Temporal Changes in the Genetic Diversity of Plasmodium vivax Merozoite Surface Protein-1 in Myanmar
Abstract
:1. Introduction
2. Materials and Methods
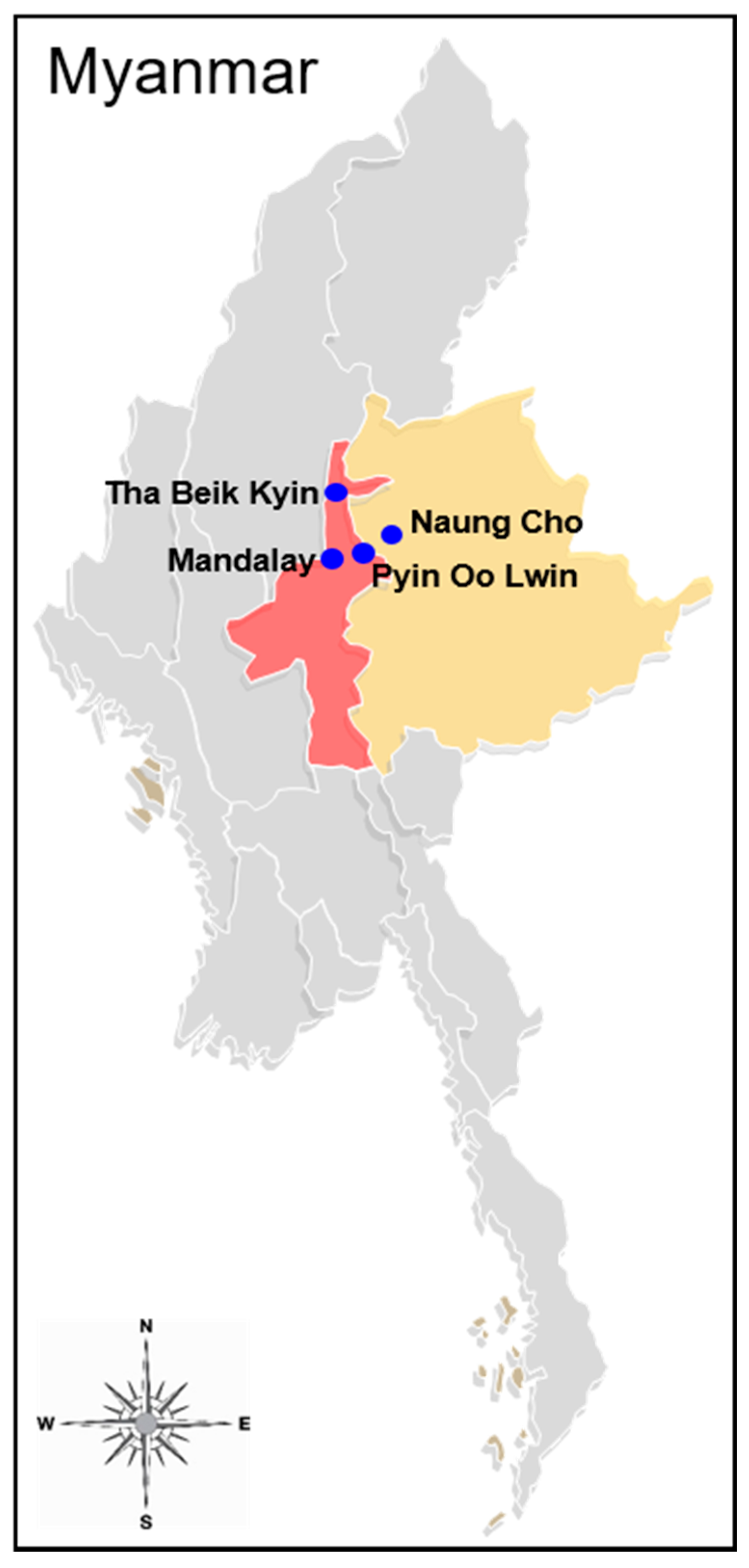
2.1. Blood Sample Collection
2.2. Genomic DNA Extraction
2.3. Amplification and Sequence Analysis of pvmsp-1 ICB 5-6
2.4. Temporal Change of Genetic Diversity in Myanmar pvmsp-1 ICB 5-6
3. Results
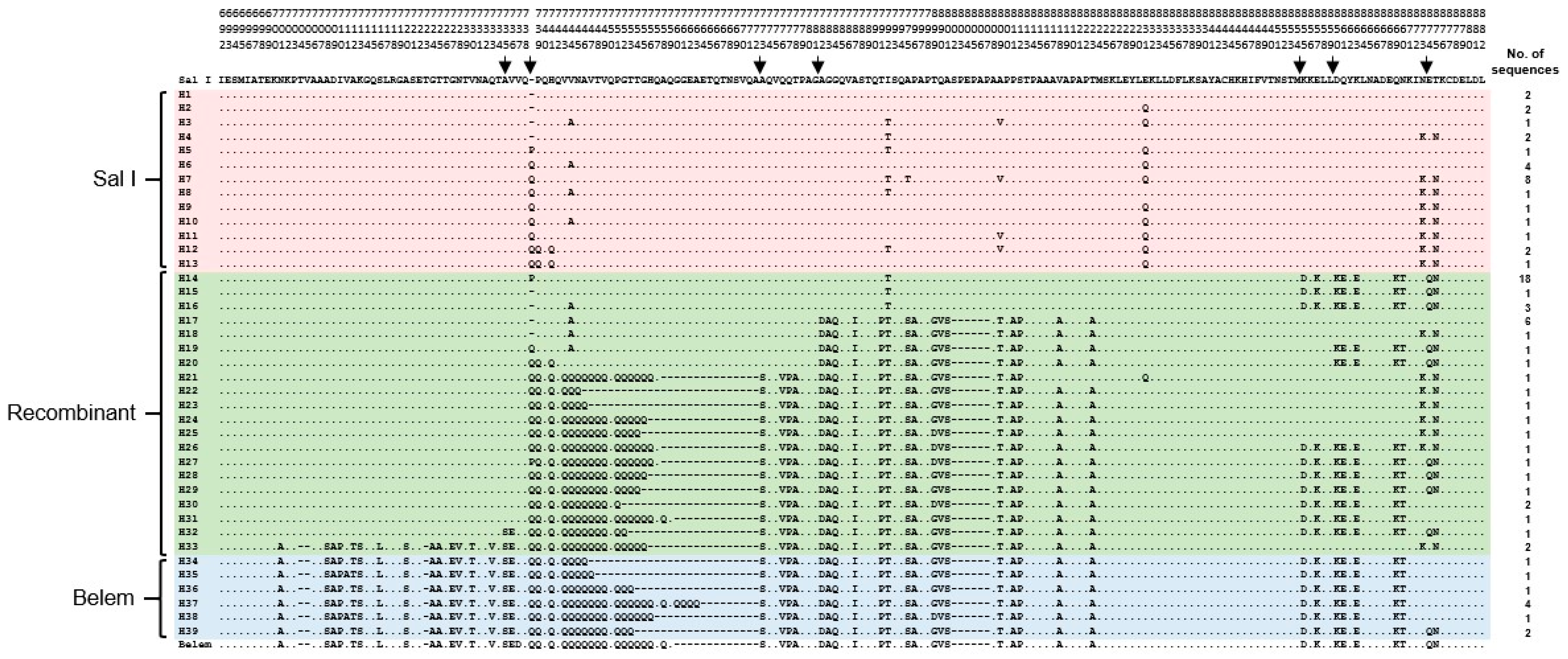
3.1. Sequence Analysis of Myanmar pvmsp-1 ICB 5-6
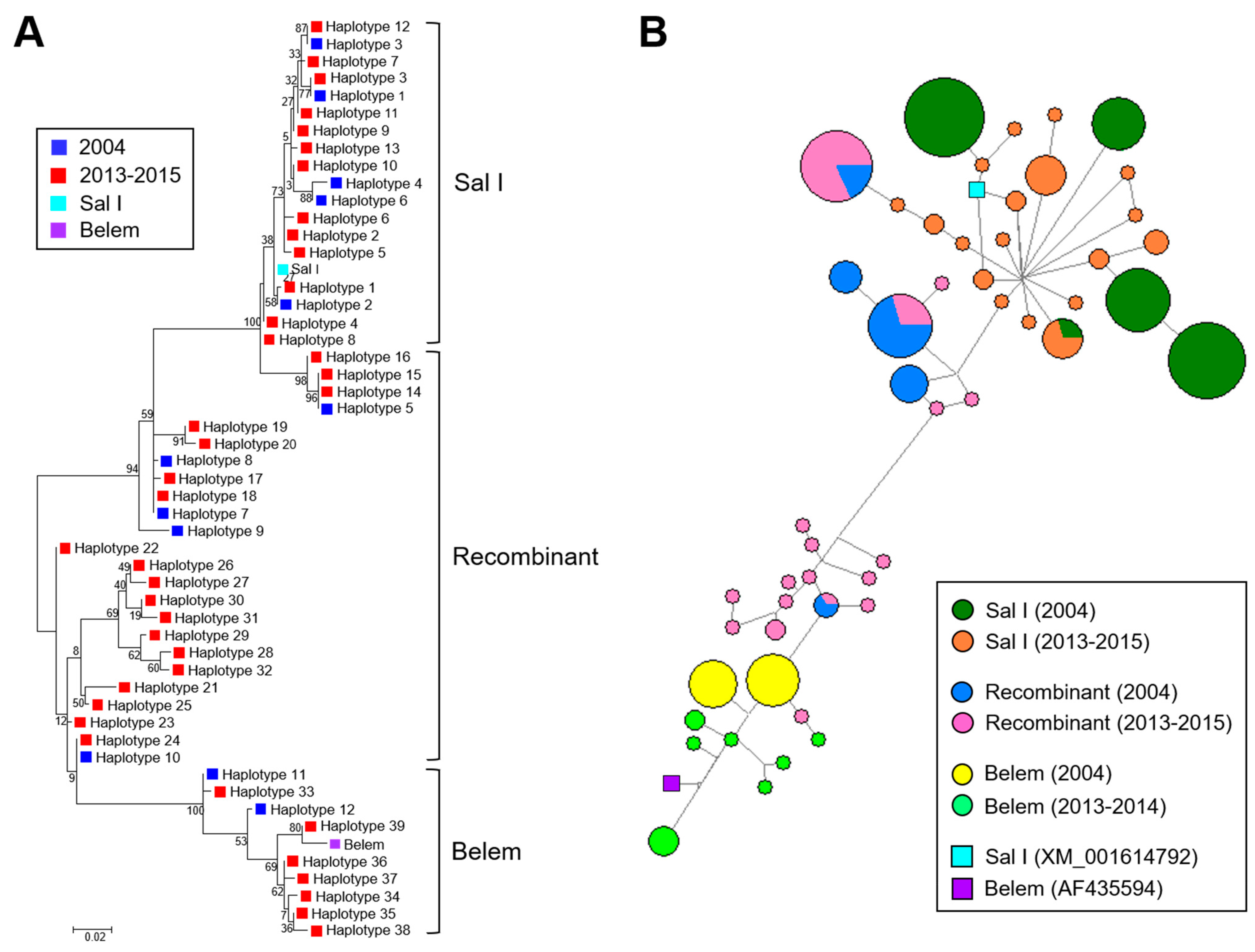
3.2. Temporal Changes in Myanmar pvmsp-1 ICB 5-6 in 2004 and in 2013–2015
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2020: 20 Years of Global Progress and Challenges; World Health Organization: Geneva, Switzerland, 2020; ISBN 9789240015791. [Google Scholar]
- World Health Organization. World Malaria Report 2016; World Health Organization: Geneva, Switzerland, 2017; ISBN 9789241565158. [Google Scholar]
- Mu, T.T.; Sein, A.A.; Kyi, T.T.; Min, M.; Aung, N.M.; Anstey, N.M.; Kyaw, M.P.; Soe, C.; Kyi, M.M.; Hanson, J. Malaria incidence in Myanmar 2005–2014: Steady but fragile progress towards elimination. Malar. J. 2016, 15, 503. [Google Scholar] [CrossRef] [Green Version]
