Abstract
Beta-hemolytic streptococci cause a variety of infectious diseases associated with high morbidity and mortality. A key factor for successful infection is host colonization, which can be difficult in a multispecies environment. Secreting bacteriocins can be beneficial during this process. Bacteriocins are small, ribosomally produced, antimicrobial peptides produced by bacteria to inhibit the growth of other, typically closely related, bacteria. In this systematic review, bacteriocin production and regulation of beta-hemolytic streptococci was surveyed. While Streptococcus pyogenes produces eight different bacteriocins (Streptococcin A-FF22/A-M49, Streptin, Salivaricin A, SpbMN, Blp1, Blp2, Streptococcin A-M57), only one bacteriocin of Streptococcus agalactiae (Agalacticin = Nisin P) and one of Streptococcus dysgalactiae subsp. equisimilis (Dysgalacticin) has been described. Expression of class I bacteriocins is regulated by a two-component system, typically with autoinduction by the bacteriocin itself. In contrast, a separate quorum sensing system regulates expression of class II bacteriocins. Both identified class III bacteriocins are plasmid-encoded and regulation has not been elucidated.
1. Introduction
Bacteriocins are small antimicrobial peptides or proteins of bacterial origin. They inhibit the growth of other, often closely related, bacteria, resulting in a colonization advantage of the producer [1]. The mostly cationic nature of these peptides leads to an efficient interaction with the negatively charged bacterial membrane, enabling membrane permeabilization, the main mechanism of action. Typical bacteriocin features like high thermal and pH stability in connection with antimicrobial activity provide interesting characteristics for an application in food preservation. Due to their antimicrobial effects, bacteriocins are also investigated in clinical settings to explore their potential as alternative therapeutics for infectious diseases [2]. Other clinical applications of bacteriocins include oral care, treatment of peptic ulcers and skin care [3]. Additionally, bacteriocins are applied in food preservation and plant growth promotion. Bacteriocins of Gram-positive bacterial species are subdivided into four distinct classes, with each class having different characteristics [4]. Class I bacteriocins are small, heat-stable peptides. The mature peptides are formed by post-translational modifications that generate non-proteinogenic amino acids such as lanthionine, β-methyl lanthionine, and dehydrated amino acids [5]. Class II bacteriocins are typically smaller than 10 kDa and rarely harbor any post-translational modifications [6]. Bacteriocins belonging to class III are heat-labile proteins with a molecular weight of around 30 kDa. Class I and II bacteriocins typically cause bacterial membrane permeabilization via direct interactions, whereas some class III bacteriocins can also exert enzymatic functions, such as the degradation of cell wall structures [7]. Class IV bacteriocins are large complex proteins incorporating either lipid or carbohydrate moieties. However, some publications re-classified these macromolecules as bacteriolysins (hydrolytic polypeptides), leaving only three distinct bacteriocin classes [4,8].
Regulation of bacteriocin expression is often dependent on cell density [9,10], which is monitored by quorum sensing (QS) systems to control intra- and interspecies communication [11]. Signaling peptides of QS systems are peptide pheromones or autoinducing peptides that are secreted into the extracellular environment. At the threshold concentration of the autoinducing peptide bacterial group behavior is triggered, such as the production of antimicrobial peptides. In general, two QS activation pathways exist [9]. One activation pathway involves an autoinducing peptide and a cytoplasmic transcriptional regulator. In this case, the transcriptional cytoplasmic regulator typically belongs to the RRNPP family (Rap, Rgg, NprR, PlcR, and PrgX) and is able to modulate gene expression after a direct interaction with the autoinducing peptide [12]. The second system consists of an autoinducing peptide and a two-component signal transduction system. The autoinducing peptide is exported and sensed by the transmembrane histidine kinase of the two-component system. The response regulator is then phosphorylated and acts as a transcription regulator, ultimately leading to an alteration in gene expression. A prominent example of such a system is the bacteriocin-like peptide (blp) region in Streptococcus pneumoniae. The peptide pheromone BlpC is exported via the ATP-binding cassette transporter BlpAB and sensed by the histidine kinase BlpH, which phosphorylates the transcription activator BlpR [13]. Phosphorylated BlpR turns on the expression of blp and bacteriocin-genes. Secretion of BlpC is induced by the competence-stimulating peptide CSP and couples bacteriocin production to competence [14]. The blp locus is highly variable and at least 16 bacteriocin encoding genes have been described in connection to the blp locus of S. pneumoniae [15,16]. An association of bacteriocin production with the Blp system has also been reported in other streptococci, including Streptococcus mutans, Streptococcus thermophilus, and Streptococcus salivarius [17,18,19,20].
