google Scholar] [CrossRef]
- Ramadan, H.K.-A.; Mahmoud, M.A.; Aburahma, M.Z.; Elkhawaga, A.A.; El-Mokhtar, M.A.; Sayed, I.M.; Hosni, A.; Hassany, S.M.; Medhat, M.A. Predictors of Severity and Co-Infection Resistance Profile in COVID-19 Patients: First Report from Upper Egypt. Infect. Drug Resist. 2020, 13, 3409–3422. [Google Scholar] [CrossRef]
- Nebreda-Mayoral, T.; Miguel-Gómez, M.A.; March-Rosselló, G.A.; Puente-Fuertes, L.; Cantón-Benito, E.; Martínez-García, A.M.; Muñoz-Martín, A.B.; Orduña-Domingo, A. Infección bacteriana/fúngica en pacientes con COVID-19 ingresados en un hospital de tercer nivel de Castilla y León, España. Enferm. Infecc. Microbiol. Clin. 2020, 3. [Google Scholar] [CrossRef]
- Sharifipour, E.; Shams, S.; Esmkhani, M.; Khodadadi, J.; Fotouhi-Ardakani, R.; Koohpaei, A.; Doosti, Z.; EJ Golzari, S. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect. Dis. 2020, 20, 646. [Google Scholar] [CrossRef]
- Yang, S.; Hua, M.; Liu, X.; Du, C.; Pu, L.; Xiang, P.; Wang, L.; Liu, J. Bacterial and fungal co-infections among COVID-19 patients in intensive care unit. Microbes Infect. 2021, 104806. [Google Scholar] [CrossRef] [PubMed]
- Durán-Manuel, E.M.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Bravata-Alcantará, J.C.; Sosa-Hernández, O.; Delgado-Balbuena, L.; León-García, G.; Cortés-Ortíz, I.A.; Cureño-Díaz, M.A.; Castro-Escarpulli, G.; et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J. Infect. Dev. Ctries 2021, 15, 58–68. [Google Scholar] [CrossRef]
- Karruli, A.; Boccia, F.; Gagliardi, M.; Patauner, F.; Ursi, M.P.; Sommese, P.; De Rosa, R.; Murino, P.; Ruocco, G.; Corcione, A.; et al. Multidrug-Resistant Infections and Outcome of Critically Ill Patients with Coronavirus Disease 2019: A Single Center Experience. Microb. Drug Resist. 2021. [Google Scholar] [CrossRef]
- Wu, N.; Dai, J.; Guo, M.; Li, J.; Zhou, X.; Li, F.; Gao, Y.; Qu, H.; Lu, H.; Jin, J.; et al. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microbes Infect. 2021, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Innes, G.K.; Walters, M.S.; Mehr, J.; Arias, J.; Greeley, R.; Chew, D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February–July 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, M.A.; Ozer, E.A.; Hauser, A.R. Utility of Whole-Genome Sequencing in Characterizing Acinetobacter Epidemiology and Analyzing Hospital Outbreaks. J. Clin. Microbiol. 2016, 54, 593–612. [Google Scholar] [CrossRef] [Green Version]
- Koeleman, J.G.M.; Stoof, J.; Van Der Bijl, M.W.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Identification of Epidemic Strains of Acinetobacter baumannii by Integrase Gene PCR. J. Clin. Microbiol. 2001, 39, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.-L.; Ma, L.; Chang, J.-C.; Su, L.-H.; Chu, C.; Leu, H.-S.; Siu, L.K. Variable Resistance Patterns of Integron-Associated Multidrug-Resistant Acinetobacter baumannii Isolates in a Surgical Intensive Care Unit. Microb. Drug Resist. 2004, 10, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Kaufmann, M.E.; Glover, J.; Coelho, J.M.; Warner, M.; Pike, R.; Pitt, T.L. Detection and Typing of Integrons in Epidemic Strains of Acinetobacter baumannii Found in the United Kingdom. J. Clin. Microbiol. 2005, 43, 3074–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaur, A.; Prakash, P.; Anupurba, S.; Mohapatra, T.M. Possible role of integrase gene polymerase chain reaction as an epidemiological marker: Study of multidrug-resistant Acinetobacter baumannii isolated from nosocomial infections. Int. J. Antimicrob. Agents 2007, 29, 446–450. [Google Scholar] [CrossRef]
