Identification of a Novel Papillomavirus Type (MfoiPV1) Associated with Acrochordon in a Stone Marten (Martes foina)
Abstract
1. Introduction
2. Results
2.1. Case Description
2.2. Histopathology of the Skin Tumor
2.3. PCR, NGS and Genome Analysis
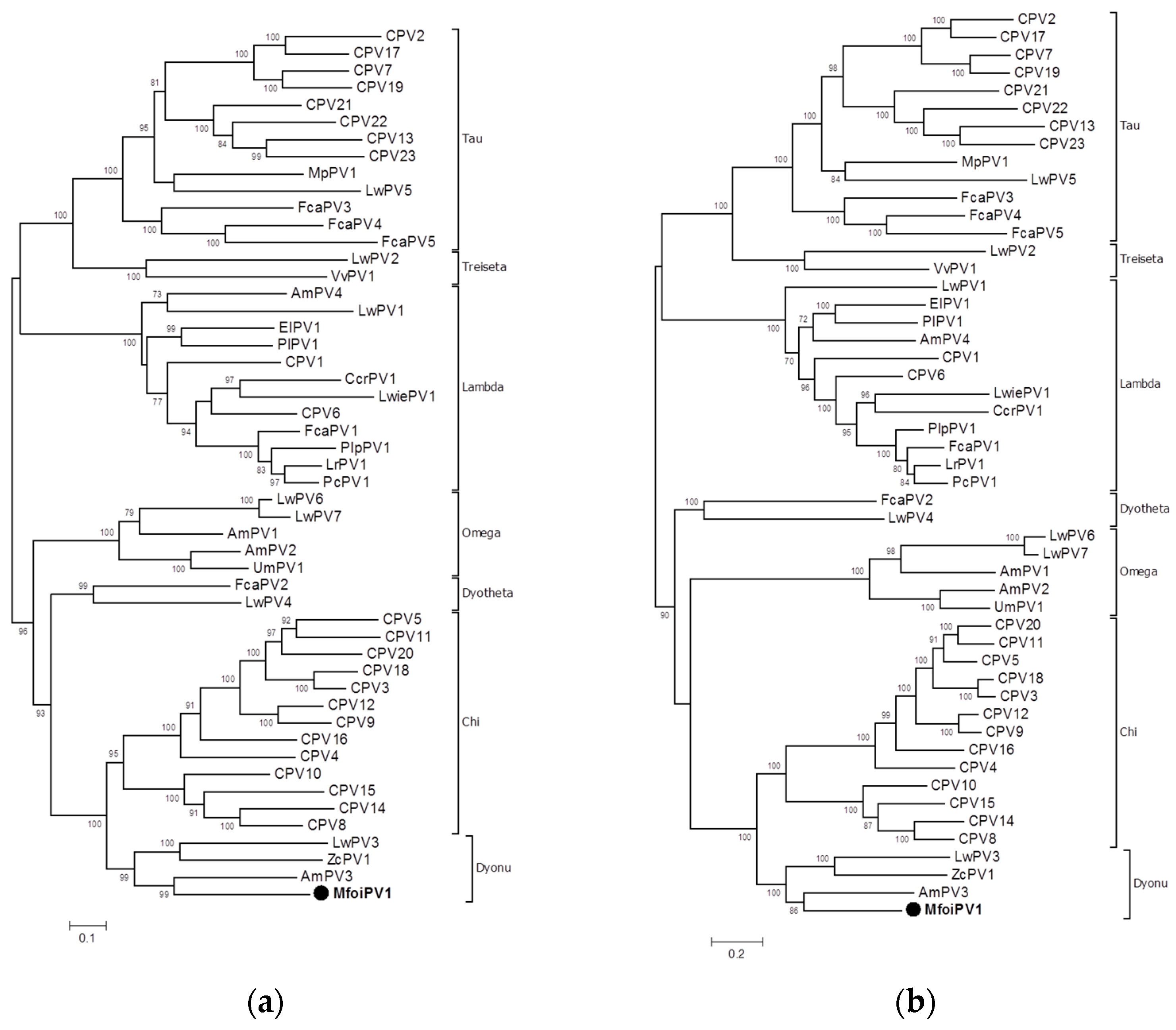
2.4. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Case Description and Sample Collection
4.2. Papillomavirus PCR
4.3. Whole-Genome Sequencing, Bioinformatics Analysis of NGS Data and Genome Annotation
4.4. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Doorslaer, K. Evolution of the papillomaviridae. Virology 2013, 445, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Chen, Z.; Bernard, H.U.; Chan, P.K.S.; DeSalle, R.; Dillner, J.; Forslund, O.; Haga, T.; McBride, A.A.; Villa, L.L.; et al. ICTV Virus Taxonomy Profile: Papillomaviridae. J Gen. Virol. 2018, 99, 989–990. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.U.; Burk, R.D.; Chen, Z.; van Doorslaer, K.; zur Hausen, H.; de Villiers, E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.M.; Baker, C.C. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 2006, 11, 2286–2302. [Google Scholar] [CrossRef]