- Lin, K. Malaria Control in Myanmar; Kano, S., Tongol-Rivera, P., Eds.; Asian Parasitology, Malaria in Asia; The Federation of Asian Parasitologists: Chiba, Japan, 2005; pp. 123–134. [Google Scholar]
- Kang, J.M.; Cho, P.Y.; Moe, M.; Lee, J.; Jun, H.; Lee, H.W.; Ahn, S.K.; Kim, T.I.; Pak, J.H.; Myint, M.K.; et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar. J. 2017, 16, 119. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Zhang, L.; Xue, J.B.; Zhou, H.N.; Thi, A.; Zhang, J.; Zhou, S.S.; Xia, Z.G.; Zhou, X.N. From control to elimination: A spatial-temporal analysis of malaria along the China-Myanmar border. Infect. Dis. Poverty 2020, 9, 158. [Google Scholar] [CrossRef]
- Valderrama-Aguirre, A.; Quintero, G.; Gómez, A.; Castellanos, A.; Pérez, Y.; Méndez, F.; Arévalo-Herrera, M.; Herrera, S. Antigenicity, immunogenicity, and protective efficacy of Plasmodium vivax MSP1 Pv200L: A potential malaria vaccine subunit. Am. J. Trop. Med. Hyg. 2005, 73, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Herrera, S.; Corradin, G.; Arévalo-Herrera, M. An update on the search for a Plasmodium vivax vaccine. Trends Parasitol. 2007, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Versiani, F.G.; Almeida, M.E.; Mariuba, L.A.; Orlandi, P.P.; Nogueira, P.A. N-terminal Plasmodium vivax merozoite surface protein-1, a potential subunit for malaria vivax vaccine. Clin. Dev. Immunol. 2013, 2013, 965841. [Google Scholar] [CrossRef] [Green Version]
- Putaporntip, C.; Jongwutiwes, S.; Sakihama, N.; Ferreira, M.U.; Kho, W.G.; Kaneko, A.; Kanbara, H.; Hattori, T.; Tanabe, K. Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc. Natl. Acad. Sci. USA 2002, 99, 16348–16353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnott, A.; Barry, A.E.; Reeder, J.C. Understanding the population genetics of Plasmodium vivax is essential for malaria control and elimination. Malar. J. 2012, 11, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.U.; Lee, H.W.; Kim, J.Y.; Na, B.K.; Cho, S.H.; Lin, K.; Sohn, W.M.; Kim, T.S. High frequency of genetic diversity of Plasmodium vivax field isolates in Myanmar. Acta Trop. 2009, 109, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Del Portillo, H.A.; Longacre, S.; Khouri, E.; David, P.H. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc. Natl. Acad. Sci. USA 1991, 88, 4030–4034. [Google Scholar] [CrossRef] [Green Version]
- Ruan, W.; Zhang, L.L.; Feng, Y.; Zhang, X.; Chen, H.L.; Lu, Q.Y.; Yao, L.N.; Hu, W. Genetic diversity of Plasmodium vivax revealed by the merozoite surface protein-1 icb5-6 fragment. Infect. Dis. Poverty 2017, 6, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, A.A.; Kain, K.C. Molecular analysis of strains of Plasmodium vivax from paired primary and relapse infections. J. Infect. Dis. 1996, 174, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Snounou, G.; Viriyakosol, S.; Jarra, W.; Thaithong, S.; Brown, K.N. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol. Biochem. Parasitol. 1993, 58, 283–292. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- National Strategic Plan: Intensifying Malaria Control and Accelerating Progress towards Malaria Elimination (2016–2020); Ministry of Health and Sports: Nay Pyi Taw, Myanmar, 2016; Available online: https://apmen.org/sites/default/files/all_resources/National%20Strategic%20Plan_Myanmar_2016-2020.pdf (accessed on 12 October 2020).
- Lê, H.G.; Kang, J.M.; Jun, H.; Lee, J.; Thái, T.L.; Myint, M.K.; Aye, K.S.; Sohn, W.M.; Shin, H.J.; Kim, T.S.; et al. Changing pattern of the genetic diversities of Plasmodium falciparum merozoite surface protein-1 and merozoite surface protein-2 in Myanmar isolates. Malar. J. 2019, 18, 241. [Google Scholar] [CrossRef]
- Koepfli, C.; Ross, A.; Kiniboro, B.; Smith, T.A.; Zimmerman, P.A.; Siba, P.; Mueller, I.; Felger, I. Multiplicity and diversity of Plasmodium vivax infections in a highly endemic region in papua New Guinea. PLoS Negl. Trop. Dis. 2011, 5, e1424. [Google Scholar] [CrossRef]
- Gunawardena, S.; Ferreira, M.U.; Kapilananda, G.M.G.; Wirth, D.F.; Karunaweera, N.D. The Sri Lankan paradox: High genetic diversity in Plasmodium vivax populations despite decreasing levels of malaria transmission. Parasitology 2014, 141, 880–890. [Google Scholar] [CrossRef]
- Li, Y.C.; Wang, G.Z.; Meng, F.; Zeng, W.; He, C.H.; Hu, X.M.; Wang, S.Q. Genetic diversity of Plasmodium vivax population before elimination of malaria in Hainan Province, China. Malar. J. 2015, 14, 78. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.M.; Lee, J.; Cho, P.Y.; Kim, T.I.; Sohn, W.M.; Park, J.W.; Kim, T.S.; Na, B.K. Dynamic changes of Plasmodium vivax population structure in South Korea. Infect. Genet. Evol. 2016, 45, 90–94. [Google Scholar] [CrossRef]
- Vallejo, A.F.; Chaparro, P.E.; Benavides, Y.; Álvarez, Á.; Quintero, J.P.; Padilla, J.; Arévalo-Herrera, M.; Herrera, S. High prevalence of sub-microscopic infections in Colombia. Malar. J. 2015, 14, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipmontree, R.; Fungladda, W.; Kaewkungwal, J.; Tempongko, M.A.; Schelp, F.P. Migrants and malaria risk factors: A study of the Thai-Myanmar border. Southeast Asian J. Trop. Med. Public Health 2009, 40, 1148–1157. [Google Scholar]
- Li, Y.; Hu, Y.; Zhao, Y.; Wang, Q.; Ngassa Mbenda, H.G.; Kittichai, V.; Lawpoolsri, S.; Sattabongkot, J.; Menezes, L.; Liu, X.; et al. Dynamics of Plasmodium vivax populations in border areas of the Greater Mekong sub-region during malaria elimination. Malar. J. 2020, 19, 145. [Google Scholar] [CrossRef] [Green Version]
- Kolakovich, K.A.; Ssengoba, A.; Wojcik, K.; Tsuboi, T.; Al-Yaman, F.; Alpers, M.; Adams, J.H. Plasmodium vivax: Favored gene frequen cies of the merozoite surface protein-1 and the multiplicity of infection in a malaria endemic region. Exp. Parasitol. 1996, 83, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Fola, A.A.; Harrison, G.L.A.; Hazairin, M.H.; Barnadas, C.; Hetzel, M.W.; Iga, J.; Siba, P.M.; Mueller, I.; Barry, A.E. Higher complexity of infection and genetic diversity of Plasmodium vivax than Plasmodium falciparum across all malaria transmission zones of Papua New Guinea. Am. J. Trop. Med. Hyg. 2017, 96, 630–641. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.L.; Zhou, H.N.; Liu, Q.; Yang, Y.M. Genetic polymorphism of merozoite surface proteins 1 and 2 of Plasmodium falciparum in the China-Myanmar border region. Malar. J. 2019, 18, 367. [Google Scholar] [CrossRef] [PubMed]
- Võ, T.C.; Lê, H.G.; Kang, J.M.; Naw, H.; Fan, C.K.; Trinh, N.T.M.; Quang, H.H.; Na, B.K. Molecular surveillance of malaria in the Central Highlands, Vietnam. Parasitol. