Cerebral Organoids Derived from a Parkinson’s Patient Exhibit Unique Pathogenesis from Chikungunya Virus Infection When Compared to a Non-Parkinson’s Patient
Abstract
1. Introduction
2. Results
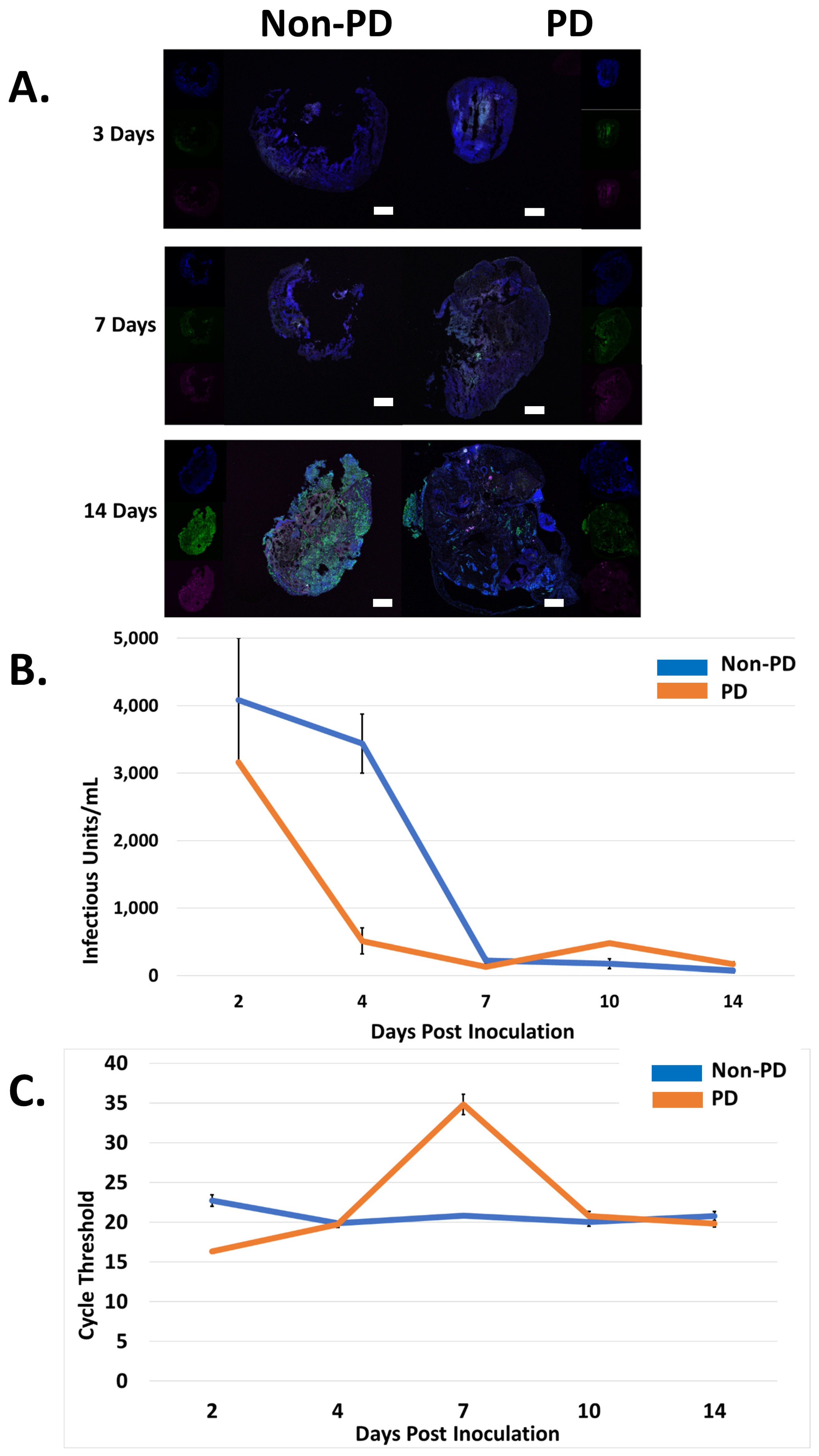
2.1. Size and Morphology Do Not Differ between Organoids from Non-PD and Diseased Backgrounds in Response to CHIKV Infection
2.2. Non-PD and PD Organoids Exhibit Unique Responses to CHIKV Infection
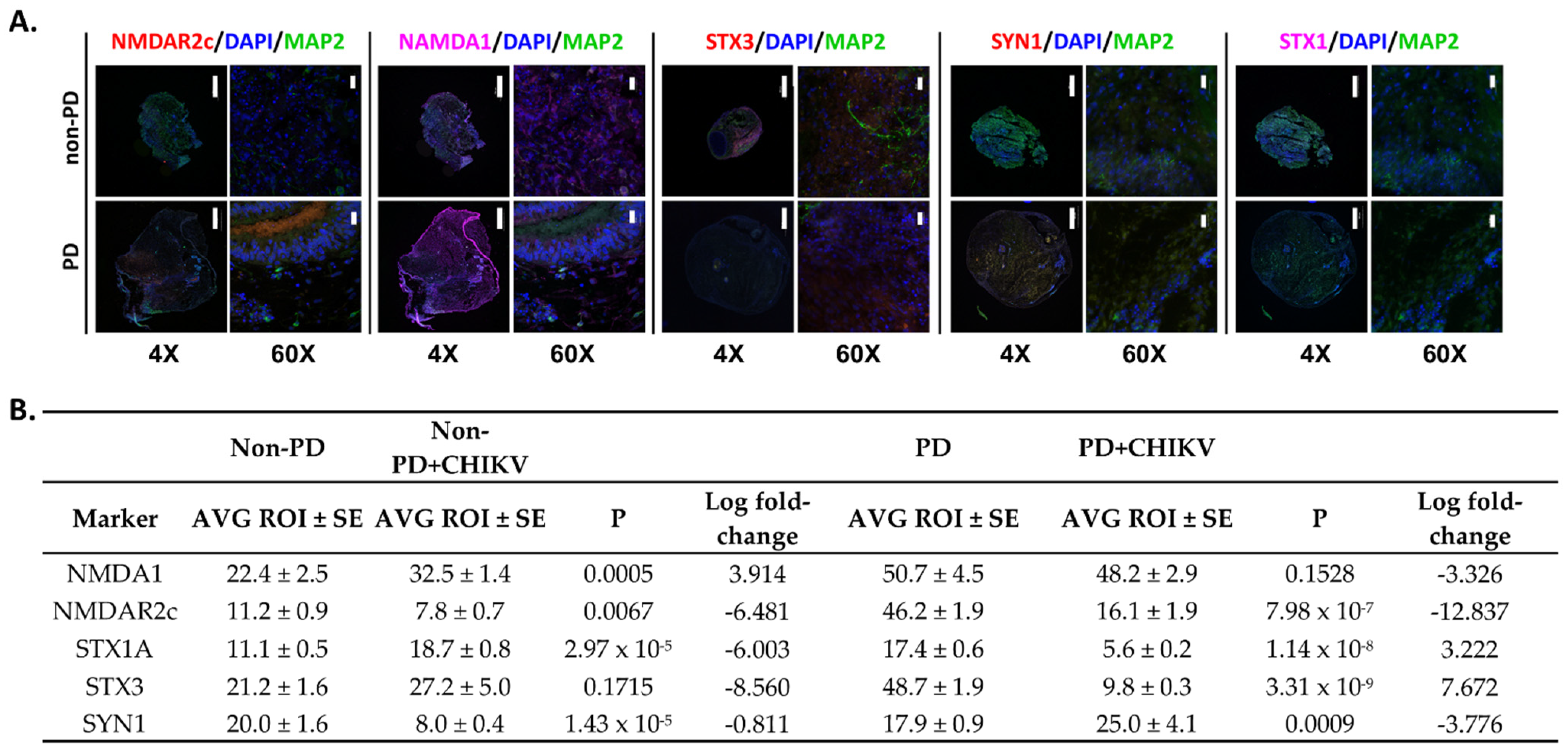
2.3. Neurotransmission Is Reduced in Parkinson’s Organoids
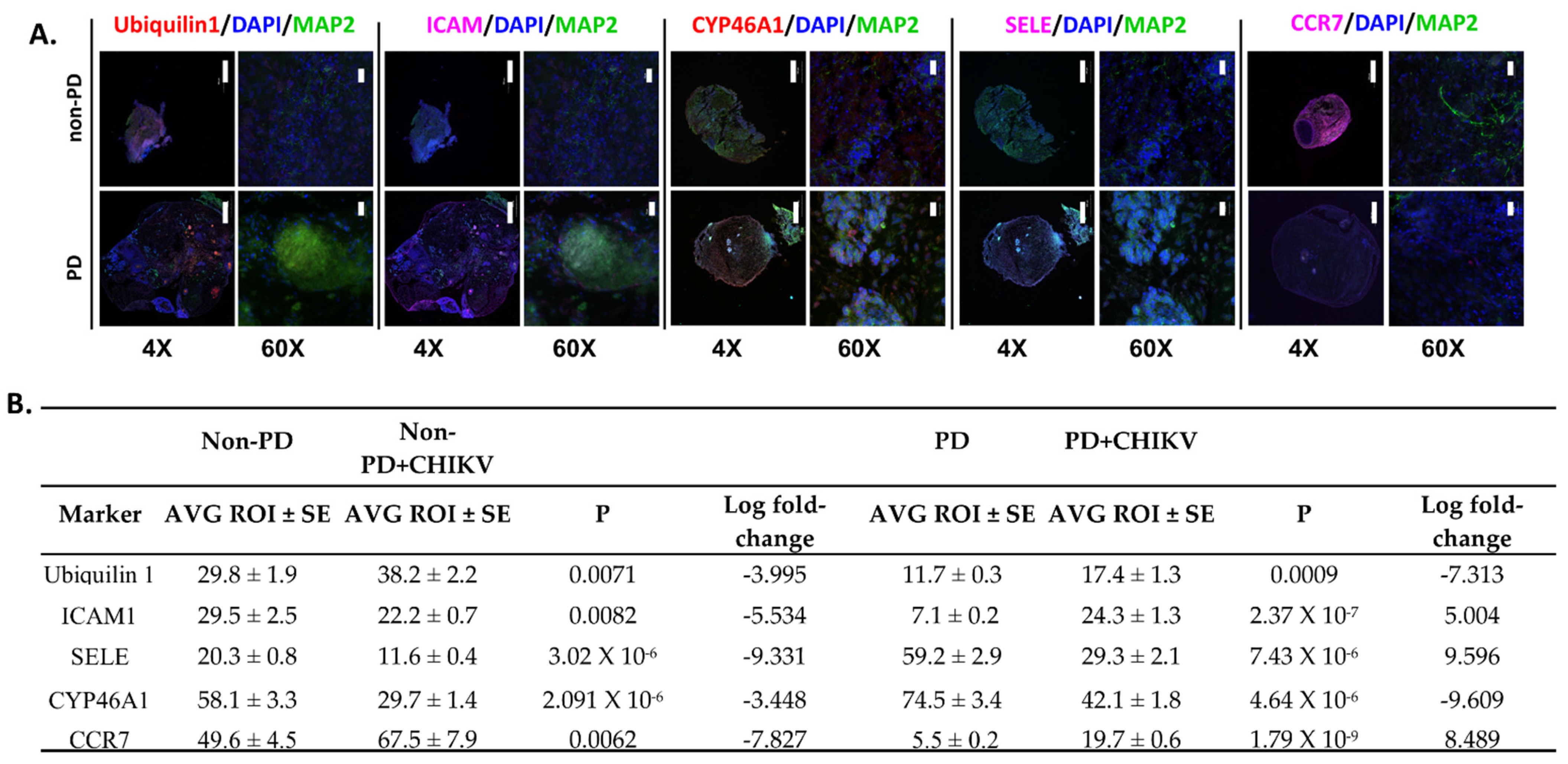
2.4. Alterations in Immune Regulation
2.5. Expression for other Markers
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Virus Propagation
4.2. Generation and Infection of Human Cerebral Organoids
4.3. Viral Quantification
4.4. Cerebral Organoid mRNA Extraction and Gene Expression
4.5. Cerebral Organoid Size Measurements
4.6. Immunofluorescence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jang, H.; Boltz, D.A.; Webster, R.G.; Smeyne, R.J. Viral parkinsonism. Biochim. Biophys. Acta 2009, 1792, 714–721. [Google Scholar] [CrossRef]
- Weaver, S.C.; Winegar, R.; Manger, I.D.; Forrester, N.L. Alphaviruses: Population genetics and determinants of emergence. Antivir. Res. 2012, 94, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Rocklov, J.; Quam, M.B.; Sudre, B.; German, M.; Kraemer, M.U.G.; Brady, O.; Bogoch, I.; Liu-Helmersson, J.; Wilder-Smith, A.; Semenza, J.C.; et al. Assessing Seasonal Risks for the Introduction and Mosquito-borne Spread of Zika Virus in Europe. EBioMedicine 2016, 9, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Sooryanarain, H.; Elankumaran, S. Environmental role in influenza virus outbreaks. Annu. Rev. Anim. Biosci. 2015, 3, 347–373. [Google Scholar] [CrossRef]
- Grassly, N.C.; Fraser, C. Seasonal infectious disease epidemiology. Proc. R. Soc. B Biol. Sci. 2006, 273, 2541–2550. [Google Scholar] [CrossRef]
- Barr, K.L.; Khan, E.; Farooqi, J.Q.; Imtiaz, K.; Prakoso, D.; Malik, F.; Lednicky, J.A.; Long, M.T. Evidence of Chikungunya Virus Disease in Pakistan Since 2015 With Patients Demonstrating Involvement of the Central Nervous System. Front. Public Health 2018, 6, 186. [Google Scholar] [CrossRef]
- Barr, K.L.; Vaidhyanathan, V. Chikungunya in Infants and Children: Is Pathogenesis Increasing? Viruses 2019, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Tandel, K.; Dash, P.K.; Parida, M. Zika virus: A public health threat. J. Med. Virol. 2017, 89, 1693–1699. [Google Scholar] [CrossRef]
- Gregianini, T.S.; Ranieri, T.; Favreto, C.; Nunes, Z.M.A.; Tumioto Giannini, G.L.; Sanberg, N.D.; da Rosa, M.T.M.; da Veiga, A.B.G. Emerging arboviruses in Rio Grande do Sul, Brazil: Chikungunya and Zika outbreaks, 2014–2016. Rev. Med. Virol. 2017, 27, e1943. [Google Scholar] [CrossRef] [PubMed]
- Brault, A.C. Changing patterns of West Nile virus transmission: Altered vector competence and host susceptibility. Vet. Res. 2009, 40, 1–19. [Google Scholar] [CrossRef]
- Bakonyi, T.; Erdélyi, K.; Ursu, K.; Ferenczi, E.; Csörgo, T.; Lussy, H.; Chvala, S.; Bukovsky, C.; Meister, T.; Weissenböck, H.; et al. Emergence of Usutu virus in Hungary. J. Clin. Microbiol. 2007, 45, 3870–3874. [Google Scholar] [CrossRef]
- Wang, H.; Liu, X.; Tan, C.; Zhou, W.; Jiang, J.; Peng, W.; Zhou, X.; Mo, L.; Chen, L. Bacterial, viral, and fungal infection-related risk of Parkinson’s disease: Meta-analysis of cohort and case-control studies. Brain Behav. 2020, 10, e01549. [Google Scholar] [CrossRef]
- Abdoli, A.; Taghipour, A.; Pirestani, M.; Mofazzal Jahromi, M.A.; Roustazadeh, A.; Mir, H.; Ardakani, H.M.; Kenarkoohi, A.; Falahi, S.; Karimi, M. Infections, inflammation, and risk of neuropsychiatric disorders: The neglected role of “co-infection”. Heliyon 2020, 6, e05645. [Google Scholar] [CrossRef] [PubMed]
