Could Antigen Presenting Cells Represent a Protective Element during SARS-CoV-2 Infection in Children?
Abstract
1. Introduction
2. Innate Immunity Response to SARS-CoV-2
2.1. Cytokines
2.2. Type I IFN Response and SARS-CoV-2
2.3. Cytokine Storm in SARS-CoV-2
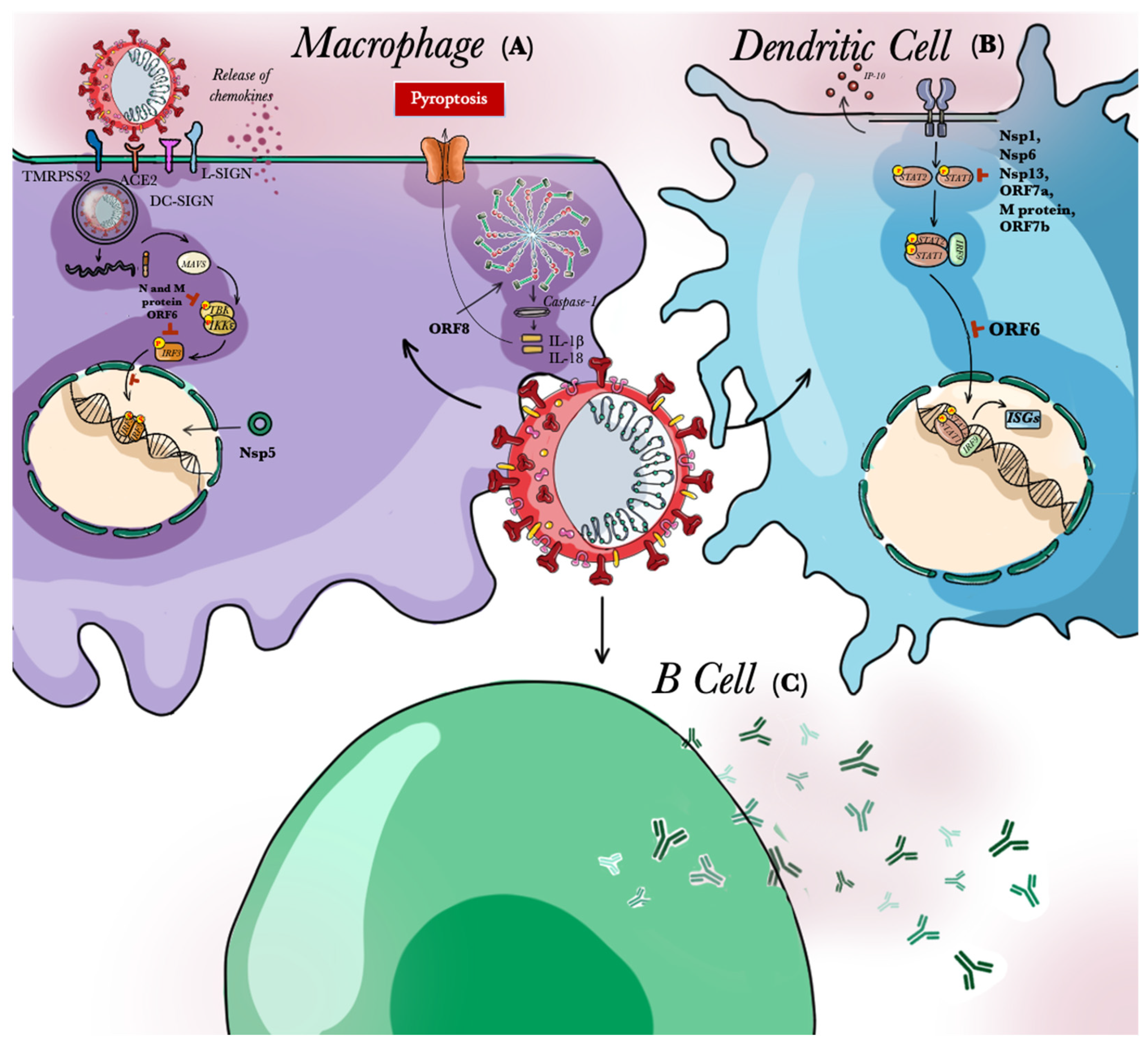
3. Role of APCs in SARS-CoV-2
4. APCs and Children Response to SARS-CoV-2: Any Relation?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Wormser, G.P.; Prasad, A.; Neuhaus, E.; Joshi, S.; Nowakowski, J.; Nelson, J.; Mittleman, A.; Aguero-Rosenfeld, M.; Topal, J.; Krause, J.P. Emergence of resistance to azithromycin-atovaquone in immunocompromised patients with Babesia microti infection. Clin. Infect. Dis. 2010, 50, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, P.W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Li, G.; Fan, Y.; Lai, Y.; Han, T.; Li, Z.; Zhou, P.; Pan, P.; Wang, W.; Hu, D.; Liu, X.; et al. Coronavirus infections and immune responses. J. Med. Virol. 2020, 92, 424–432. [Google Scholar] [CrossRef]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef]
- Wilson, L.; McKinlay, C.; Gage, P.; Ewart, G. SARS coronavirus E protein forms cation-selective ion channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Verdiá-Báguena, G.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef]
- Shah, A. Novel Coronavirus-Induced NLRP3 Inflammasome Activation: A Potential Drug Target in the Treatment of COVID-19. Front. Immunol. 2020, 11, 1021. [Google Scholar] [CrossRef] [PubMed]
- Bowie, A.; Kiss-Toth, E.; Symons, J.A.; Smith, G.L.; Dower, S.K.; O’Neill, L.A.J. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 2000, 97, 10162–10167. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. Virol. J. 2007, 81, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lai, D.-Y.; Zhang, H.-N.; Jiang, H.-W.; Tian, X.; Ma, M.-L.; Meng, Q.F.; Guo, S.J.; Wu, Y. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 1095–1097. [Google Scholar] [CrossRef]
- Nguyen-Contant, P.; Embong, A.K.; Kanagaiah, P.; Chaves, F.A.; Yang, H.; Branche, A.R.; Topham, D.J.; Sangster, M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. mBio 2020, 11, 20. [Google Scholar] [CrossRef]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes. Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef]
- Weisberg, S.P.; Connors, T.J.; Zhu, Y.; Baldwin, M.R.; Lin, W.H.; Wontakal, S.; Szabo, P.A.; Wells, S.B.; Dogra, P.; Gray, J.; et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021, 22, 25–31. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- McGettrick, A.F.; O’Neill, L.A. The expanding family of MyD88-like adaptors in Toll-like receptor signal transduction. Mol. Immunol. 2004, 41, 577–582. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, A.; Hakeem, A.R.; Radhakrishnan, S.; Reddy, M.S.; Rela, M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology 2021, 29, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Sa Ribero, M.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020, 16, 1008737. [Google Scholar] [CrossRef] [PubMed]