- Chen, J.; Li, H.; Yang, J.; Zhan, R.; Chen, A.; Yan, Y. Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study. Int. J. Env. Res. Public Health 2015, 12, 10093–10105. [Google Scholar] [CrossRef] [Green Version]
- Deylam Salehi, M.; Ferdosi-Shahandashti, E.; Yahyapour, Y.; Khafri, S.; Pournajaf, A.; Rajabnia, R. Integron-Mediated Antibiotic Resistance in Acinetobacter baumannii Isolated from Intensive Care Unit Patients, Babol, North of Iran. Biomed. Res. Int. 2017, 2017, 7157923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nepka, M.; Perivolioti, E.; Kraniotaki, E.; Politi, L.; Tsakris, A.; Pournaras, S. In Vitro Bactericidal Activity of Trimethoprim-Sulfamethoxazole Alone and in Combination with Colistin against Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates. Antimicrob. Agents Chemother. 2016, 60, 6903–6906. [Google Scholar] [CrossRef] [Green Version]
| Mechanism of Resistance | Element Name | Resistance | Element Symbol (Gene) | Protein Products | |
|---|---|---|---|---|---|
| Class A beta lactamases | class A broad-spectrum beta-lactamase TEM-1 | extended-spectrum | blaTEM-1 | WP_000027057.1 and others | |
| class A extended-spectrum beta-lactamase SHV-5 | blaSHV-5 | WP_011117369.1 and others | |||
| class A extended-spectrum beta-lactamase SHV-12 | blaSHV-12 | WP_002904004.1 and others | |||
| carbapenem-hydrolyzing class A beta-lactamase GES-5 | blaGES-5 | WP_012658785.1 | |||
| class A extended-spectrum beta-lactamase GES-11 | blaGES-11 | WP_001211000.1 and others | |||
| class A beta-lactamase GES-12 | blaGES-12 | WP_063860500.1 and others | |||
| inhibitor-resistant class A extended-spectrum beta-lactamase PER-1 | blaPER-1 | WP_001100753.1 and others | |||
| class A extended-spectrum beta-lactamase PER-7 | blaPER-7 | WP_032495440.1 and others | |||
| class A extended-spectrum beta-lactamase VEB-1 | blaVEB-1 | WP_000706731.1 and others | |||
| class A extended-spectrum beta-lactamase CTX-M-15 | blaCTX-M-15 | WP_000239590.1 and others | |||
| class A extended-spectrum beta-lactamase CTX-M-55 | blaCTX-M-55 | WP_015387340.1 | |||
| class A extended-spectrum beta-lactamase CTX-M-115 | blaCTX-M-115 | WP_035895532.1 | |||
| carbapenem-hydrolyzing class A beta-lactamase KPC-2 | blaKPC-2 | WP_004199234.1 and others | |||
| Class B metallo-beta-lactamases | subclass B1 metallo-beta-lactamase NDM-1 | all (except monobactams) | blaNDM-1 | WP_004201164.1 and others | |
| subclass B1 metallo-beta-lactamase IMP-1 | blaIMP-1 | WP_003159548.1 | |||
| subclass B1 metallo-beta-lactamase IMP-4 | blaIMP-4 | WP_015060105.1 | |||
| subclass B1 metallo-beta-lactamase IMP-14 | blaIMP-14 | WP_039819893.1 | |||
| subclass B1 metallo-beta-lactamase IMP-16 | blaIMP-16 | WP_063860576.1 | |||
| Class C beta-lactamases | class C extended-spectrum beta-lactamase ADC-11 | extended-spectrum | blaADC-11 | WP_001211205.1 and others | |
| class C beta-lactamase ADC-25 | blaADC-25 | WP_001211217.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-26 | blaADC-26 | WP_001211238.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-30 | blaADC-30 | WP_001211218.1 and others | |||
| cefepime-hydrolyzing class C extended-spectrum beta-lactamase ADC-33 | blaADC-33 | WP_001211220.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-52 | blaADC-52 | WP_001211232.1 and others | |||