- McLoone, P.; Dyussupov, O.; Nurtlessov, Z.; Kenessariyev, U.; Kenessary, D. The effect of exposure to crude oil on the immune system. Health implications for people living near oil exploration activities. Int. J. Environ. Health Res. 2019, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Zubcevic, J.; Jun, J.Y.; Kim, S.; Perez, P.D.; Afzal, A.; Shan, Z.; Li, W.; Santisteban, M.M.; Yuan, W.; Febo, M.; et al. Altered inflammatory response is associated with an impaired autonomic input to the bone marrow in the spontaneously hypertensive rat. Hypertension 2014, 63, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Göke, J.; Cukuroglu, E.; Dranias, M.R.; VanDongen, A.M.J.; Stanton, L.W. Molecular Features Underlying Neurodegeneration Identified through In Vitro Modeling of Genetically Diverse Parkinson’s Disease Patients. Cell Rep. 2016, 15, 2411–2426. [Google Scholar] [CrossRef]
- Biller-Takahashi, J.D.; Takahashi, L.S.; Mingatto, F.E.; Urbinati, E.C. The immune system is limited by oxidative stress: Dietary selenium promotes optimal antioxidative status and greatest immune defense in pacu Piaractus mesopotamicus. Fish. Shellfish Immunol. 2015, 47, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Phani, S.; Loike, J.D.; Przedborski, S. Neurodegeneration and Inflammation in Parkinson’s disease. Parkinsonism Relat. Disord. 2012, 18, S207–S209. [Google Scholar] [CrossRef]
- Guo, M.-L.; Buch, S. Neuroinflammation & pre-mature aging in the context of chronic HIV infection and drug abuse: Role of dysregulated autophagy. Brain Res. 2019, 1724, 146446. [Google Scholar] [CrossRef]
- Cvejic, E.; Lemon, J.; Hickie, I.B.; Lloyd, A.R.; Vollmer-Conna, U. Neurocognitive disturbances associated with acute infectious mononucleosis, Ross River fever and Q fever: A preliminary investigation of inflammatory and genetic correlates. Brain Behav. Immun. 2014, 36, 207–214. [Google Scholar] [CrossRef]
- Piraino, B.; Vollmer-Conna, U.; Lloyd, A.R. Genetic associations of fatigue and other symptom domains of the acute sickness response to infection. Brain Behav. Immun. 2012, 26, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Vollmer-Conna, U.; Piraino, B.F.; Cameron, B.; Davenport, T.; Hickie, I.; Wakefield, D.; Lloyd, A.R. Cytokine polymorphisms have a synergistic effect on severity of the acute sickness response to infection. Clin. Infect. Dis. 2008, 47, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Bopeththa, B.; Ralapanawa, U. Post encephalitic parkinsonism following dengue viral infection. BMC Res. Notes 2017, 10, 655. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.L.; Shahida, S.; Madan, N.; Rao, S.; Khardori, N. Transient parkinsonism in West Nile virus encephalitis. Am. J. Med. 2003, 115, 252–253. [Google Scholar] [CrossRef]
- Ogata, A.; Tashiro, K.; Pradhan, S. Parkinsonism due to predominant involvement of substantia nigra in Japanese encephalitis. Neurology 2000, 55, 602. [Google Scholar] [CrossRef]
- Cerna, F.; Mehrad, B.; Luby, J.P.; Burns, D.; Fleckenstein, J.L. St. Louis encephalitis and the substantia nigra: MR imaging evaluation. AJNR Am. J. Neuroradiol. 1999, 20, 1281–1283. [Google Scholar] [PubMed]
- Caggiu, E.; Arru, G.; Hosseini, S.; Niegowska, M.; Sechi, G.; Zarbo, I.R.; Sechi, L.A. Inflammation, Infectious Triggers, and Parkinson’s Disease. Front. Neurol. 2019, 10, 122. [Google Scholar] [CrossRef]
- Schwamborn, J.C. Is Parkinson’s Disease a Neurodevelopmental Disorder and Will Brain Organoids Help Us to Understand It? Stem Cells Dev. 2018, 27, 968–975. [Google Scholar] [CrossRef]
- Sison, S.L.; Vermilyea, S.C.; Emborg, M.E.; Ebert, A.D. Using Patient-Derived Induced Pluripotent Stem Cells to Identify Parkinson’s Disease-Relevant Phenotypes. Curr. Neurol. Neurosci. Rep. 2018, 18, 84. [Google Scholar] [CrossRef]
- Nugraha, B.; Buono, M.F.; von Boehmer, L.; Hoerstrup, S.P.; Emmert, M.Y. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther. 2019, 105, 79–85. [Google Scholar] [CrossRef]
- Nugraha, B.; Buono, M.F.; Emmert, M.Y. Modelling human cardiac diseases with 3D organoid. Eur. Heart J. 2018, 39, 4234–4237. [Google Scholar] [CrossRef]