- Birra, D.; Benucci, M.; Landolfi, L.; Merchionda, A.; Loi, G.; Amato, P.; Licata, G.; Quartuccion, L.; Triggiani, M.; Moscato, P. COVID 19: A clue from innate immunity. Immunol. Res. 2020, 68, 161–168. [Google Scholar] [CrossRef]
- Totura, A.L.; Whitmore, A.; Agnihothram, S.; Schafer, A.; Katze, M.G.; Heise, M.T.; Baric, R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2015, 6, 00638–15. [Google Scholar] [CrossRef]
- Hornung, V.; Barchet, W.; Schlee, M.; Hartmann, G. RNA Recognition via TLR7 and TLR8. In Handbook of Experimental Pharmacology; Springer: Berlin, Heidelberg, Germany, 2008; pp. 71–86. [Google Scholar]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. Virol. J. 2005, 79, 14614–14621. [Google Scholar] [CrossRef]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Fujita, T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef]
- Li, K.; Hao, Z.; Zhao, X.; Du, J.; Zhou, Y. SARS-CoV-2 infection-induced immune responses: Friends or foes? Scand. J. Immunol. 2020, 92, 12895. [Google Scholar] [CrossRef] [PubMed]
- Ramana, C.V.; Gil, M.P.; Han, Y.; Ransohoff, R.M.; Schreiber, R.D.; Stark, G.R. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 2001, 98, 6674–6679. [Google Scholar] [CrossRef] [PubMed]
- Torrado, E.; Cooper, A.M. Cytokines in the balance. Nat. Immunol. 2019, 20, 1557. [Google Scholar]
- Yuen, C.K.; Lam, J.-Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.P.; Jin, D.Y.; To, K.K.W.; Chan, J.F.W.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes. Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Lichtman, A.; Pillai, S. Cellular and Molecular Immunology, 9th ed.; Elsevier: Philadelphia, PA, USA, 2018; p. 565. [Google Scholar]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodriguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Ank, N.; West, H.; Bartholdy, C.; Eriksson, K.; Thomsen, A.R.; Paludan, S.R. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. Virol. J. 2006, 80, 4501–4509. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Billiau, A.; Matthys, P. Interferon-gamma: A historical perspective. Cytokine Growth Factor Rev. 2009, 20, 97–113. [Google Scholar] [CrossRef]
- Akdis, M.; Burgler, S.; Crameri, R.; Eiwegger, T.; Fujita, H.; Gomez, E.; Klunker, S.; Meyer, N.; O’Mahony, L.; Plaomares, O. Interleukins, from 1 to 37, and interferon-gamma: Receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2011, 127, 701–721. [Google Scholar] [CrossRef]
- Zhang, J.M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Sims, J.E.; Smith, D.E. The IL-1 family: Regulators of immunity. Nat. Rev. Immunol. 2010, 10, 89–102. [Google Scholar] [CrossRef]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta 2014, 1843, 2563–2582. [Google Scholar] [CrossRef] [PubMed]