In regulators of bacteriocin production, cross-reactivity of the autoinducing peptide between different species can occur [21]. For class I bacteriocins, the quorum sensing system is normally part of the bacteriocin gene cluster, and the structural gene of the bacteriocin functions as an autoinducer, as it was described for Nisin [22]. In class II bacteriocins, a separate QS system is typically present, which is induced by a peptide pheromone and controls bacteriocin production. In this review, bacteriocin production and its regulation through quorum sensing systems were surveyed for beta-hemolytic streptococci.
Beta-hemolytic streptococci relevant for human infections mainly comprise Streptococcus pyogenes (Group A streptococci, GAS), Streptococcus agalactiae (Group B streptococci, GBS), and Streptococcus dysgalactiae subsp. equisimilis (SDSE). These species cause a variety of infectious diseases and are associated with high levels of morbidity and mortality. Colonization often precedes streptococcal infections and the production of bacteriocins provides a colonization advantage for the producing organism. Understanding the determinants of streptococcal colonization may thus provide the knowledge required for interfering with the development of streptococcal infections. Several bacteriocins of S. pyogenes have been described already. In contrast, only very few aspects are known about bacteriocin production in SDSE and S. agalactiae. An overview is given in Table 1 and Figure 1.

Table 1.
Schematic overview of bacteriocins produced by beta-hemolytic streptococci. N.D. indicates not determined.
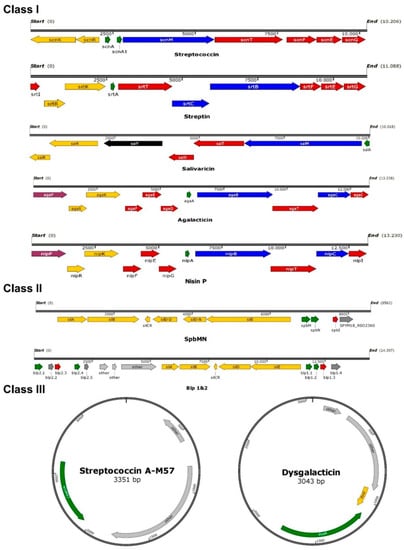
Figure 1.
Schematic overview of bacteriocin operons described in beta-hemolytic streptococci. Overview was constructed using SnapGene. Green indicates the structural gene, blue stands for modification, yellow marks regulation genes, genes for transport and immunity are colored red, purple indicates a peptidase, black genes have unknown functions, and grey genes are not part of the bacteriocin gene cluster. To obtain operon organization and DNA sequences of the published bacteriocins, the accession numbers provided by authors were used (Streptococcin A-FF22: AF026542; Streptin: AB030831; Salivaricin A: AE014074; Streptococcin A-M57: AY648561; SpbMN: AE009949; Blp1&2: CP021640; Dysgalacticin: AY907345; Agalacticin: NZ_GL636070; Nisin P: WIDP00000000).
2. Streptococcus pyogenes
So far, eight different bacteriocins have been identified and characterized for S. pyogenes. Bacteriocin production of S. pyogenes was first described in 1971 with the identification of Streptococcin A-FF22 [23]. This 26-amino-acid (aa) peptide is a lantibiotic and belongs to type AII bacteriocins. Type AII lantibiotics are characterized by a ring structure and the typical leader peptide cleavage. Specific genes for modification (scnM), transport (scnT), immunity (scnFEG), and regulation (scnKR) of the bacteriocin are present in the A-FF22 gene cluster. Two structural genes with scnA and scnA’ are also part of the scn gene cluster [33]. The role of scnA’ is however unclear since no actual peptide appears to be produced. Pore formation and the subsequent disruption of the membrane potential leads to the death of sensitive target bacteria [34]. Furthermore, the complete gene cluster seems to be part of a mobile genetic element, which may indicate an acquisition of these genes through horizontal gene transfer. The locus can be found in 9% (13 of 144) of S. pyogenes strains [35]. In a different S. pyogenes background, a minor variant of A-FF22 exists, Streptococcin A-M49. It is encoded by two slightly different structural genes, scnA’ and scnA´´, while the mature bacteriocin A-M49 is identical to mature Streptococcin A-FF22 [36]. ScnA´ has the same aa sequence as Streptococcin A-FF22, whereas scnA´´ differs in four aa of the leader peptide sequence. Streptococcin A-M49 is produced by 33% (7 of 21) of M-type 49 strains under anaerobic conditions and shows the same spectrum of antimicrobial activity as A-FF22 [24]. In addition to the sequence similarity of the bacteriocin itself, adjacent genetic regions are identical between Streptococcin A-M49 and Streptococcin A-FF22. A scnA-like gene can furthermore be found in S. salivarius, S. dysgalactiae and S. macedonicus. Regulation of Streptococcin A-FF22 is dependent on a two-component system formed by the response regulator gene scnR and the histidine kinase gene scnK. An inactivation of one of these genes leads to a loss of Streptococcin A-FF22 production [33]. However, in contrast to other class I bacteriocins, production of Streptococcin A-FF22 does not seem to be autoinducible [35]. A-FF22 has a wide spectrum of antimicrobial activity against Gram-positive species. It inhibits the growth of Staphylococcus citreus, Micrococcus spp., Bacillus spp., Corynebacterium spp., and various streptococci, including S. pyogenes, Streptococcus. agalactiae, Streptococcus equi, and Streptococcus cremoris [37].