| cefepime-hydrolyzing class C extended-spectrum beta-lactamase ADC-56 | blaADC-56 | WP_031973850.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-73 | blaADC-73 | WP_001211219.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-74 | blaADC-74 | WP_001211203.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-76 | blaADC-76 | WP_001211237.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-79 | blaADC-79 | WP_001159760.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-80 | blaADC-80 | WP_029424536.1 and others | |||
| class C extended-spectrum beta-lactamase ADC-82 | blaADC-82 | WP_001211216.1 and others | |||
| class C beta-lactamase ADC-152 | CEPH | blaADC-152 | WP_001211233.1 and others | ||
| class C beta-lactamase ADC-156 | blaADC-156 | WP_024436624.1 and others | |||
| class C beta-lactamase ADC-162 | blaADC-162 | WP_031980335.1 and others | |||
| class C beta-lactamase ADC-176 | blaADC-176 | WP_001159761.1 and others | |||
| class C beta-lactamase ADC-182 | blaADC-182 | WP_057691114.1 and others | |||
| class C beta-lactamase ADC-212 | blaADC-212 | WP_031975357.1 and others | |||
| class C beta-lactamase ADC-222 | blaADC-222 | WP_031960999.1 and others | |||
| Class D beta-lactamases (oxicillinases) | carbapenem-hydrolyzing class D beta-lactamase OXA-23 | CAR | blaOXA-23 | WP_001046004.1 and others | |
| OXA-23 family carbapenem-hydrolyzing class D beta-lactamase OXA-239 | blaOXA-239 | WP_063862190.1 and others | |||
| carbapenem-hydrolyzing class D beta-lactamase OXA-24 | CAR | blaOXA-24 | WP_012754353.1 and others | ||
| OXA-24 family carbapenem-hydrolyzing class D beta-lactamase OXA-72 | blaOXA-72 | WP_000713530.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-51 | CAR | blaOXA-51 | WP_002033109.1 and others | ||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-64 | blaOXA-64 | WP_001021788.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-65 | blaOXA-65 | WP_001021782.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-66 | blaOXA-66 | WP_001021792.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-68 | blaOXA-68 | WP_001021775.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-69 | blaOXA-69 | WP_001021779.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-71 | blaOXA-71 | WP_001021785.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-82 | blaOXA-82 | WP_001021793.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-90 | blaOXA-90 | WP_001021781.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-91 | blaOXA-91 | WP_001021776.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-94 | blaOXA-94 | WP_029424390.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-95 | blaOXA-95 | WP_031960432.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-98 | blaOXA-98 | WP_001021777.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-100 | blaOXA-100 | WP_001021795.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-104 | blaOXA-104 | WP_024433915.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-120 | blaOXA-120 | WP_004738885.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-223 | blaOXA-223 | WP_001022758.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-259 | blaOXA-259 | WP_001021784.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-371 | blaOXA-371 | WP_063862738.1 and others | |||
| OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-402 | blaOXA-402 | WP_001021789.1 and others | |||
| carbapenem-hydrolyzing class D beta-lactamase OXA-58 | CAR | blaOXA-58 | WP_002002480.1 and others | ||
| OXA-58 family carbapenem-hydrolyzing class D beta-lactamase OXA-96 | blaOXA-96 | WP_063864543.1 | |||