- Araldi, R.P.; Assaf, S.M.R.; Carvalho, R.F.; Carvalho, M.A.C.R.; Souza, J.M.; Magnelli, R.F.; Módolo, D.G.; Roperto, F.P.; Stocco, R.C.; Beçak, W. Papillomaviruses: A systematic review. Genet. Mol. Biol. 2017, 40, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Rector, A.; Van Ranst, M. Animal papillomaviruses. Virology 2013, 445, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Daudt, C.; Da Silva, F.R.C.; Lunardi, M.; Alves, C.B.D.T.; Weber, M.N.; Cibulski, S.P.; Alfieri, A.F.; Alfieri, A.A.; Canal, C.W. Papillomaviruses in ruminants: An update. Transbound. Emerg. Dis. 2018, 65, 1381–1395. [Google Scholar] [CrossRef]
- Gil da Costa, R.M.; Peleteiro, M.C.; Pires, M.A.; DiMaio, D. An Update on Canine, Feline and Bovine Papillomaviruses. Transbound. Emerg. Dis. 2017, 64, 1371–1379. [Google Scholar] [CrossRef]
- Savini, F.; Dal Molin, E.; Gallina, L.; Casà, G.; Scagliarini, A. Papillomavirus in healthy skin and mucosa of wild ruminants in the Italian Alps. J. Wildl. Dis. 2016, 52, 82–87. [Google Scholar] [CrossRef]
- Borzacchiello, G.; Roperto, F. Bovine papillomaviruses, papillomas and cancer in cattle. Vet. Res. 2008, 39, 45. [Google Scholar] [CrossRef] [PubMed]
- Campo, M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002, 89, 249–261. [Google Scholar] [CrossRef]
- Munday, J.S. Bovine and human papillomaviruses: A comparative review. Vet. Pathol. 2014, 51, 1063–1075. [Google Scholar] [CrossRef]
- PaVE. The Papillomavirus Knowledge Source. Available online: https://pave.niaid.nih.gov (accessed on 8 February 2021).