- Kalliolias, G.D.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Blaser, H.; Mak, T.W. Regulation of tumour necrosis factor signalling: Live or let die. Nat. Rev. Immunol. 2015, 15, 362–374. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yosif, A.S.; Bals, J.; Hauser, B.M. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.C.; Zhong, G.; Huang, I.-C.; Farzan, M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef] [PubMed]
- Hamana, A.; Takahashi, Y.; Uchida, T.; Nishikawa, M.; Imamura, M.; Chayama, K.; Takakura, Y. Evaluation of antiviral effect of type I, II, and III interferons on direct-acting antiviral-resistant hepatitis C virus. Antivir. Res. 2017, 146, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.; de Haan, C.A.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [PubMed]
- Odendall, C.; Dixit, E.; Stavru, F.; Bierne, H.; Franz, K.M.; Fieggen, A.; Boulant, S.; Gehrke, L.; Cossart, P.; Kagan, J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014, 15, 717–726. [Google Scholar] [CrossRef]
- O’Brien, T.R.; Prokunina-Olsson, L.; Donnelly, R.P. IFN-lambda4: The paradoxical new member of the interferon lambda family. J. Interferon Cytokine Res. 2014, 34, 829–838. [Google Scholar] [CrossRef]
- Wack, A.; Terczynska-Dyla, E.; Hartmann, R. Guarding the frontiers: The biology of type III interferons. Nat. Immunol. 2015, 16, 802–809. [Google Scholar] [CrossRef]
- Bolen, C.R.; Ding, S.; Robek, M.D.; Kleinstein, S.H. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology 2014, 59, 1262–1272. [Google Scholar] [CrossRef]
- Andreakos, E.; Zanoni, I.; Galani, I.E. Lambda interferons come to light: Dual function cytokines mediating antiviral immunity and damage control. Curr. Opin. Immunol. 2019, 56, 67–75. [Google Scholar] [CrossRef]
- Tseng, C.T.; Huang, C.; Newman, P.; Wang, N.; Narayanan, K.; Watts, D.M.; Makino, S.; Packard, M.M.; Zaki, S.R.; Chan, T.S.; et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. Virol. J. 2007, 81, 1162–1173. [Google Scholar] [CrossRef]
- Galani, I.E.; Triantafyllia, V.; Eleminiadou, E.E.; Koltsida, O.; Stavropoulos, A.; Manioudaki, M.; Thanos, D.; Doyle, S.E.; Kotenko, S.V.; Thanopoulou, K.; et al. Interferon-lambda Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity 2017, 46, 875–890.e6. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 84. [Google Scholar] [CrossRef]
- Jauregui, A.R.; Savalia, D.; Lowry, V.K.; Farrell, C.M.; Wathelet, M.C. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS ONE 2013, 8, 62416. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qui, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Lau, Y.L.; Peiris, J.S. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 2005, 17, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Proinflammatory cytokines. Chest 2000, 118, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Braciale, T.J.; Hahn, Y.S. Immunity to viruses. Immunol. Rev. 2013, 255, 5–12. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Rezaei, M.; Mansouri, N.; Marjani, M.; Mansouri, D. Immunologic Features in Coronavirus Disease 2019: Functional Exhaustion of T Cells and Cytokine Storm. J. Clin. Immunol. 2020, 40, 974–976. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhou, Y.; Binqing, F.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Aberrant Pathogenic GM-CSF+ T Cells and Inflammatory CD14+CD16+ Monocytes in Severe Pulmonary Syndrome Patients of a New Coronavirus. BioRxiv 2020. [Google Scholar] [CrossRef]