Another lantibiotic, called Streptin, has only been described for S. pyogenes. It self-regulates its expression via QS [38]. Two forms of Streptin are known, with Streptin 2 being an unprocessed and differently dehydrated form of Streptin 1. The Streptin gene cluster shows high homology to Nisin and Subtilin gene clusters. SrtA, the structural gene for Streptin, is part of a 10-gene operon [25]. Similar to Nisin and Subtilin, the cluster contains genes important for modification (srtBC), regulation (srtRK), and transport and immunity (srtT, srtI, srtEFG). The genes srtRK appear to constitute a two-component system that is activated by Streptin [39]. The gene cluster was found to be present in 69% (40 of 58) of S. pyogenes strains. However, only 10 of these strains were able to produce active Streptin [38]. In some cases, the srt locus showed mutations, but several strains had an intact srt locus without producing Streptin. Concerning its spectrum of antibacterial activity, the bacteriocin inhibits the growth of S. pyogenes strains and Micrococcus luteus.
A third lantibiotic expressed by S. pyogenes is Salivaricin A. It is a type IIA lantibiotic that was first described in S. salivarius, where it can be found in approximately 10% of the strains [40]. In S. pyogenes, Salivaricin A seems to be exclusively expressed by M4-isolates, even though the majority of analyzed strains contain the sal locus [21,41]. The production of Salivaricin A has been reported for at least six different streptococcal species, among them S. dysgalactiae and S. agalactiae [41]. Furthermore, five different variants have been described (salA1-5) [41]. Expression of salA is autoinduced and all genes necessary for quorum sensing (salTKR) as well as for modification (salM) and immunity (salXY) are part of one gene cluster [11,29,30]. Autoinduction capacity is not limited to certain subtypes, in fact, all subtypes are able to induce the production of the other subtypes [21]. Even though a weak interaction of Salivaricin A2 with lipid II exists, the mechanism of action has not been elucidated [42]. Salivaricin A of S. salivarius is active against S. pyogenes, M. luteus, S. pneumoniae and Corynebacterium spp. [42]. To our knowledge, the activity of Salivaricin A produced by S. pyogenes has not been investigated.
In contrast to class I bacteriocins of S. pyogenes, class II bacteriocins are regulated by an independent quorum sensing mechanism. In this context, only the streptococcal invasion locus (sil) has been described. The sil locus was first identified by Hidalgo-Grass and has been demonstrated to be important for the virulence of S. pyogenes [43]. Furthermore, this locus shows high homologies to the competence system of S. pneumoniae, which is linked to bacteriocin production [13,43]. The sil locus consists of a two-component system (SilAB), an autoinducing peptide (SilCR), and an ATP-binding cassette transporter system (SilDE) [43,44]. The histidine kinase SilB senses extracellular SilCR and phosphorylates the response regulator SilA. Phosphorylated SilA alters gene expression and induces SilCR production, which is subsequently processed and excreted by SilDE.
While 16% of clinical S. pyogenes isolates contain a sil system, only 9% seem to have a functional system, since the other strains carry relevant mutations of this locus [45]. One possible explanation for this finding may be that sil originally provided an advantage to S. pyogenes strains for survival in a polymicrobial environment, but degenerated during adaptation to the host [27]. However, even possessing parts of sil may be beneficial. Strains carrying an incomplete sil locus can still produce immunity proteins, which provide protection against bacteriocins of competitor strains. Essential in this context is an intact sensing system (SilAB) enabling a reaction to SilCR present in the environment.