- Van Doorslaer, K.; Tan, Q.; Xirasagar, S.; Bandaru, S.; Gopalan, V.; Mohamoud, Y.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 2012, 41, D571–D578. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef]
- Bravo, I.G.; de Sanjosé, S.; Gottschling, M. The clinical importance of understanding the evolution of papillomaviruses. Trends Microbiol. 2010, 18, 432–438. [Google Scholar] [CrossRef]
- Gottschling, M.; Göker, M.; Stamatakis, A.; Bininda-Emonds, O.R.; Nindl, I.; Bravo, I.G. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol. Biol. Evol. 2011, 28, 2101–2113. [Google Scholar] [CrossRef]
- Frias-de-Diego, A.; Jara, M.; Escobar, L.E. Papillomavirus in Wildlife. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef]
- López-Bueno, A.; Mavian, C.; Labella, A.M.; Castro, D.; Borrego, J.J.; Alcami, A.; Alejo, A. Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream. J. Virol. 2016, 90, 8768–8779. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Sidi, A.O.; Zanier, K.; Rybin, V.; Deryckère, F.; Rector, A.; Burk, R.D.; Lienau, E.K.; van Ranst, M.; Travé, G. Identification of unusual E6 and E7 proteins within avian papillomaviruses: Cellular localization, biophysical characterization, and phylogenetic analysis. J. Virol. 2009, 83, 8759–8770. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Kraberger, S.; Jennings, S.; Porzig, E.L.; Julian, L.; Massaro, M.; Pollard, A.; Ballard, G.; Ainley, D.G. A novel papillomavirus in Adélie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J. Gen. Virol. 2014, 95, 1352–1365. [Google Scholar] [CrossRef]
- Canuti, M.; Munro, H.J.; Robertson, G.J.; Kroyer, A.N.K.; Roul, S.; Ojkic, D.; Whitney, H.G.; Lang, A.S. New Insight Into Avian Papillomavirus Ecology and Evolution From Characterization of Novel Wild Bird Papillomaviruses. Front. Microbiol. 2019, 10, 701. [Google Scholar] [CrossRef] [PubMed]
- Herbst, L.H.; Lenz, J.; Van Doorslaer, K.; Chen, Z.; Stacy, B.A.; Wellehan, J.F.; Manire, C.A.; Burk, R.D. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology 2009, 383, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.E.; Favrot, C.; Ackermann, M.; Gull, J.; Vetsch, E.; Tobler, K. Novel snake papillomavirus does not cluster with other non-mammalian papillomaviruses. Virol. J. 2011, 8, 436. [Google Scholar] [CrossRef]
- da Silva, F.R.; Cibulski, S.P.; Daudt, C.; Weber, M.N.; Guimarães, L.L.; Streck, A.F.; Mayer, F.Q.; Roehe, P.M.; Canal, C.W. Novel Bovine Papillomavirus Type Discovered by Rolling-Circle Amplification Coupled with Next-Generation Sequencing. PLoS ONE 2016, 11, e0162345. [Google Scholar] [CrossRef] [PubMed]
- Altan, E.; Seguin, M.A.; Leutenegger, C.M.; Phan, T.G.; Deng, X.; Delwart, E. Nasal virome of dogs with respiratory infection signs include novel taupapillomaviruses. Virus Genes 2019, 55, 191–197. [Google Scholar] [CrossRef]