- Wong, R.S.; Wu, A.; To, K.F.; Lee, N.; Lam, C.W.K.; Wong, C.K.; Chan, P.K.S.; Ng, M.H.L.; Yu, L.M.; Hui, D.S.; et al. Haematological manifestations in patients with severe acute respiratory syndrome: Retrospective analysis. BMJ 2003, 326, 1358–1362. [Google Scholar] [CrossRef]
- Li, T.; Qui, Z.; Zhang, L.; Han, Y.; He, W.; Liu, Z.; Ma, X.; Fan, H.; Lu, W.; Xie, J.; et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J. Infect. Dis. 2004, 189, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; To, K.K.W.; Wong, Y.C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.Y.; Lau, T.T.K.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877. [Google Scholar] [CrossRef] [PubMed]
- Page, C.; Giocochea, L.; Matthews, K.; Zhang, Y.; Klover, P.; Holtzman, M.J.; Hennighausen, L.; Freiman, M. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. Virol. J. 2012, 86, 13334–13349. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chu, H.; Hou, Y.; Chai, Y.; Shuai, H.; Lee, A.C.Y.; Zhang, X.; Wang, Y.; Hu, B.; Huang, X. Attenuated Interferon and Proinflammatory Response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated with Viral Antagonism of STAT1 Phosphorylation. J. Infect. Dis. 2020, 222, 734–745. [Google Scholar] [CrossRef]
- Trinchieri, G. Physiology of the immune system. Minerva Anestesiol. 1978, 44, 673–678. [Google Scholar] [PubMed]
- Law, H.K.; Cheung, C.Y.; Ng, H.Y.; Sia, S.F.; Chan, Y.O.; Luk, W.; Nicholls, J.M.; Peiris, J.S.M.; Lau, Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 2005, 106, 2366–2374. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, C.; Li, J.; Yuan, J.; Wei, J.; Huang, F.; Wang, F.; Li, G.; Li, Y.; Xing, L.; et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020, 146, 119–127. [Google Scholar] [CrossRef]
- Yao, X.H.; Li, T.Y.; He, Z.C.; Ping, Y.F.; Liu, H.W.; Yu, S.C.; Mou, H.M.; Wang, L.H.; Zhang, H.R.; Fu, W.J.; et al. A pathological report of three COVID-19 cases by minimal invasive autopsies. Chin. J. Pathol. 2020, 49, 411–417. [Google Scholar]
- Yang, Z.Y.; Huang, Y.; Ganesh, L.; Leung, K.; Kong, W.P.; Schwartz, O.; Subbarao, K.; Nabel, G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. Virol. J. 2004, 78, 5642–5650. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Fang, W.F.; Chen, Y.M.; Lin, C.Y.; Huang, H.L.; Yeh, H.; Chang, Y.T.; Huang, K.T.; Lin, M.C. Histone deacetylase 2 (HDAC2) attenuates lipopolysaccharide (LPS)-induced inflammation by regulating PAI-1 expression. J. Inflamm. (Lond.) 2018, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Fang, M.; Li, P.; Fang, F.; Xu, Y. HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells. J. Mol. Cell. Cardiol. 2009, 46, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Jamilloux, Y.; Henry, T.; Belot, A.; Veil, S.; Fauter, M.; El Jamamal, T.; Walzer, T.; Francois, B.; Seve, P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020, 19, 102567. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, F.; Wang, R.; Yange, M.; Guan, K.; Jiang, T.; Xu, G.; Sun, J.; Chang, C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020, 109, 102434. [Google Scholar] [CrossRef]