| OXA-134 family carbapenem-hydrolyzing class D beta-lactamase OXA-235 | CAR | blaOXA-235 | WP_000854009.1 and others | ||
| OXA-134 family carbapenem-hydrolyzing class D beta-lactamase OXA-237 | blaOXA-237 | WP_000854010.1 and others | |||
| carbapenem-hydrolyzing class D beta-lactamase OXA-143 | CAR | blaOXA-143 | WP_063861042.1 | ||
| OXA-143 family carbapenem-hydrolyzing class D beta-lactamase OXA-253 | blaOXA-253 | WP_032495764.1 | |||
| Efflux pumps | multidrug efflux RND transporter AdeABC outer membrane channel subunit AdeC | CEPH, CAR | adeC | WP_000047249.1 and others | |
| Acinetobacter baumannii efflux resistant AdeR | adeR_A91V | WP_039198290.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeR | adeR_P56S | WP_088753133.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeR | adeR_P116L | WP_111853508.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeS | adeS_G336S, adeS_N125K | WP_031975145.1, WP_057691178.1 and others | |||
| Acinetobacter baumannii efflux resistant AdeS | adeS_H189Y | WP_119491814.1, WP_000837466.1, WP_046882653.1 | |||
| Penicillin-binding proteins | Acinetobacter baumannii carbapenem resistant FtsI | CAR | ftsI_A515V | WP_000227939.1 and others (penicillin-binding protein PBP3) | |
| Element Name and Symbol | Resistance | Gene | Protein Products | |
|---|---|---|---|---|
| Aminoglycoside acetyltransferases | Aminoglycoside 2’-N-acetyltransferase AAC(2’)-Ib | GEN, TOB, DIB, NET [101] | aac(2′)-Ib | WP_001159732.1 † |
| Aminoglycoside 3-N-acetyltransferase | GEN | aac(3) | WP_195206917.1 | |
| AAC(3)-I family aminoglycoside 3-N-acetyltransferase | GEN | aac(3)-I | HAV6561382.1, WP_069597335.1 | |
| Aminoglycoside N-acetyltransferase AAC(3)-Ia | AST, GEN, SIS [102] | aac(3)-Ia | WP_002089484.1, and others | |
| Aminoglycoside N-acetyltransferase AAC(3)-IId | GEN | aac(3)-IId | WP_000557454.1, WP_126562472.1 | |
| Aminoglycoside N-acetyltransferase AAC(3)-IIe | GEN | aac(3)-IIe | WP_000557452.1, WP_002063884.1, WP_033107705.1, WP_095530619.1, and others | |
| Aminoglycoside N-acetyltransferase AAC(3)-Iva | APR, GEN, TOB | aac(3)-IVa | WP_001199192.1 | |
| Aminoglycoside 6’-N-acetyltransferase | all | aac(6’) | HAV4276337.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-31 | all | aac(6’)-31 | WP_044424439.1 | |
| Aminoglycoside 6’-N-acetyltransferase AAC(6’)-33 | all | aac(6’)-33 | WP_015059044.1 | |
| AAC(6’)-Ia family aminoglycoside 6’-N-acetyltransferase | AMI, KAN, TOB, putatively against all | aac(6’) | EGY2236091.1, EGY5968849.1, WP_088756823.1, WP_088774065.1, WP_140976846.1 | |
| AAC(6’)-Ia family aminoglycoside 6’-N-acetyltransferase AacA16 | all | aacA16 | WP_001109644.1 | |
| Aminoglycoside 6’-N-acetyltransferase AacA34 | all | aacA34 | WP_052285801.1 | |
| AAC(6’)-Ia family aminoglycoside 6’-N-acetyltransferase AacA43 | KAN, TOB | aacA43 | WP_024437351.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ian | AMI, KAN, TOB, putatively against all | aac(6’)-Ian or aacA57-2 | WP_000960976.1 | |
| AAC(6’)-Ib family aminoglycoside 6’-N-acetyltransferase | AMI, DIB, GEN, ISE, KAN, NET, SIS, TOB [102] | aac(6’)-Ib | WP_063840280.1, and others | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ib’ | GEN | aac(6’)-Ib’ | WP_014454105.1 | |
| AAC(6’)-Ighjkrstuvwx family aminoglycoside N-acetyltransferase | AMI, KAN, TOB | aac(6’)-I | WP_169109636.1, WP_150956588.1, WP_005288246.1, WP_005243483.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ib3 | AMI, KAN, TOB | aac(6’)-Ib3 | WP_032488579.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Ib4 | GEN | aac(6’)-Ib4 | WP_003159191.1 | |
| aminoglycoside N-acetyltransferase AAC(6’)-Ih | AMI, KAN, TOB | aac(6’)-Ih | WP_016541245.1 † | |
| Aminoglycoside N-acetyltransferase AAC(6’)-Il | AMI, KAN, TOB | aac(6’)-Il | WP_156193962.1 | |
| AAC(6’)-II family aminoglycoside 6’-N-acetyltransferase AacA35 | GEN, KAN, TOB | aacA35 | WP_024437054.1 | |
| Aminoglycoside N-acetyltransferase AAC(6’)-IIc | GEN, KAN, TOB | aac(6’)-IIc | WP_149959345.1, WP_149938250.1 | |
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr | AMI, KAN, TOB, QUI | aac(6’)-Ib-cr | WP_185936887.1 | |
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr5 | AMI, KAN, TOB, QUI | aac(6’)-Ib-cr5 | WP_063840321.1 | |
| Aminoglycoside adenyltransferases | Aminoglycoside nucleotidyltransferase ANT(2’’)-Ia | DIB, GEN, KAN, SIS, TOB [102] | ant(2’’)-Ia | WP_000381802.1, and others |
| ANT(3’’)-I family aminoglycoside nucleotidyltransferase | STR, SPE | ant(3’’) | WP_038350223.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA | STR, SPE | ant(3’’)-Ia | WP_001205725.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA1 | STR | aadA1 | WP_001206316.1, and others | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA2 | STR | aadA2 | WP_001206356.1, WP_001261740.1, and others | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA5 | STR | aadA5 | WP_000503573.1, WP_000503574.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA11 | STR | aadA11 | WP_048608579.1, HAV4466908.1 | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA13 | STR | aadA13 | WP_001424636.1 † | |
| ANT(3’’)-Ia family aminoglycoside nucleotidyltransferase AadA16 | STR | aadA16 | WP_001749984.1, WP_185936919.1 | |
| ANT(3’’)-II family aminoglycoside nucleotidyltransferase | STR, SPE | ant(3’’)-II | WP_005240470.1 | |
| Aminoglycoside nucleotidyltransferase ANT(3’’)-IIa | STR, SPE | ant(3’’)-IIa | WP_001279062.1, WP_001279061.1,WP_001112625.1, and others | |
| Aminoglycoside nucleotidyltransferase ANT(3’’)-IIc | STR, SPE | ant(3’’)-IIc | WP_005281276.1 | |
| Aminoglycoside phosphotransferases | APH(3’) family aminoglycoside O-phosphotransferase | all | aph(3’) | WP_196077463.1 |
| Aminoglycoside O-phosphotransferase APH(3’)-Ia | GEN, KAN, NEO, PAR, LIV, RIB [102] | aph(3’)-Ia | WP_000018326.1, and others | |
| APH(3’)-II family aminoglycoside O-phosphotransferase | KAN | aph(3’)-II | WP_000262467.1 | |
| Aminoglycoside O-phosphotransferase APH(3’)-IIa | KAN | aph(3’)-IIa | WP_000572405.1, WP_171502934.1, and others | |
| APH(3’)-VI family aminoglycoside O-phosphotransferase | AMI, KAN | aph(3’)-VI | WP_014386410.1, and others | |
| Aminoglycoside O-phosphotransferase APH(3’)-VIa | AMI, KAN | aph(3’)-VIa | WP_000422636.1, and others | |
| Aminoglycoside O-phosphotransferase APH(3’)-VIb | AMI, KAN | aph(3’)-VIb | WP_000422633.1, WP_000422632.1, and others | |
| Aminoglycoside O-phosphotransferase APH(3’’)-Ib | STR | aph(3’’)-Ib | WP_001082319.1, and others | |
| Aminoglycoside O-phosphotransferase APH(4)-Ia | HYG | aph(4)-Ia | WP_185218783.1 | |
| Aminoglycoside O-phosphotransferase APH(6)-Id | STR | aph(6)-Id | WP_000480968.1, and others | |
| Target mutation: 16S rRNA methylase genes | ArmA family 16S rRNA (guanine(1405)-N(7))-methyltransferase | GEN | armA | WP_000359986.1, and others |
| 16S rRNA (guanine(1405)-N(7))-methyltransferase RmtB1 | all | rmtB and rmtB1 | WP_012372818.1 | |
| RmtE family 16S rRNA (guanine(1405)-N(7))-methyltransferase | all | rmtE | WP_120494548.1 | |
| Efflux pump overactivity | Multidrug efflux MFS transporter AmvA | Putatively against all | amvA | WP_001170321.1, and others |
| Multidrug efflux RND transporter AdeABC outer membrane channel subunit AdeC | adeC | WP_000047249.1, and others | ||
| Multidrug efflux RND transporter periplasmic adaptor subunit AdeD | adeD | WP_002119008.1, WP_039254548.1 | ||
| Multidrug efflux RND transporter permease subunit AdeE | adeE | WP_002118518.1, WP_039254549.1 | ||
| Efflux system DNA-binding response regulator transcription factor AdeR | adeR | WP_032002707.1, EGY8404952.1,WP_020752724.1 | ||
| Two-component sensor histidine kinase AdeS two-component sensor histidine kinase | adeS | WP_031975145.1, and others | ||
| Multidrug efflux SMR transporter EmrE | emrE | WP_109847152.1 |
| Element name and symbol | Resistance | Gene | Protein products |
|---|---|---|---|
| Tetracycline efflux MFS transporter Tet(39) | DOX, TET [110,111] | tet(39) | WP_004856455.1, and others |
| Tetracycline efflux MFS transporter Tet(A) | DOX, MIN, TET, TIG * | tet(A) | WP_000804064.1, and others |
| tetracycline efflux MFS transporter Tet(B) | DOX, MIN, TET [111] | tet(B) | WP_001089072.1, and others |
| Tetracycline efflux MFS transporter Tet(C) | TET [112] | tet(C) | WP_000841448.1 |
| Tetracycline efflux MFS transporter Tet(D) | TET [113] | tet(D) | WP_024436252.1 |
| Tetracycline efflux MFS transporter Tet(G) | DOX, MIN, TET | tet(G) | WP_001257840.1 |
| Tetracycline efflux MFS transporter Tet(H) | OXY, TET [114] | tet(H) | WP_006248867.1 |
| Tetracycline resistance ribosomal protection protein Tet(M) | TET, MIN | tet(M) | WP_000691727.1 |
| Tetracycline-inactivating monooxygenase Tet(X) | all | tet(X) | WP_024160783.1, and others |
| Element Name and Symbol | Resistance | Gene | Protein Products |
|---|---|---|---|
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr | CIP, NOR | aac(6’)-Ib-cr | WP_185936887.1 |
| Fluoroquinolone-acetylating aminoglycoside 6’-N-acetyltransferase AAC(6’)-Ib-cr5 | CIP, NOR | aac(6’)-Ib-cr5 | WP_063840321.1 |
| A. baumannii quinolone resistant GyrA (DNA gyrase subunit A) | CIP, LEV | gyrA_S81L | WP_000116444.1, and others |
| A. baumannii quinolone resistant ParC (DNA topoisomerase IV subunit A) | CIP, LEV | parC_E88K | WP_000202265.1, and others |
| A. baumannii quinolone resistant ParC (DNA topoisomerase IV subunit A) | CIP, LEV | parC_S84F | WP_000202250.1, and others |
| A. baumannii quinolone resistant ParC (DNA topoisomerase IV subunit A) | CIP, LEV | parC_S84L | WP_000202252.1, and others |
| QnrA family quinolone resistance pentapeptide repeat protein | CIP | qnrA | HAV5951840.1 |
| QnrB family quinolone resistance pentapeptide repeat protein | CIP | qnrB | WP_185936934.1 |
| Quinolone resistance pentapeptide repeat protein QnrB19 | CIP | qnrB19 | WP_012954666.1 |
| QnrS family quinolone resistance pentapeptide repeat protein | CIP, * | qnrS | WP_147508156.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. https://doi.org/10.3390/pathogens10030373
Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens. 2021; 10(3):373. https://doi.org/10.3390/pathogens10030373
Chicago/Turabian StyleKyriakidis, Ioannis, Eleni Vasileiou, Zoi Dorothea Pana, and Athanasios Tragiannidis. 2021. "Acinetobacter baumannii Antibiotic Resistance Mechanisms" Pathogens 10, no. 3: 373. https://doi.org/10.3390/pathogens10030373
APA StyleKyriakidis, I., Vasileiou, E., Pana, Z. D., & Tragiannidis, A. (2021). Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens, 10(3), 373. https://doi.org/10.3390/pathogens10030373