Adjacent to the sil region, two class II bacteriocins have been described. Armstrong et al. investigated the bacteriocin (spb) genes of S. pyogenes strain MGAS8232 [26]. In this strain, however, the sil system does not seem to be functional. SpbM and spbN encode putative bacteriocins, which share homologies to blpMN of S. pneumoniae. Both peptides have a double-glycine-leader peptide, which is necessary for export of the mature peptide [46]. Furthermore, SpbM and SpbN contain GxxxG motifs. These motifs are important for the interaction of the antimicrobial peptides with each other and furthermore for insertion into the bacterial membrane of sensitive strains [6]. When tested in a deferred antagonism assay, only the combination of SpbM and SpbN was able to inhibit target strains, establishing that SpbMN is a two-component bacteriocin. Antimicrobial activity was demonstrated for several S. pyogenes strains as well as for Streptococcus dysgalactiae, Streptococcus uberis, Micrococcus luteus, and Lactococcus lactis.
A second and third class II bacteriocin encoded by a genetic locus adjacent to sil were described by Hertzog et al., Blp1 and Blp2 [27]. The study investigated S. pyogenes strain JS12, which, in contrast to other S. pyogenes strains, encodes a functional sil system. Hallmark traits of class II bacteriocins including a double-glycine-leader peptide and a GxxxG motif can be detected in the peptides Blp1.1 and Blp1.2 that are encoded in the blp1 region. These two peptides work as a two-component bacteriocin (Blp1). Another functional bacteriocin Blp2 is encoded in the blp2 region. Both of these bacteriocins are produced by the strain J12 and inhibit other S. pyogenes strains. Analyzing publicly available genomes of S. pyogenes strains harboring a sil system revealed that putative bacteriocin genes are commonly found adjacent to sil. Bioinformatic tools are available to efficiently identify and characterize these bacteriocins [47].
A class III bacteriocin designated Streptococcin A-M57 is exclusively produced by M-type 57 S. pyogenes strains [24,36,48]. Streptococcin A-M57 is plasmid-encoded and sensitive towards heat and proteases. It does not inhibit the growth of other S. pyogenes strains but targets several other Gram-positive bacteria such as Micrococcus luteus, Lactococcus lactis, Listeria spp., Bacillus megaterium, and Staphylococcus simulans. Due to structural similarities of Streptococcin A-M57 to Dysgalacticin (see SDSE), a similar mode of action, including impaired glucose uptake and disturbed membrane potential, is proposed.
3. Streptococcus dysgalactiae Subspecies equisimilis
While the sil locus was first described in S. pyogenes, it is much more prevalent in SDSE with 82% of the strains harboring this genetic region [49]. Like in S. pyogenes, a class II bacteriocin blpM gene cluster can be found adjacent to this locus. However, the functionality of these genes has not been investigated. Nevertheless, at a descriptive level, there are reports of SDSE showing inhibitory activity against other streptococci and Corynebacterium spp., but no link to known bacteriocins or bacteriocin genes has been established [50,51,52].
One bacteriocin of SDSE has been characterized at a functional level. It is Dysgalacticin, which belongs to the class III bacteriocins and shows similarities in secondary structure to Streptococcin A-M57 of S. pyogenes. It is a 21.5kDa large protein encoded on an indigenous plasmid [29]. Of its 220 aa, the first 28 aa seem to be a leader peptide, causing an export via the sec dependent pathway, as described for Zoocin A [53].
Dysgalacticin was shown to target the phosphoenolpyruvate-dependent glucose and mannose phosphotransferase system (PTS) of S. pyogenes, leading to a starvation of the bacterial cell [30]. Furthermore, Dysgalacticin causes a disturbed membrane potential and the increased membrane permeability leads to a leakage of intracellular ions. Adjacent to the structural gene dysA, the gene dysI, encoding an immunity factor, is found [54]. DysI is a membrane-associated protein, protecting producing cells from the detrimental effects of Dysgalacticin by a putative interaction with the glucose and mannose PTS, which constitutes the Dysgalacticin receptor of the target cell. The regulation of Dysgalacticin production in SDSE has not been explored. In contrast to other streptococcal bacteriocins, Dysgalacticin has a narrow spectrum of activity that appears to be limited to S. pyogenes strains.