- Chen, I.Y.; Moriyama, M.; Chang, M.F.; Ichinohe, T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019, 10, 50. [Google Scholar] [CrossRef]
- Nielsen, S.C.A.; Yang, F.; Hoh, R.A.; Jackson, K.J.L.; Poeltgen, K.; Lee, J.Y.; Rustagi, A.; Rogers, A.J.; Powell, A.E.; Kim, P.S.; et al. B cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Dong, X.; Cao, Y.Y.; Lu, X.X.; Zhang, J.J.; Du, H.; Yan, Y.Q.; Akdis, C.A.; Gao, Y.D. Eleven faces of coronavirus disease 2019. Allergy 2020, 75, 1699–1709. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.C.; Goh, H.P.; Koh, D. Treatment of COVID-19: Old tricks for new challenges. Crit. Care 2020, 24, 91. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Jackson, D.J.; Busse, W.W.; Bacharier, L.B.; Kattan, M.; O’Connor, G.T.; Wood, R.A.; Visness, C.M.; Durham, S.R.; Larson, D.; Esnault, S.; et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J. Allergy Clin. Immunol. 2020, 146, 203–206.e3. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Do, A.; Vicencio, A. Nasal Gene Expression of Angiotensin-Converting Enzyme 2 in Children and Adults. JAMA 2020, 323, 2427–2429. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.I.; Hu, Y.L.; Chen, P.Y.; Huang, Y.C.; Hsueh, P.R. Are children less susceptible to COVID-19? J. Microbiol. Immunol. Infect. 2020, 53, 371–372. [Google Scholar] [CrossRef] [PubMed]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Joosten, L.A.B.; Ifrim, D.C.; Saeed, S.; Jacobs, C.; van Loenhout, J.; de Jong, D.; Stunnenberg, H.G.; et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 17537–17542. [Google Scholar] [CrossRef]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef]
- Kolodny, O.; Berger, M.; Feldman, M.W.; Ram, Y. A new perspective for mitigation of SARS-CoV-2 infection: Priming the innate immune system for viral attack. Open Biol. 2001, 10, 7. [Google Scholar]
- Singh, A.K.; Gillies, C.L.; Singh, R.; Singh, A.; Chudasama, Y.; Coles, B.; Seidu, S.; Zaccardi, F.; Davies, M.J.; Khunti, K. Prevalence of co-morbidities and their association with mortality in patients with COVID-19: A systematic review and meta-analysis. Diabetes Obes. Metab. 2020, 22, 1915–1924. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, E.; Bamford, A.; Kenny, J.; Karforou, M.; Jones, C.; Shah, P.; Pamnarayan, P.; Fraisse, A.; Miller, O.; Davies, P.; et al. Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-CoV-2. JAMA 2020, 324, 259–269. [Google Scholar] [CrossRef]
- Feldstein, L.R.; Rose, E.B.; Horowitz, S.M.; Collins, J.P.; Newhams, M.M.; Son, M.B.F.; Newburger, J.W.; Kleinman, L.C.; Heidemann, S.M. Multisystem Inflammatory Syndrome in U.S. Children and Adolescents. N. Engl. J. Med. 2020, 383, 334–346. [Google Scholar] [CrossRef]