- Munday, J.S.; Dittmer, K.E.; Thomson, N.A.; Hills, S.F.; Laurie, R.E. Genomic characterisation of Felis catus papillomavirus type 5 with proposed classification within a new papillomavirus genus. Vet. Microbiol. 2017, 207, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Carrai, M.; Van Brussel, K.; Shi, M.; Li, C.X.; Chang, W.S.; Munday, J.S.; Voss, K.; McLuckie, A.; Taylor, D.; Laws, A.; et al. Identification of A Novel Papillomavirus Associated with Squamous Cell Carcinoma in A Domestic Cat. Viruses 2020, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.E.; Vetsch, E.; Ackermann, M.; Favrot, C.; Tobler, K. Four novel papillomavirus sequences support a broad diversity among equine papillomaviruses. J. Gen. Virol. 2013, 94, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.S.; Hardcastle, M.R.; Sim, M. Detection of a Putative Novel Papillomavirus Type within a Large Exophytic Papilloma on the Fetlock of a Horse. Pathogens 2020, 9, 816. [Google Scholar] [CrossRef]
- Alberti, A.; Pirino, S.; Pintore, F.; Addis, M.F.; Chessa, B.; Cacciotto, C.; Cubeddu, T.; Anfossi, A.; Benenati, G.; Coradduzza, E.; et al. Ovis aries Papillomavirus 3: A prototype of a novel genus in the family Papillomaviridae associated with ovine squamous cell carcinoma. Virology 2010, 407, 352–359. [Google Scholar] [CrossRef]
- Tore, G.; Cacciotto, C.; Anfossi, A.G.; Dore, G.M.; Antuofermo, E.; Scagliarini, A.; Burrai, G.P.; Pau, S.; Zedda, M.T.; Masala, G.; et al. Host cell tropism, genome characterization, and evolutionary features of OaPV4, a novel Deltapapillomavirus identified in sheep fibropapilloma. Vet. Microbiol. 2017, 204, 151–158. [Google Scholar] [CrossRef]
- Dolz, G.; Lecis, R.; Solórzano-Morales, A.; Aguilar-Vargas, F.; Solórzano-Scott, T.; Peña, R.; Zobba, R.; Tore, G.; Pittau, M.; Alberti, A. Leopardus wiedii Papillomavirus type 1, a novel papillomavirus species in the tree ocelot, suggests Felidae Lambdapapillomavirus polyphyletic origin and host-independent evolution. Infect. Genet. Evol. 2020, 81, 104239. [Google Scholar] [CrossRef]
- Cortés-Hinojosa, G.; Subramaniam, K.; Wellehan, J.F.X.; Ng, T.F.F.; Delwart, E.; McCulloch, S.D.; Goldstein, J.D.; Schaefer, A.M.; Fair, P.A.; Reif, J.S.; et al. Genomic sequencing of a virus representing a novel type within the species Dyopipapillomavirus 1 in an Indian River Lagoon bottlenose dolphin. Arch. Virol. 2019, 164, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Knowles, S.; Windels, S.K.; Adams, M.; Hall, J.S. Lambdapapillomavirus 2 in a Gray Wolf ( Canis lupus ) from Minnesota, USA with Oral Papillomatosis and Sarcoptic Mange. J. Wildl. Dis. 2017, 53, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Mengual-Chuliá, B.; Wittstatt, U.; Bravo, I.G. The First Papillomavirus Isolated from Vulpes vulpes (VvulPV1) Is Basal to the Gammapapillomavirus Genus. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Everest, D.J.; Dastjerdi, A.; Inglese, N.; Barlow, A.; Couper, D. First report of papillomatosis due to papillomavirus in a Eurasian badger (Meles meles). Vet. Rec. 2019, 7, e000912. [Google Scholar] [CrossRef]
- Liu, P.; Qiu, Y.; Xing, C.; Zhou, J.H.; Yang, W.H.; Wang, Q.; Li, J.Y.; Han, X.; Zhang, Y.Z.; Ge, X.Y. Detection and genome characterization of two novel papillomaviruses and a novel polyomavirus in tree shrew (Tupaia belangeri chinensis) in China. Virol. J. 2019, 16, 35. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, S.; Shan, T.; Hou, R.; Liu, Z.; Li, W.; Guo, L.; Wang, Y.; Chen, P.; Wang, X.; et al. Virome comparisons in wild-diseased and healthy captive giant pandas. Microbiome 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Erdélyi, K.; Bálint, A.; Dencso, L.; Dán, A.; Ursu, K. Characterisation of the first complete genome sequence of the roe deer (Capreolus capreolus) papillomavirus. Virus Res. 2008, 135, 307–311. [Google Scholar] [CrossRef]
- Erdélyi, K.; Gál, J.; Sugár, L.; Ursu, K.; Forgách, P.; Szeredi, L.; Steineck, T. Papillomavirus-associated fibropapillomas of red deer (Cervus elaphus). Acta Vet. Hung. 2009, 57, 337–344. [Google Scholar] [CrossRef]
- Garcês, A.; Pires, I.; Savini, F.; Scagliarini, A.; Gallina, L. Cutaneous Fibropapilloma in a Red Deer (Cervus elaphus) Associated with Cervus elaphus Papillomavirus in Portugal. J. Wildl. Dis. 2020, 56, 636–639. [Google Scholar] [CrossRef]
- Scagliarini, A.; Gallina, L.; Battilani, M.; Turrini, F.; Savini, F.; Lavazza, A.; Chiari, M.; Coradduzza, E.; Peli, A.; Erdélyi, K.; et al. Cervus elaphus papillomavirus (CePV1): New insights on viral evolution in deer. Vet. Microbiol. 2013, 165, 252–259. [Google Scholar] [CrossRef]
- Gallina, L.; Savini, F.; Casà, G.; Bertoletti, I.; Bianchi, A.; Gibelli, L.R.; Lelli, D.; Lavazza, A.; Scagliarini, A. Epitheliotropic Infections in Wildlife Ruminants From the Central Alps and Stelvio National Park. Front. Vet. Sci. 2020, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Kmetec, J.; Kuhar, U.; Fajfar, A.G.; Vengušt, D.; Vengušt, G. A Comprehensive Study of Cutaneous Fibropapillomatosis in Free-Ranging Roe Deer. Viruses 2020, 12, 1001. [Google Scholar] [CrossRef] [PubMed]
- Genovesi, P.; Secchi, M.; Boitani, L. Diet of stone martens: An example of ecological flexibility. J. Zool. 1996, 238, 545–555. [Google Scholar] [CrossRef]
- Kryštufek, B. Distribution of martens (gen. Martes Pinel, 1792, Carnivora, Mammalia) in Slovenia. Biološki Vestn. 1984, 32, 21–26. [Google Scholar]