- Dutta, D.; Clevers, H. Organoid culture systems to study host-pathogen interactions. Curr. Opin. Immunol. 2017, 48, 15–22. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Ghatak, S.; Dolatabadi, N.; Trudler, D.; Zhang, X.; Wu, Y.; Mohata, M.; Ambasudhan, R.; Talantova, M.; Lipton, S.A. Mechanisms of hyperexcitability in Alzheimer’s disease hiPSC-derived neurons and cerebral organoids vs isogenic controls. eLife 2019, 8, e50333. [Google Scholar] [CrossRef] [PubMed]
- Nickels, S.L.; Modamio, J.; Mendes-Pinheiro, B.; Monzel, A.S.; Betsou, F.; Schwamborn, J.C. Reproducible generation of human midbrain organoids for in vitro modeling of Parkinson’s disease. Stem Cell Res. 2020, 46, 101870. [Google Scholar] [CrossRef]
- Smits, L.M.; Schwamborn, J.C. Midbrain Organoids: A New Tool to Investigate Parkinson’s Disease. Front. Cell Dev. Biol. 2020, 8, 359. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.; Anastacio, H.; Bardy, C. Genetic predispositions of Parkinson’s disease revealed in patient-derived brain cells. NPJ Parkinson’s Dis. 2020, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Kouroupi, G.; Taoufik, E.; Vlachos, I.S.; Tsioras, K.; Antoniou, N.; Papastefanaki, F.; Chroni-Tzartou, D.; Wrasidlo, W.; Bohl, D.; Stellas, D.; et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E3679–E3688. [Google Scholar] [CrossRef]
- Galet, B.; Cheval, H.; Ravassard, P. Patient-Derived Midbrain Organoids to Explore the Molecular Basis of Parkinson’s Disease. Front. Neurol. 2020, 11, 1005. [Google Scholar] [CrossRef]
- Krokidis, M.G. Identification of biomarkers associated with Parkinson’s disease by gene expression profiling studies and bioinformatics analysis. AIMS Neurosci. 2019, 6, 333–345. [Google Scholar] [CrossRef]
- Klettner, A.; Richert, E.; Kuhlenbäumer, G.; Nölle, B.; Bhatia, K.P.; Deuschl, G.; Roider, J.; Schneider, S.A. Alpha synuclein and crystallin expression in human lens in Parkinson’s disease. Mov. Disord. 2016, 31, 600–601. [Google Scholar] [CrossRef]
- Nagatsu, T.; Nakashima, A.; Ichinose, H.; Kobayashi, K. Human tyrosine hydroxylase in Parkinson’s disease and in related disorders. J. Neural Transm. 2019, 126, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Gan-Or, Z.; Liong, C.; Alcalay, R.N. GBA-Associated Parkinson’s Disease and Other Synucleinopathies. Curr. Neurol. Neurosci. Rep. 2018, 18, 44. [Google Scholar] [CrossRef]
- Goureau, O.; Orieux, G. Photoreceptor cell transplantation for future treatment of retinitis pigmentosa. Med. Sci. 2020, 36, 600–606. [Google Scholar] [CrossRef]
- Youhanna, S.; Lauschke, V.M. The Past, Present and Future of Intestinal In Vitro Cell Systems for Drug Absorption Studies. J. Pharm. Sci. 2020, 110, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid Models of Tumor Immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Cortesi, M.; Zamagni, A.; Arienti, C.; Pignatta, S.; Tesei, A. Modeling neoplastic disease with spheroids and organoids. J. Hematol. Oncol. 2020, 13, 97. [Google Scholar] [CrossRef]
- Oltra, J.A.E. Improving Therapeutic Interventions of Schizophrenia with Advances in Stem Cell Technology. Clin. Psychopharmacol. Neurosci. 2020, 18, 352–361. [Google Scholar] [CrossRef]
- Makovoz, B.; Moeller, R.; Zebitz Eriksen, A.; tenOever, B.R.; Blenkinsop, T.A. SARS-CoV-2 Infection of Ocular Cells from Human Adult Donor Eyes and hESC-Derived Eye Organoids. Soc. Sci. Res. Netw. 2020, 3650574. [Google Scholar] [CrossRef]
- Wong, Y.C.; Lau, S.Y.; Wang To, K.K.; Mok, B.W.Y.; Li, X.; Wang, P.; Deng, S.; Woo, K.F.; Du, Z.; Li, C.; et al. Natural transmission of bat-like SARS-CoV-2 PRRA variants in COVID-19 patients. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Smedley, R.C.; Patterson, J.S.; Miller, R.; Massey, J.P.; Wise, A.G.; Maes, R.K.; Wu, P.; Kaneene, J.B.; Kiupel, M. Sensitivity and specificity of monoclonal and polyclonal immunohistochemical staining for West Nile virus in various organs from American crows (Corvus brachyrhynchos) 120. BMC Infect. Dis. 2007, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Armah, H.B.; Wang, G.; Omalu, B.I.; Tesh, R.B.; Gyure, K.A.; Chute, D.J.; Smith, R.D.; Dulai, P.; Vinters, H.V.; Kleinschmidt-DeMasters, B.K.; et al. Systemic Distribution of West Nile Virus Infection: Postmortem Immunohistochemical Study of Six Cases. Brain Pathol. 2007, 17, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Sá, P.K.O.; Nunes, M.M.; Leite, I.R.; Campelo, M.D.G.L.; Leão, C.F.R.; Souza, J.R.; Castellano, L.R.; Fernandes, A.I.V. Chikungunya virus infection with severe neurologic manifestations: Report of four fatal cases. Rev. Soc. Bras. Med. Trop 2017, 50, 265–268. [Google Scholar] [CrossRef]