- Consiglio, C.R.; Cotugno, N.; Sardh, F.; Pou, C.; Amodio, D.; Rodriguez, L.; Tan, Z.; Zicari, S.; Ruggiero, A.; Pascucci, G.R.; et al. The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell 2020, 183, 968–981. [Google Scholar] [CrossRef]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, 27. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Balleseros, O.; Stralin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, N.E.; Pasztoi, M.; Schraml, B.U. Understanding the Functional Properties of Neonatal Dendritic Cells: A Doorway to Enhance Vaccine Effectiveness? Front. Immunol. 2018, 9, 3123. [Google Scholar] [CrossRef] [PubMed]
- Velilla, P.A.; Rugeles, M.T.; Chougnet, C.A. Defective antigen-presenting cell function in human neonates. Clin. Immunol. 2006, 121, 251–259. [Google Scholar] [CrossRef]
- De Wit, D.; Tonon, S.; Olaslagers, V.; Goriely, S.; Boutriaux, M.; Goldman, M.; Willems, F. Impaired responses to toll-like receptor 4 and toll-like receptor 3 ligands in human cord blood. J. Autoimmun. 2003, 21, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.R.; Quin, G.; Byers, D.M.; Stadnyk, A.W.; Al-Hertani, W.; Bortolussi, R. Role of MyD88 in diminished tumor necrosis factor alpha production by newborn mononuclear cells in response to lipopolysaccharide. Infect. Immun. 2004, 72, 1223–1229. [Google Scholar] [CrossRef]
- De Kleer, I.; Willems, F.; Lambrecht, B.; Goriely, S. Ontogeny of myeloid cells. Front. Immunol. 2014, 5, 423. [Google Scholar] [CrossRef]
- He, Y.M.; Li, X.; Perego, M.; Nefedova, Y.; Kossenkov, A.; Jensen, E.A.; Kagan, V.; Liu, Y.F.; Fu, S.Y.; Ye, Q.J.; et al. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat. Med. 2018, 24, 224–231. [Google Scholar] [CrossRef]
- Hoeffel, G.; Chen, J.; Lavin, Y.; Low, D.; Almeida, F.F.; See, P.; Beaudin, A.E.; Lum, J.; Low, I.; Forsberg, E.C.; et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 2015, 42, 665–678. [Google Scholar] [CrossRef]
- Ginhoux, F.; Guilliams, M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Chen, Y.C.; Chen, C.L.; Chiu, C.H. SARS-CoV-2 infection in children: Transmission dynamics and clinical characteristics. J. Formos. Med. Assoc. 2020, 119, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Lin, D.J.; Xiao, M.F.; Wang, J.C.; Wei, Y.; Lei, Z.X.; Zeng, Z.Q.; Li, L.; Li, H.A.; Xiang, W. 2019-novel coronavirus infection in a three-month-old baby. Zhonghua Er Ke Za Zhi 2020, 58, 6. [Google Scholar]
- Carsetti, R.; Quintarelli, C.; Quinti, I.; Piano Mortari, E.; Zumla, A.; Ippolito, G.; Locatelli, F. The immune system of children: The key to understanding SARS-CoV-2 susceptibility? Lancet Child Adolesc. Health 2020, 4, 414–416. [Google Scholar] [CrossRef]