4. Streptococcus agalactiae
In 1983, the production of antimicrobial inhibitory substances was described for Group B streptococcal strains, isolated from humans and animals [51]. All in all, only 5% of the S. agalactiae isolates showed any inhibition of other bacteria. However, characteristics of a class III bacteriocin (large, heat-labile protein) and of a small, heat-resistant bacteriocin were described, but in the years following the initial observation, no further molecular genetic studies were reported on these findings.
Just recently, a first detailed molecular description of a bacteriocin was provided for S. agalactiae [31]. Agalacticin was identified by genome analysis with the data mining tool BAGEL3 and is produced by the S. agalactiae-type strain ATCC 13813 [31]. It shows high similarity to Nisin, including the leader peptide. All genes necessary for modification (agaBC), transport (agaT), immunity (agaI and agaFEG), regulation (agaRK), and leader peptidase (agaP) are present within the gene cluster. Agalacticin shows antimicrobial activity against several Gram-positive species (Enterococcus faecalis, Bacillus cereus, Staphylococcus aureus MRSA, Micrococcus flavus, and Listeria monocytogenes). However, regulation of the expression of Agalacticin has not yet been investigated. The similarity to the Nisin gene cluster as well as the presence of lanRK, which encodes a putative two-component regulatory system, point in the direction of a quorum sensing system autoregulated by Agalacticin. Another bacteriocin described for S. agalactiae is Nisin P, which was identified in S. agalactiae strain DPC7040 [32]. Nisin P production required an initial induction with Nisin A but, over time, self-induction occurred. Additionally, a cross reactivity of the Nisin A promotor to Nisin P was observed, even though higher concentrations of Nisin P are needed to activate the promotor. Several Gram-positive species, especially lactobacilli and staphylococci, were inhibited by Nisin P. Since peptide sequence, operon organization and the size of Agalacticin and Nisin P are identical, the same peptide may have been described in both publications under different names. Furthermore, DNA sequence analysis revealed 99.96% of sequence identity.
5. Conclusions
Beta-hemolytic streptococcal species are part of polymicrobial environments that require competing with the commensal microbiota. Interestingly, while for S. pyogenes a variety of bacteriocins have been described, only a few are reported for SDSE and S. agalactiae. The identified bacteriocins are mainly active against other Gram-positive bacteria and closely related streptococcal species, providing a colonization advantage to bacteriocin producers. Understanding the regulation of these peptides might help in elucidating the role they play in microbial communities and their importance for host colonization.
Author Contributions
V.V. researched and wrote the manuscript, and B.S. designed, modified, and reviewed it. All authors have read and agreed to the published version of the manuscript.
Funding
V.V. was supported by the International Graduate School in Molecular Medicine Ulm. The work of B.S. was funded by the German Research Foundation (DFG) within the CRC 1279.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the members of the Streptococci laboratory at University Ulm Medical Center for helpful input and discussions. V.V. thanks the International Graduate School in Molecular Medicine Ulm for the support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin Production Augments Niche Competition by Enterococci in the Mammalian Gastrointestinal Tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial Activities of Bacteriocins: Application in Foods and Pharmaceuticals. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and Emerging Applications of Bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Nissen-Meyer, J.; Rogne, P.; Oppegård, C.; Haugen, H.S.; Kristiansen, P.E. Structure-Function Relationships of the Non-Lanthionine-Containing Peptide (Class II) Bacteriocins Produced by Gram-Positive Bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a Cell Wall-Degrading Bacteriocin from Enterococcus Faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- Kareb, O.; Aïder, M. Quorum Sensing Circuits in the Communicating Mechanisms of Bacteria and Its Implication in the Biosynthesis of Bacteriocins by Lactic Acid Bacteria: A Review. Probiotics Antimicrob. Proteins 2020, 12, 5–17. [Google Scholar] [CrossRef]
- Kleerebezem, M. Quorum Sensing Control of Lantibiotic Production; Nisin and Subtilin Autoregulate Their Own Biosynthesis. Peptides 2004, 25, 1405–1414. [Google Scholar] [CrossRef]
- Atkinson, S.; Williams, P. Quorum Sensing and Social Networking in the Microbial World. J. R. Soc. Interface 2009, 6, 959–978. [Google Scholar] [CrossRef]
- Neiditch, M.B.; Capodagli, G.C.; Prehna, G.; Federle, M.J. Genetic and Structural Analyses of RRNPP Intercellular Peptide Signaling of Gram-Positive Bacteria. Annu. Rev. Genet. 2017, 51, 311–333. [Google Scholar] [CrossRef] [PubMed]
- de Saizieu, A.; Gardès, C.; Flint, N.; Wagner, C.; Kamber, M.; Mitchell, T.J.; Keck, W.; Amrein, K.E.; Lange, R. Microarray-Based Identification of a Novel Streptococcus Pneumoniae Regulon Controlled by an Autoinduced Peptide. J. Bacteriol. 2000, 182, 4696–4703. [Google Scholar] [CrossRef] [PubMed]
- Wholey, W.-Y.; Kochan, T.J.; Storck, D.N.; Dawid, S. Coordinated Bacteriocin Expression and Competence in Streptococcus Pneumoniae Contributes to Genetic Adaptation through Neighbor Predation. PLoS Pathog. 2016, 12, e1005413. [Google Scholar] [CrossRef]
- Bogaardt, C.; van Tonder, A.J.; Brueggemann, A.B. Genomic Analyses of Pneumococci Reveal a Wide Diversity of Bacteriocins-Including Pneumocyclicin, a Novel Circular Bacteriocin. BMC Genomics 2015, 16, 554. [Google Scholar] [CrossRef]
- Valente, C.; Dawid, S.; Pinto, F.R.; Hinds, J.; Simões, A.S.; Gould, K.A.; Mendes, L.A.; de Lencastre, H.; Sá-Leão, R. The Blp Locus of Streptococcus pneumoniae Plays a Limited Role in the Selection of Strains That Can Cocolonize the Human Nasopharynx. Appl. Environ. Microbiol. 2016, 82, 5206–5215. [Google Scholar] [CrossRef]
- Shanker, E.; Federle, M.J. Quorum Sensing Regulation of Competence and Bacteriocins in Streptococcus Pneumoniae and Mutans. Genes 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Mignolet, J.; Fontaine, L.; Sass, A.; Nannan, C.; Mahillon, J.; Coenye, T.; Hols, P. Circuitry Rewiring Directly Couples Competence to Predation in the Gut Dweller Streptococcus Salivarius. Cell Rep. 2018, 22, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, L.; Boutry, C.; Guédon, E.; Guillot, A.; Ibrahim, M.; Grossiord, B.; Hols, P. Quorum-Sensing Regulation of the Production of Blp Bacteriocins in Streptococcus Thermophilus. J. Bacteriol. 2007, 189, 7195–7205. [Google Scholar] [CrossRef] [PubMed]
- van der Ploeg, J.R. Regulation of Bacteriocin Production in Streptococcus Mutans by the Quorum-Sensing System Required for Development of Genetic Competence. J. Bacteriol. 2005, 187, 3980–3989. [Google Scholar] [CrossRef]
- Upton, M.; Tagg, J.R.; Wescombe, P.; Jenkinson, H.F. Intra- and Interspecies Signaling between Streptococcus Salivarius and Streptococcus Pyogenes Mediated by SalA and SalA1 Lantibiotic Peptides. J. Bacteriol. 2001, 183, 3931–3938. [Google Scholar] [CrossRef]
- Kuipers, O.P.; Beerthuyzen, M.M.; de Ruyter, P.G.G.A.; Luesink, E.J.; de Vos, W.M. Autoregulation of Nisin Biosynthesis in Lactococcus Lactis by Signal Transduction. J. Biol. Chem. 1995, 270, 27299–27304. [Google Scholar] [CrossRef] [PubMed]
- Tagg, J.R.; Read, R.S.D.; McGiven, A.R. Bacteriocine Production by Group a Streptococci. Pathology 1971, 3, 277–278. [Google Scholar] [CrossRef]