- Cerny, T.; Schwarz, M.; Schwarz, U.; Lemant, J.; Gérardin, P.; Keller, E. The Range of Neurological Complications in Chikungunya Fever. Neurocrit. Care 2017, 27, 447–457. [Google Scholar] [CrossRef]
- Davenport, B.J.; Bullock, C.; McCarthy, M.K.; Hawman, D.W.; Murphy, K.M.; Kedl, R.M.; Diamond, M.S.; Morrison, T.E. Chikungunya Virus Evades Antiviral CD8(+) T Cell Responses To Establish Persistent Infection in Joint-Associated Tissues. J. Virol. 2020, 94, e02036-19. [Google Scholar] [CrossRef]
- Huang, J.; Liu, F.; Tang, H.; Wu, H.; Li, L.; Wu, R.; Zhao, J.; Wu, Y.; Liu, Z.; Chen, J. Tranylcypromine Causes Neurotoxicity and Represses BHC110/LSD1 in Human-Induced Pluripotent Stem Cell-Derived Cerebral Organoids Model. Front. Neurol. 2017, 8, 626. [Google Scholar] [CrossRef]
- Dezonne, R.S.; Sartore, R.C.; Nascimento, J.M.; Saia-Cereda, V.M.; Romão, L.F.; Alves-Leon, S.V.; de Souza, J.M.; Martins-de-Souza, D.; Rehen, S.K.; Gomes, F.C. Derivation of Functional Human Astrocytes from Cerebral Organoids. Sci. Rep. 2017, 7, 45091. [Google Scholar] [CrossRef] [PubMed]
- Sofroniew, M.V.; Vinters, H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010, 119, 7–35. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.; Alberts, B.; Johnson, A.; Walter, P.; Hunt, T. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Holmes, Z.E.; Hamilton, D.J.; Hwang, T.; Parsonnet, N.V.; Rinn, J.L.; Wuttke, D.S.; Batey, R.T. The Sox2 transcription factor binds RNA. Nat. Commun. 2020, 11, 1805. [Google Scholar] [CrossRef]
- Bouma, E.M.; van de Pol, D.P.I.; Sanders, I.D.; Rodenhuis-Zybert, I.A.; Smit, J.M. Serotonergic Drugs Inhibit Chikungunya Virus Infection at Different Stages of the Cell Entry Pathway. J. Virol. 2020, 94, e00274-20. [Google Scholar] [CrossRef] [PubMed]
- Mainou, B.A.; Ashbrook, A.W.; Smith, E.C.; Dorset, D.C.; Denison, M.R.; Dermody, T.S. Serotonin Receptor Agonist 5-Nonyloxytryptamine Alters the Kinetics of Reovirus Cell Entry. J. Virol. 2015, 89, 8701–8712. [Google Scholar] [CrossRef] [PubMed]
- Celli, J.; Rappold, G.; Niesler, B. The Human Serotonin Type 3 Receptor Gene (HTR3A-E) Allelic Variant Database. Hum. Mutat. 2017, 38, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Culibrk, R.A.; Hahn, M.S. The Role of Chronic Inflammatory Bone and Joint Disorders in the Pathogenesis and Progression of Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, Á.; Molnár, K.; Nógrádi, B.; Hernádi, Z.; Nyúl-Tóth, Á.; Wilhelm, I.; Krizbai, I.A. Neurovascular Inflammaging in Health and Disease. Cells 2020, 9, 1614. [Google Scholar] [CrossRef]
- Suarez-Lopez, J.R.; Hood, N.; Suárez-Torres, J.; Gahagan, S.; Gunnar, M.R.; López-Paredes, D. Associations of acetylcholinesterase activity with depression and anxiety symptoms among adolescents growing up near pesticide spray sites. Int. J. Hyg. Environ. Health 2019, 222, 981–990. [Google Scholar] [CrossRef]
- Yanez, M.; Vina, D. Dual inhibitors of monoamine oxidase and cholinesterase for the treatment of Alzheimer disease. Curr. Top. Med. Chem. 2013, 13, 1692–1706. [Google Scholar] [CrossRef]
- Suarez-Lopez, J.R.; Himes, J.H.; Jacobs, D.R., Jr.; Alexander, B.H.; Gunnar, M.R. Acetylcholinesterase activity and neurodevelopment in boys and girls. Pediatrics 2013, 132, e1649–e1658. [Google Scholar] [CrossRef]
- Häcker, H.; Karin, M. Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, 2006, re13. [Google Scholar] [CrossRef]
- Sommer, A.; Marxreiter, F.; Krach, F.; Fadler, T.; Grosch, J.; Maroni, M.; Graef, D.; Eberhardt, E.; Riemenschneider, M.J.; Yeo, G.W.; et al. Th17 Lymphocytes Induce Neuronal Cell Death in a Human iPSC-Based Model of Parkinson’s Disease. Cell Stem Cell 2018, 23, 123–131. [Google Scholar] [CrossRef]
- Gilardini Montani, M.S.; Santarelli, R.; Granato, M.; Gonnella, R.; Torrisi, M.R.; Faggioni, A.; Cirone, M. EBV reduces autophagy, intracellular ROS and mitochondria to impair monocyte survival and differentiation. Autophagy 2019, 15, 652–667. [Google Scholar] [CrossRef]
- Santarelli, R.; Granato, M.; Faggioni, A.; Cirone, M. Interference with the Autophagic Process as a Viral Strategy to Escape from the Immune Control: Lesson from Gamma Herpesviruses. J. Immunol. Res. 2015, 2015, 546063. [Google Scholar] [CrossRef]
- Gutiérrez, T.; Simmen, T. Endoplasmic reticulum chaperones and oxidoreductases: Critical regulators of tumor cell survival and immunorecognition. Front. Oncol. 2014, 4, 291. [Google Scholar] [CrossRef]
- El Kalamouni, C.; Frumence, E.; Bos, S.; Turpin, J.; Nativel, B.; Harrabi, W.; Wilkinson, D.A.; Meilhac, O.; Gadea, G.; Desprès, P.; et al. Subversion of the Heme Oxygenase-1 Antiviral Activity by Zika Virus. Viruses 2018, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.L.; Zhang, P.; Wang, H.Q.; Li, Y.F.; Hu, J.; Jiang, J.D.; Li, Y.H. heme oxygenase-1 agonist CoPP suppresses influenza virus replication through IRF3-mediated generation of IFN-α/β. Virology 2019, 528, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Jabłonowska, E.; Wójcik, K.; Szymańska, B.; Omulecka, A.; Cwiklińska, H.; Piekarska, A. Hepatic HMOX1 expression positively correlates with Bach-1 and miR-122 in patients with HCV mono and HIV/HCV coinfection. PLoS ONE 2014, 9, e95564. [Google Scholar] [CrossRef] [PubMed]
- Santisteban, M.M.; Zubcevic, J.; Baekey, D.M.; Raizada, M.K. Dysfunctional brain-bone marrow communication: A paradigm shift in the pathophysiology of hypertension. Curr. Hypertens. Rep. 2013, 15, 377–389. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tseng, C.K.; Lin, C.K.; Wu, Y.H.; Chen, Y.H.; Chen, W.C.; Young, K.C.; Lee, J.C. Human heme oxygenase 1 is a potential host cell factor against dengue virus replication. Sci. Rep. 2016, 6, 32176. [Google Scholar] [CrossRef]
- Papa, A.; Tsergouli, K.; Çağlayık, D.Y.; Bino, S.; Como, N.; Uyar, Y.; Korukluoglu, G. Cytokines as biomarkers of Crimean-Congo hemorrhagic fever. J. Med. Virol. 2016, 88, 21–27. [Google Scholar] [CrossRef]
- Gherardi, M.M.; Ramírez, J.C.; Esteban, M. IL-12 and IL-18 act in synergy to clear vaccinia virus infection: Involvement of innate and adaptive components of the immune system. J. Gen. Virol. 2003, 84, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, T.; Dias da Costa, M.; Segenreich, D. Manic Episode After a Chikungunya Virus Infection in a Bipolar Patient Previously Stabilized With Valproic Acid. J. Clin. Psychopharmacol. 2018, 38, 395–397. [Google Scholar] [CrossRef]
- Michlmayr, D.; Pak, T.R.; Rahman, A.H.; Amir, E.D.; Kim, E.Y.; Kim-Schulze, S.; Suprun, M.; Stewart, M.G.; Thomas, G.P.; Balmaseda, A.; et al. Comprehensive innate immune profiling of chikungunya virus infection in pediatric cases. Mol. Syst. Biol. 2018, 14, e7862. [Google Scholar] [CrossRef]
- Rodriguez-Morales, A.J.; Hernandez-Moncada, A.M.; Hoyos-Guapacha, K.L.; Vargas-Zapata, S.L.; Sanchez-Zapata, J.F.; Mejia-Bernal, Y.V.; Ocampo-Serna, S.; Meneses-Quintero, O.M.; Gutierrez-Segura, J.C. Potential relationships between chikungunya and depression: Solving the puzzle with key cytokines. Cytokine 2018, 102, 161–162. [Google Scholar] [CrossRef]
- Lind, L.; Eriksson, K.; Grahn, A. Chemokines and matrix metalloproteinases in cerebrospinal fluid of patients with central nervous system complications caused by varicella-zoster virus. J. Neuroinflamm. 2019, 16, 42. [Google Scholar] [CrossRef]
- Bardina, S.V.; Brown, J.A.; Michlmayr, D.; Hoffman, K.W.; Sum, J.; Pletnev, A.G.; Lira, S.A.; Lim, J.K. Chemokine Receptor Ccr7 Restricts Fatal West Nile Virus Encephalitis. J. Virol. 2017, 91, e02409-16. [Google Scholar] [CrossRef]