- Heine, G.; Drozdenko, G.; Grun, J.R.; Chang, H.D.; Radbruch, A.; Worm, M. Autocrine IL-10 promotes human B-cell differentiation into IgM- or IgG-secreting plasmablasts. Eur. J. Immunol. 2014, 44, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Grimsholm, O.; Piano Mortari, E.; Davydov, A.N.; Shugay, M.; Obraztsova, A.S.; Bocci, C.; Marasco, E.; Marcellini, V.; Aranburu, A.; Farroni, C.; et al. The Interplay between CD27(dull) and CD27(bright) B Cells Ensures the Flexibility, Stability, and Resilience of Human B Cell Memory. Cell Rep. 2020, 30, 2963–2977. [Google Scholar] [CrossRef] [PubMed]


| Cytokine | Family | Type and Function | Cell Sources |
|---|---|---|---|
| IL-1β | IL-1 | Pro-inflammatory cytokine; Pyrogenic cytokine; Induction of Macrophages and T cells proliferation and differentiation; | Macrophages, monocytes, fibroblasts |
| IL-1Ra | IL-1 | Anti-inflammatory cytokine; Inhibition of IL-1 activity | Macrophages, monocytes, fibroblasts |
| IL-2 | IL-2 | Major growth factor and effector of T cells | T cells |
| IL-6 | IL-6 | Pro-inflammatory cytokine; Pyrogenic function; | Macrophages, T cells, endothelial cells |
| IL-10 | IL-10 | Anti-inflammatory cytokine; Inhibition of cytokine release and inflammatory response | Monocytes, T cells, B cells |
| IL-12 | IL-12 | Pro-inflammatory cytokine; Promotion of the Th1 pathway; Suppression of the Th2 pathway; Activation of NK cells | DCs, Macrophages, Neutrophils |
| IL-17 | IL-17 | Promotion of neutrophilic inflammation | Activated T cells, NK cells |
| IFN-α | IFN-I | Induction of antiviral immunity | pDCs, Monocytes, Leukocytes |
| IFN-β | IFN-I | Induction of antiviral immunity | pDCs, Monocytes, Fibroblasts |
| IFN-γ | IFN-II | Pro-inflammatory cytokine; Activation of Macrophages and NK cells | Th1 cells, CTLs, NK cells, activated B cells; |
| IFN-λ | IFN-III | Induction of antiviral immunity | Activated T cells, NK cells, DCs, Macrophages |
| TNFα | TNF | Pro-inflammatory cytokine; Pyrogenic cytokine; Induction of the expression of adhesion molecules | Macrophages, T cells, NK cells |
| IP-10 | CXCL10 | Interferon-induced chemokine; Induction of recruitment of Macrophages, NK cells and T cells | Monocytes, Endothelial cells |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauro, R.; Irrera, N.; Eid, A.H.; Bitto, A. Could Antigen Presenting Cells Represent a Protective Element during SARS-CoV-2 Infection in Children? Pathogens 2021, 10, 476. https://doi.org/10.3390/pathogens10040476
Lauro R, Irrera N, Eid AH, Bitto A. Could Antigen Presenting Cells Represent a Protective Element during SARS-CoV-2 Infection in Children? Pathogens. 2021; 10(4):476. https://doi.org/10.3390/pathogens10040476
Chicago/Turabian StyleLauro, Rita, Natasha Irrera, Ali H. Eid, and Alessandra Bitto. 2021. "Could Antigen Presenting Cells Represent a Protective Element during SARS-CoV-2 Infection in Children?" Pathogens 10, no. 4: 476. https://doi.org/10.3390/pathogens10040476
APA StyleLauro, R., Irrera, N., Eid, A. H., & Bitto, A. (2021). Could Antigen Presenting Cells Represent a Protective Element during SARS-CoV-2 Infection in Children? Pathogens, 10(4), 476. https://doi.org/10.3390/pathogens10040476