- McLaughlin, R.E.; Ferretti, J.J.; Hynes, W.L. Nucleotide Sequence of the Streptococcin A-FF22 Lantibiotic Regulon: Model for Production of the Lantibiotic SA-FF22 by Strains of Streptococcus Pyogenes. FEMS Microbiol. Lett. 1999, 175, 171–177. [Google Scholar] [CrossRef]
- Jack, R.; Benz, R.; Tagg, J.; Sahl, H.-G. The Mode of Action of SA-FF22, a Lantibiotic Isolated from Streptococcus Pyogenes Strain FF22. Eur. J. Biochem. 1994, 219, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Dyet, K.H.; Dierksen, K.P.; Power, D.A.; Jack, R.W.; Burton, J.P.; Inglis, M.A.; Wescombe, A.L.; Tagg, J.R. Salivaricin G32, a Homolog of the Prototype Streptococcus Pyogenes Nisin-Like Lantibiotic SA-FF22, Produced by the Commensal Species Streptococcus Salivarius. Int. J. Microbiol. 2012, 2012, 738503. [Google Scholar] [CrossRef] [PubMed]
- Tagg, J.R.; Skjold, S.A. A Bacteriocin Produced by Certain M-Type 49 Streptococcus Pyogenes Strains When Incubated Anaerobically. J. Hyg. 1984, 93, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Hynes, W.L.; Friend, V.L.; Ferretti, J.J. Duplication of the Lantibiotic Structural Gene in M-Type 49 Group A Streptococcus Strains Producing Streptococcin A-M49. Appl. Environ. Microbiol. 1994, 60, 4207–4209. [Google Scholar] [CrossRef]
- Papadelli, M.; Karsioti, A.; Anastasiou, R.; Georgalaki, M.; Tsakalidou, E. Characterization of the Gene Cluster Involved in the Biosynthesis of Macedocin, the Lantibiotic Produced by Streptococcus Macedonicus. FEMS Microbiol. Lett. 2007, 272, 75–82. [Google Scholar] [CrossRef]
- Tagg, J.R.; Read, R.S.D.; McGiven, A.R. Bacteriocin of a Group A Streptococcus: Partial Purification and Properties. Antimicrob. Agents Chemother. 1973, 4, 214–221. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Tagg, J.R. Purification and Characterization of Streptin, a Type A1 Lantibiotic Produced by Streptococcus Pyogenes. Appl. Environ. Microbiol. 2003, 69, 2737–2747. [Google Scholar] [CrossRef]
- Karaya, K.; Shimizu, T.; Taketo, A. New Gene Cluster for Lantibiotic Streptin Possibly Involved in Streptolysin S Formation. J. Biochem. 2001, 129, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Quadri, L.E. Peptide Pheromone-Dependent Regulation of Antimicrobial Peptide Production in Gram-Positive Bacteria: A Case of Multicellular Behavior. Peptides 2001, 22, 1579–1596. [Google Scholar] [CrossRef]
- Ross, K.F.; Ronson, C.W.; Tagg, J.R. Isolation and Characterization of the Lantibiotic Salivaricin A and Its Structural Gene SalA from Streptococcus Salivarius 20P3. Appl. Environ. Microbiol. 1993, 59, 2014–2021. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Upton, M.; Dierksen, K.P.; Ragland, N.L.; Sivabalan, S.; Wirawan, R.E.; Inglis, M.A.; Moore, C.J.; Walker, G.V.; Chilcott, C.N.; et al. Production of the Lantibiotic Salivaricin A and Its Variants by Oral Streptococci and Use of a Specific Induction Assay to Detect Their Presence in Human Saliva. Appl. Environ. Microbiol. 2006, 72, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Geng, M.; Austin, F.; Shin, R.; Smith, L. Covalent Structure and Bioactivity of the Type AII Lantibiotic Salivaricin A2. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Eran, Y.; Getter, Y.; Baruch, M.; Belotserkovsky, I.; Padalon, G.; Mishalian, I.; Podbielski, A.; Kreikemeyer, B.; Hanski, E. Transcriptional Regulation of the Sil Locus by the SilCR Signalling Peptide and Its Implications on Group A Streptococcus Virulence. Mol. Microbiol. 2007, 63, 1209–1222. [Google Scholar] [CrossRef]
- Armstrong, B.D.; Herfst, C.A.; Tonial, N.C.; Wakabayashi, A.T.; Zeppa, J.J.; McCormick, J.K. Identification of a Two-Component Class IIb Bacteriocin in Streptococcus Pyogenes by Recombinase-Based in Vivo Expression Technology. Sci. Rep. 2016, 6, 36233. [Google Scholar] [CrossRef]
- Hertzog, B.B.; Kaufman, Y.; Biswas, D.; Ravins, M.; Ambalavanan, P.; Wiener, R.; Angeli, V.; Chen, S.L.; Hanski, E. A Sub-Population of Group A Streptococcus Elicits a Population-Wide Production of Bacteriocins to Establish Dominance in the Host. Cell Host Microbe 2018, 23, 312–323.e6. [Google Scholar] [CrossRef]
- Simpson, W.J.; Tagg, J.R. M-Type 57 Group A Streptococcus Bacteriocin. Can. J. Microbiol. 1983, 29, 1445–1451. [Google Scholar] [CrossRef]
- Heng, N.C.K.; Ragland, N.L.; Swe, P.M.; Baird, H.J.; Inglis, M.A.; Tagg, J.R.; Jack, R.W. Dysgalacticin: A Novel, Plasmid-Encoded Antimicrobial Protein (Bacteriocin) Produced by Streptococcus Dysgalactiae Subsp. Equisimilis. Microbiol. Read. Engl. 2006, 152, 1991–2001. [Google Scholar] [CrossRef]
- Swe, P.M.; Cook, G.M.; Tagg, J.R.; Jack, R.W. Mode of Action of Dysgalacticin: A Large Heat-Labile Bacteriocin. J. Antimicrob. Chemother. 2009, 63, 679–686. [Google Scholar] [CrossRef]
- van Heel, A.J.; Kloosterman, T.G.; Montalban-Lopez, M.; Deng, J.; Plat, A.; Baudu, B.; Hendriks, D.; Moll, G.N.; Kuipers, O.P. Discovery, Production and Modification of Five Novel Lantibiotics Using the Promiscuous Nisin Modification Machinery. ACS Synth. Biol. 2016, 5, 1146–1154. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; O’Connor, P.M.; Saalbach, G.; Walsh, C.J.; Hegarty, J.W.; Guinane, C.M.; Mayer, M.J.; Narbad, A.; Cotter, P.D. First Evidence of Production of the Lantibiotic Nisin P. Sci. Rep. 2020, 10, 3738. [Google Scholar] [CrossRef] [PubMed]
- Plainvert, C.; Dinis, M.; Ravins, M.; Hanski, E.; Touak, G.; Dmytruk, N.; Fouet, A.; Poyart, C. Molecular Epidemiology of Sil Locus in Clinical Streptococcus Pyogenes Strains. J. Clin. Microbiol. 2014, 52, 2003–2010. [Google Scholar] [CrossRef]
- van Belkum, M.J.; Worobo, R.W.; Stiles, M.E. Double-Glycine-Type Leader Peptides Direct Secretion of Bacteriocins by ABC Transporters: Colicin V Secretion in Lactococcus Lactis. Mol. Microbiol. 1997, 23, 1293–1301. [Google Scholar] [CrossRef]
- Yount, N.Y.; Weaver, D.C.; de Anda, J.; Lee, E.Y.; Lee, M.W.; Wong, G.C.L.; Yeaman, M.R. Discovery of Novel Type II Bacteriocins Using a New High-Dimensional Bioinformatic Algorithm. Front. Immunol. 2020, 11, 1873. [Google Scholar] [CrossRef]
- Hidalgo-Grass, C.; Ravins, M.; Dan-Goor, M.; Jaffe, J.; Moses, A.E.; Hanski, E. A Locus of Group A Streptococcus Involved in Invasive Disease and DNA Transfer. Mol. Microbiol. 2002, 46, 87–99. [Google Scholar] [CrossRef]
- Michael-Gayego, A.; Dan-Goor, M.; Jaffe, J.; Hidalgo-Grass, C.; Moses, A.E. Characterization of Sil in Invasive Group A and G Streptococci: Antibodies against Bacterial Pheromone Peptide SilCR Result in Severe Infection. Infect. Immun. 2013, 81, 4121–4127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ciszewski, M.; Szewczyk, E.M. Potential Factors Enabling Human Body Colonization by Animal Streptococcus Dysgalactiae Subsp. Equisimilis Strains. Curr. Microbiol. 2017, 74, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Schofield, C.R.; Tagg, J.R. Bacteriocin-like Activity of Group B and Group C Streptococci of Human and of Animal Origin. J. Hyg. 1983, 90, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Tagg, J.R.; Wong, H.K. Inhibitor Production by Group-G Streptococci of Human and of Animal Origin. J. Med. Microbiol. 1983, 16, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, R.S.; Naidoo, J.; Jones, C.L.; Tagg, J.R. The Streptococcal Bacteriocin-like Inhibitory Substance, Zoocin A, Reduces the Proportion of Streptococcus Mutans in an Artificial Plaque. Microb. Ecol. Health Dis. 1995, 8, 281–292. [Google Scholar] [CrossRef][Green Version]
- Swe, P.M.; Heng, N.C.K.; Cook, G.M.; Tagg, J.R.; Jack, R.W. Identification of DysI, the Immunity Factor of the Streptococcal Bacteriocin Dysgalacticin. Appl. Environ. Microbiol. 2010, 76, 7885–7889. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).