- SiStat. Republic of Slovenia—Statistical Office—Hunting (number), Slovenia, Annually. 2019. Available online: https://pxweb.stat.si/SiStatData/pxweb/en/Data/-/1673150S.px/ (accessed on 4 March 2021).
- de Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Bidaut, A.P.; Gross, T.L.; Noli, C.; Welle, M.; Suter, M.M. Acrochordonous plaques in two Bulldogs and a Pug dog. Vet. Dermatol. 2003, 14, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.A.; Friesen, R.; Oliveira, J.K.; Teixeira, W.T.; Sillas, T.; Werner, J.; Castro, J.L.C.; Dornbusch, P.T. Acrochordonous plaque in a Doberman Pinscher dog. Open Vet. J. 2019, 9, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Pezeshkpoor, F.; Jafarian, A.H.; Ghazvini, K.; Yazdanpanah, M.J.; Sadeghian, A.; Esmaili, H.; Karrabi, M.; Rohani, F.; Joushan, B. An association of human papillomaviruses low risk and high risk subtypes with skin tag. Iran. J. Basic Med. Sci. 2012, 15, 840–844. [Google Scholar] [PubMed]
- Dianzani, C.; Calvieri, S.; Pierangeli, A.; Imperi, M.; Bucci, M.; Degener, A.M. The detection of human papillomavirus DNA in skin tags. Br. J. Dermatol. 1998, 138, 649–651. [Google Scholar] [CrossRef]
- Sallam, M.A.; Kamel, M.M.; El Missiry, A.G.; Helal, M.F. Detection of human papillomavirus DNA in skin tags. Sci J. Al-Azhar Med. 2003, 24, 311–317. [Google Scholar]
- Gross, T.L.; Ihrke, P.J.; Walder, E.J.; Affolter, V.K. Fibrous Tumors. In Skin Diseases of the Dog and Cat: Clinical and Histopathological Diagnosis, 2nd ed.; Gross, T.L., Ihrke, P.J., Eds.; Blackwell Science: Oxford, UK, 2005; pp. 710–734. [Google Scholar]
- Ng, T.F.; Miller, M.A.; Kondov, N.O.; Dodd, E.M.; Batac, F.; Manzer, M.; Ives, S.; Saliki, J.T.; Deng, X.; Delwart, E. Oral papillomatosis caused by Enhydra lutris papillomavirus 1 (ElPV-1) in southern sea otters (Enhydra lutris nereis) in California, USA. J. Wildl. Dis. 2015, 51, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Smits, S.L.; Raj, V.S.; Oduber, M.D.; Schapendonk, C.M.; Bodewes, R.; Provacia, L.; Stittelaar, K.J.; Osterhaus, A.D.; Haagmans, B.L. Metagenomic analysis of the ferret fecal viral flora. PLoS ONE 2013, 8, e71595. [Google Scholar] [CrossRef] [PubMed]
- Rivera, R.; Robles-Sikisaka, R.; Hoffman, E.M.; Stacy, B.A.; Jensen, E.D.; Nollens, H.H.; Wellehan, J.F. Characterization of a novel papillomavirus species (ZcPV1) from two California sea lions (Zalophus californianus). Vet. Microbiol. 2012, 155, 257–266. [Google Scholar] [CrossRef]
- Smeele, Z.E.; Burns, J.M.; Van Doorsaler, K.; Fontenele, R.S.; Waits, K.; Stainton, D.; Shero, M.R.; Beltran, R.S.; Kirkham, A.L.; Berngartt, R.; et al. Diverse papillomaviruses identified in Weddell seals. J. Gen. Virol. 2018, 99, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Forslund, O.; Antonsson, A.; Nordin, P.; Stenquist, B.; Göran Hansson, B. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 1999, 80 Pt 9, 2437–2443. [Google Scholar] [CrossRef]