- Cheng, W.; Zhao, Q.; Xi, Y.; Li, C.; Xu, Y.; Wang, L.; Niu, X.; Wang, Z.; Chen, G. IFN-β inhibits T cells accumulation in the central nervous system by reducing the expression and activity of chemokines in experimental autoimmune encephalomyelitis. Mol. Immunol. 2015, 64, 152–162. [Google Scholar] [CrossRef]
- Russell, C.D.; Unger, S.A.; Walton, M.; Schwarze, J. The Human Immune Response to Respiratory Syncytial Virus Infection. Clin. Microbiol. Rev. 2017, 30, 481–502. [Google Scholar] [CrossRef]
- Zhang, F.; Qi, L.; Li, T.; Li, X.; Yang, D.; Cao, S.; Ye, J.; Wei, B. PD1(+)CCR2(+)CD8(+) T Cells Infiltrate the Central Nervous System during Acute Japanese Encephalitis Virus Infection. Virol. Sin. 2019, 34, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Cheng, B.; Cheng, J.; Wang, D.; Li, H.; He, X. CCR5 blockade promotes M2 macrophage activation and improves locomotor recovery after spinal cord injury in mice. Inflammation 2015, 38, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, B.; Kim, H.; Meyer, K.; Bose, S.K.; Di Bisceglie, A.M.; Ray, R.B.; Ray, R. Hepatitis C virus proteins inhibit C3 complement production. J. Virol. 2012, 86, 2221–2228. [Google Scholar] [CrossRef] [PubMed]
- Avirutnan, P.; Hauhart, R.E.; Marovich, M.A.; Garred, P.; Atkinson, J.P.; Diamond, M.S. Complement-mediated neutralization of dengue virus requires mannose-binding lectin. mBio 2011, 2, e00276-11. [Google Scholar] [CrossRef]
- Fuchs, A.; Lin, T.Y.; Beasley, D.W.; Stover, C.M.; Schwaeble, W.J.; Pierson, T.C.; Diamond, M.S. Direct Complement Restriction of Flavivirus Infection Requires Glycan Recognition by Mannose-Binding Lectin. Cell Host Microbe 2010, 8, 186–195. [Google Scholar] [CrossRef]
- Zietek, T.; Giesbertz, P.; Ewers, M.; Reichart, F.; Weinmüller, M.; Urbauer, E.; Haller, D.; Demir, I.E.; Ceyhan, G.O.; Kessler, H.; et al. Organoids to Study Intestinal Nutrient Transport, Drug Uptake and Metabolism-Update to the Human Model and Expansion of Applications. Front. Bioeng. Biotechnol. 2020, 8, 577656. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, K.; Maurer, C.; Peschke, K.; Teller, S.; Schuck, K.; Steiger, K.; Engleitner, T.; Öllinger, R.; Nomura, A.; Wirges, N.; et al. Mesenchymal plasticity regulated by Prrx1 drives aggressive pancreatic cancer biology. Gastroenterology 2020, 160, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, P.; Wang, Y.; Yin, Y.; de Ruiter, P.E.; Verstegen, M.M.A.; Peppelenbosch, M.P.; van der Laan, L.J.W.; Pan, Q. Rotavirus Infection and Cytopathogenesis in Human Biliary Organoids Potentially Recapitulate Biliary Atresia Development. mBio 2020, 11, e01968-20. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Herrero, L.J.; Rudd, P.A.; Mahalingam, S. Mouse models of alphavirus-induced inflammatory disease. J. Gen. Virol 2015, 96, 221–238. [Google Scholar] [CrossRef]
- Gerardin, P.; Samperiz, S.; Ramful, D.; Boumahni, B.; Bintner, M.; Alessandri, J.L.; Carbonnier, M.; Tiran-Rajaoefera, I.; Beullier, G.; Boya, I.; et al. Neurocognitive outcome of children exposed to perinatal mother-to-child Chikungunya virus infection: The CHIMERE cohort study on Reunion Island. PLoS Negl. Trop. Dis. 2014, 8, e2996. [Google Scholar] [CrossRef]
- Siegel, G.; Kluba, T.; Hermanutz-Klein, U.; Bieback, K.; Northoff, H.; Schäfer, R. Phenotype, donor age and gender affect function of human bone marrow-derived mesenchymal stromal cells. BMC Med. 2013, 11, 146. [Google Scholar] [CrossRef]
- Strässler, E.T.; Aalto-Setälä, K.; Kiamehr, M.; Landmesser, U.; Kränkel, N. Age Is Relative-Impact of Donor Age on Induced Pluripotent Stem Cell-Derived Cell Functionality. Front. Cardiovasc. Med. 2018, 5, 4. [Google Scholar] [CrossRef]
- Prigione, A.; Fauler, B.; Lurz, R.; Lehrach, H.; Adjaye, J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 2010, 28, 721–733. [Google Scholar] [CrossRef]
- ATCC. Neural Progenitor Cells Derived from ATCCDYS0530 Parkinson’s Disease (ATCC® ACS5001™); ATCC: Manassas, VA, USA, 2021. [Google Scholar]
- Hoarau, J.J.; Jaffar Bandjee, M.C.; Krejbich Trotot, P.; Das, T.; Li-Pat-Yuen, G.; Dassa, B.; Denizot, M.; Guichard, E.; Ribera, A.; Henni, T.; et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J. Immunol. 2010, 184, 5914–5927. [Google Scholar] [CrossRef]
- Roy, E.; Shi, W.; Duan, B.; Reid, S.P. Chikungunya Virus Infection Impairs the Function of Osteogenic Cells. mSphere 2020, 5, e00347-20. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.M.; Jones, T.J.; Barr, K.L. Antibodies for Venezuelan Equine Encephalitis Virus Protect Embryoid Bodies from Chikungunya Virus. Viruses 2020, 12, 262. [Google Scholar] [CrossRef] [PubMed]
- Gorchakov, R.; Wang, E.; Leal, G.; Forrester, N.L.; Plante, K.; Rossi, S.L.; Partidos, C.D.; Adams, A.P.; Seymour, R.L.; Weger, J.; et al. Attenuation of Chikungunya Virus Vaccine Strain 181/Clone 25 Is Determined by Two Amino Acid Substitutions in the E2 Envelope Glycoprotein. J. Virol. 2012, 86, 6084. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Clé, M.; Eldin, P.; Briant, L.; Lannuzel, A.; Simonin, Y.; Van de Perre, P.; Cabié, A.; Salinas, S. Neurocognitive impacts of arbovirus infections. J. Neuroinflamm. 2020, 17, 233. [Google Scholar] [CrossRef] [PubMed]
- Ao, Z.; Cai, H.; Havert, D.J.; Wu, Z.; Gong, Z.; Beggs, J.M.; Mackie, K.; Guo, F. One-Stop Microfluidic Assembly of Human Brain Organoids To Model Prenatal Cannabis Exposure. Anal. Chem. 2020, 92, 4630–4638. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Reality check for organoids in neuroscience. Nat. Methods 2020, 17, 961–964. [Google Scholar] [CrossRef]
- Papariello, A.; Newell-Litwa, K. Human-Derived Brain Models: Windows Into Neuropsychiatric Disorders and Drug Therapies. Assay Drug Dev. Technol. 2020, 18, 79–88. [Google Scholar] [CrossRef]
- B, S.R.; Patel, A.K.; Kabra, S.K.; Lodha, R.; Ratageri, V.H.; Ray, P. Virus load and clinical features during the acute phase of Chikungunya infection in children. PLoS ONE 2019, 14, e0211036. [Google Scholar] [CrossRef]
- Nakashima, A.; Ota, A.; Kaneko, Y.S.; Mori, K.; Nagasaki, H.; Nagatsu, T. A possible pathophysiological role of tyrosine hydroxylase in Parkinson’s disease suggested by postmortem brain biochemistry: A contribution for the special 70th birthday symposium in honor of Prof. Peter Riederer. J. Neural Transm. 2013, 120, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Reed, P.W.; Corse, A.M.; Porter, N.C.; Flanigan, K.M.; Bloch, R.J. Abnormal expression of mu-crystallin in facioscapulohumeral muscular dystrophy. Exp. Neurol. 2007, 205, 583–586. [Google Scholar] [CrossRef]
- Francelle, L.; Galvan, L.; Gaillard, M.C.; Guillermier, M.; Houitte, D.; Bonvento, G.; Petit, F.; Jan, C.; Dufour, N.; Hantraye, P.; et al. Loss of the thyroid hormone-binding protein Crym renders striatal neurons more vulnerable to mutant huntingtin in Huntington’s disease. Hum. Mol. Genet. 2015, 24, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Garwood, C.J.; Ratcliffe, L.E.; Simpson, J.E.; Heath, P.R.; Ince, P.G.; Wharton, S.B. Review: Astrocytes in Alzheimer’s disease and other age-associated dementias: A supporting player with a central role. Neuropathol. Appl. Neurobiol. 2017, 43, 281–298. [Google Scholar] [CrossRef] [PubMed]



| Non-PD | Non-PD + CHIKV | ∆∆Ct | PD | PD + CHIKV | ∆∆Ct | |
|---|---|---|---|---|---|---|
| Marker | AVG Ct ± SE | AVG Ct ± SE | AVG Ct ± SE | AVG Ct ± SE | ||
| GFAP | 29.7 ± 0.2 | 28.1 ± 0.5 | 29.4 ± 0.5 | 32.1 ± 0.6 | ||
| Iba1 | 26.2 ± 0.05 | 26.4 ± 0.03 | 24.2 ± 0.01 | 27.9 ± 0.01 | ||
| CCR5 | 31.3 ± 0.01 | 26.2 ± 0.1 | −3.403 | 30.1 ± 0.2 | 35.7 ± 1.3 | 2.824 |
| DRD1 | 27.6 ± 0.2 | 27.0 ± 0.7 | 28.5 ± 0.07 | 30.5 ± 0.1 | ||
| TH | 25.2 ± 0.1 | 28.5 ± 0.2 | 27.6 ± 0.2 | 30.2 ± 0.5 | ||
| CRYM | 25.6 ± 0.01 | 26.1 ± 0.0 | 27.9 ± 0.04 | 31.4 ± 0.0 | ||
| GBA | 22 ± 0.06 | 24.4 ± 0.02 | 25.1 ± 0.1 | 26.6 ± 0.03 | ||
| Aldh1l1 | 21.4 ± 0.1 | 20.9 ± 0.2 | 21.5 ± 0.1 | 25.6 ± 0.4 | ||
| IL-1a | 23.4 ± 0.1 | 22.1 ± 0.1 | −11.452 | 26.1 ± 0.1 | 27.7 ± 0.2 | 6.585 |
| IL-10 | 30.9 ± 0.1 | 25.0 ± 0.1 | 8.618 | 29.6 ± 0.3 | 33.9 ± 0.9 | −2.733 |
| CXCL10 | 24.5 ± 0.1 | 21.4 ± 0.1 | −2.368 | 26.6 ± 0.2 | 23.4 ± 0.02 | −4.98 |
| HTR3B | 29.5 ± 0.1 | 25.9 ± 0.1 | −6.522 | 29.3 ± 0.1 | 34.2 ± 1.3 | 0.944 |
| SLC6A4 | 24.9 ± 0.01 | 24.3 ± 0.04 | 13.521 | 28.8 ± 0.5 | 32.7 ± 0.7 | −3.885 |
| Gene | Accension # | Forward Primer 5′-3′ | Reverse Primer 5′-3′ |
|---|---|---|---|
| GFAP | NG_008401 | TACCCTTCTCTGTTTGCTGTG | CCTCCCAAAGTGCTAGGATTAC |
| Iba1 | D86438 | CTGAAACGAATGCTGGAGAAAC | GAGAAAGTCAGGGTAGCTGAAC |
| CCR5 | AY463215 | CCCAGTGGGACTTTGGAAATA | CGATTGTCAGGAGGATGATGAA |
| DRD1 | NM_000794 | AGGGACTTCTCTGTTCGTATCC | GGAACCTGATAACGGCAGCA |
| TH | BC143611.1 | TCATCACCTGGTCACCAAGTT | GGTCGCCGTGCCTGTACT |
| CRYM | BC018061 | GAGCTGGGAGAAGTGATTAAGG | TGGCTGCAACTGTGTCTT |
| GBA | AH006907 | GCTACTCCATTCACACCTACC | GAGCTGACTCTGTCCCTTTAAT |
| SLC6A4 | NM_001045 | TGCCCTCTCTTGCAGAATAAC | ATCACCTCCGAGCTCTCTATC |
| HTR3B | NG_011483 | GTCCTGAGTTTGGAGGTAGTTG | CTGAAGGAGATGCCTGAGATAC |
| IL-1a | BC013142 | CTGAAGGAGATGCCTGAGATAC | GATGGGCAACTGATGTGAAATAG |
| CXCL10 | BC010954 | CCATTCTGATTTGCTGCCTTATC | TACTAATGCTGATGCAGGTACAG |
| IL-10 | Z30175 | CACACACACACACACACAAATC | CTGGATAGGAGGTCCCTTACTT |
| Aldh1l1 | BC027241 | GTCAACCAGCAGAGCAAAC | GGCCCATAACCAGGAACAATA |
| Antibody | Host | Type | Source | Dilution |
|---|---|---|---|---|
| Primary Antibodies | ||||
| Syntaxin 1A | Mouse | Monoclonal | Novus Biologicals | 1:1000 |
| Syntaxin 3 | Rabbit | Polyclonal | Novus Biologicals | 1:500 |
| CYP46A1 | Rabbit | Polyclonal | Invitrogen | 1:500 |
| ICAM-1/CD54 | Mouse | Monoclonal | Novus Biologicals | 1:1000 |
| SELE/CD62E | Mouse | Monoclonal | Novus Biologicals | 1:1000 |
| CCR7 | Mouse | Monoclonal | Novus Biologicals | 1:1000 |
| NMDAR2C | Rabbit | Polyclonal | Novus Biologicals | 1:1000 |
| MAP2 | Chicken | Polyclonal | Novus Biologicals | 1:5000 |
| NMDAR1 | Mouse | Monoclonal | Novus Biological | 1:1000 |
| Synapsin 1 | Rabbit | Polyclonal | Invitrogen | 1:1000 |
| SOX2 | Mouse | Polyclonal | EMD Millipore | 1:1000 |
| Tuj1 | Rabbit | Monoclonal | EMD Millipore | 1:1000 |
| Neurofilament | Mouse | Monoclonal | EMD Millipore | 1:1000 |
| Ubiquilin | Rabbit | Polyclonal | Novus Biologicals | 1:1000 |
| Capsase 3 | Rabbit | Polyclonal | Novus Biologicals | 1:1000 |
| GFAP | Rabbit | Polyclonal | Novus Biologicals | 1:1000 |
| CHIKV CHK-48 | Mouse | Monoclonal | BEI Resources | 1:1000 |
| Secondary Antibodies | ||||
| Anti-Chicken | Goat | Alexa fluor 488 | Novus Biologicals | 1:3000 |
| Anti-Rabbit | Goat | Alexa fluor 594 | Novus Biologicals | 1:1000 |
| Anti-Mouse | Goat | Alexa fluor 647 | Novus Biologicals | 1:2000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schultz, E.M.; Jones, T.J.; Xu, S.; Dean, D.D.; Zechmann, B.; Barr, K.L. Cerebral Organoids Derived from a Parkinson’s Patient Exhibit Unique Pathogenesis from Chikungunya Virus Infection When Compared to a Non-Parkinson’s Patient. Pathogens 2021, 10, 913. https://doi.org/10.3390/pathogens10070913
Schultz EM, Jones TJ, Xu S, Dean DD, Zechmann B, Barr KL. Cerebral Organoids Derived from a Parkinson’s Patient Exhibit Unique Pathogenesis from Chikungunya Virus Infection When Compared to a Non-Parkinson’s Patient. Pathogens. 2021; 10(7):913. https://doi.org/10.3390/pathogens10070913
Chicago/Turabian StyleSchultz, Emily M., TyAnthony J. Jones, Sibei Xu, Dana D. Dean, Bernd Zechmann, and Kelli L. Barr. 2021. "Cerebral Organoids Derived from a Parkinson’s Patient Exhibit Unique Pathogenesis from Chikungunya Virus Infection When Compared to a Non-Parkinson’s Patient" Pathogens 10, no. 7: 913. https://doi.org/10.3390/pathogens10070913
APA StyleSchultz, E. M., Jones, T. J., Xu, S., Dean, D. D., Zechmann, B., & Barr, K. L. (2021). Cerebral Organoids Derived from a Parkinson’s Patient Exhibit Unique Pathogenesis from Chikungunya Virus Infection When Compared to a Non-Parkinson’s Patient. Pathogens, 10(7), 913. https://doi.org/10.3390/pathogens10070913