- Lange, C.E.; Tobler, K.; Brandes, K.; Breithardt, K.; Ordeix, L.; Von Bomhard, W.; Favrot, C. Canine inverted papillomas associated with DNA of four different papillomaviruses. Vet. Dermatol. 2010, 21, 287–291. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]

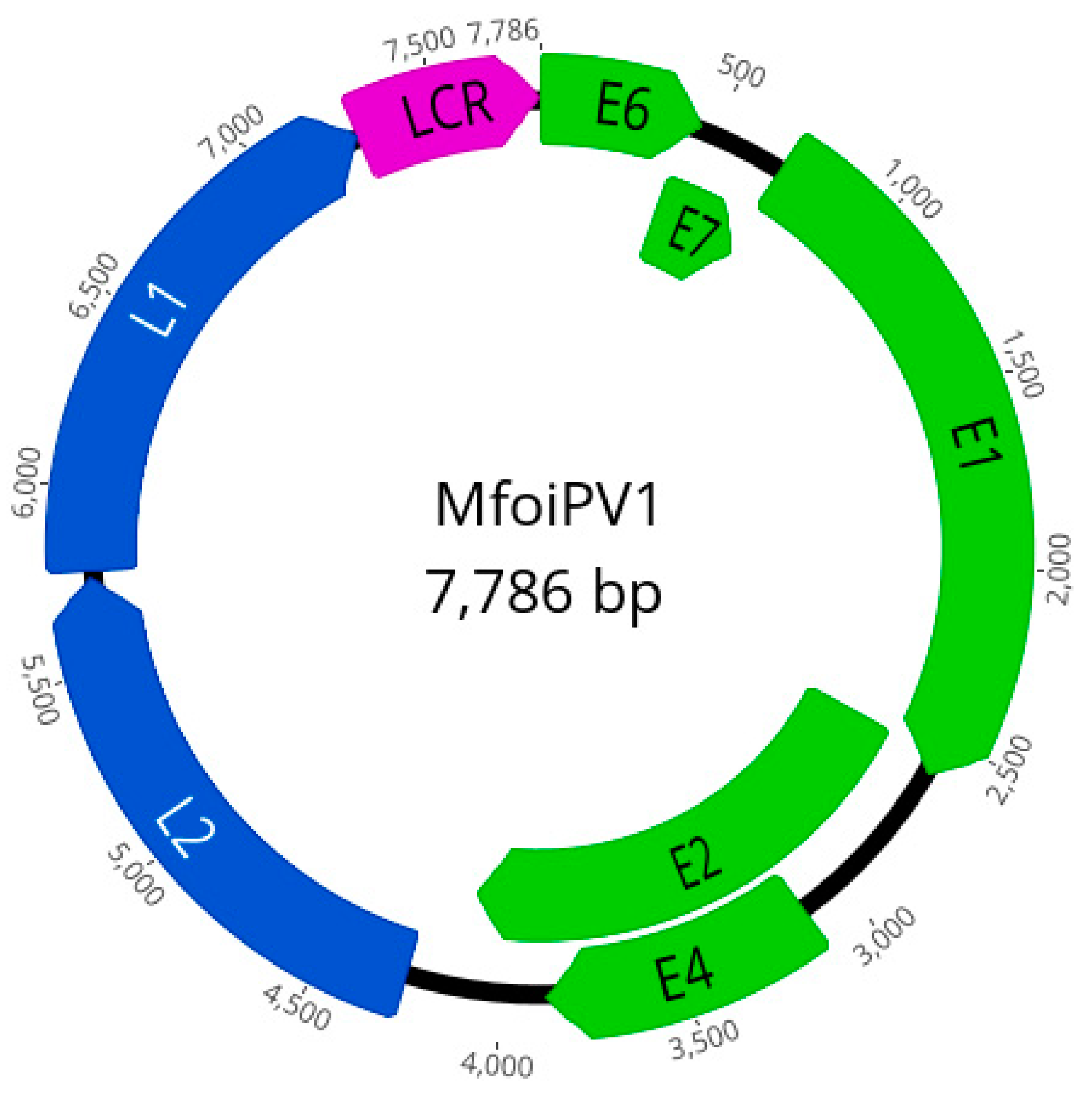
| ORF | Predicted aa Features | ||||
|---|---|---|---|---|---|
| Name | Position in Genome | Aa Length | Name | Sequence | Aa Start Position in ORF |
| E6 | 1–456 | 152 | Zinc-binding domains | CXXCX(29)CXXC | 25, 98 |
| E7 | 416–712 | 99 | RB protein binding domain | LXCXE; in MFoiPV1: LWCDE | 23 |
| Zink-binding domains | CXXCX(29)CXXC | 51 | |||
| E1 | 702–2603 | 634 | ATP-dependent helicase motif | GXXXXGK[TS]; in MFoiPV1: GPPNTGKS | 461 |
| Cyclin interaction motifs | RXL | 56, 113, 278, 398, 446, 561 | |||
| E2 | 2545–4116 | 524 | / | ||
| E4 | 3116–3874 | 253 | / | ||
| L2 | 4268–5761 | 498 | / | ||
| L1 | 5777–7273 | 499 | Bipartite nuclear localization signal | RKFLAQSSATARPVKRRAPP | 467 |
| Predicted nt features | |||||
| Name | Name | Sequence | Nt Start Position in Genome | ||
| LCR | E2 binding sites | ACCGNNNNCGGT; | 7714, 7638, 7578, 7545, 7336 | ||
| LCR | TATA-like box | TATA[AT]A[AT]; in MFoiPV1: TATATATA | 7748 | ||
| LCR | SP1 binding site | GGCGGG | 7772 | ||
| E2 | NF1 binding site | CGGAAA | 3767 | ||
| E1, E2, L2, L1 | Polyadenylation signal sites | ATAAA | 1434, 3038, 4359, 6443, 6858 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhar, U.; Žele Vengušt, D.; Jamnikar-Ciglenečki, U.; Vengušt, G. Identification of a Novel Papillomavirus Type (MfoiPV1) Associated with Acrochordon in a Stone Marten (Martes foina). Pathogens 2021, 10, 539. https://doi.org/10.3390/pathogens10050539
Kuhar U, Žele Vengušt D, Jamnikar-Ciglenečki U, Vengušt G. Identification of a Novel Papillomavirus Type (MfoiPV1) Associated with Acrochordon in a Stone Marten (Martes foina). Pathogens. 2021; 10(5):539. https://doi.org/10.3390/pathogens10050539
Chicago/Turabian StyleKuhar, Urška, Diana Žele Vengušt, Urška Jamnikar-Ciglenečki, and Gorazd Vengušt. 2021. "Identification of a Novel Papillomavirus Type (MfoiPV1) Associated with Acrochordon in a Stone Marten (Martes foina)" Pathogens 10, no. 5: 539. https://doi.org/10.3390/pathogens10050539
APA StyleKuhar, U., Žele Vengušt, D., Jamnikar-Ciglenečki, U., & Vengušt, G. (2021). Identification of a Novel Papillomavirus Type (MfoiPV1) Associated with Acrochordon in a Stone Marten (Martes foina). Pathogens, 10(5), 539. https://doi.org/10.3390/pathogens10050539







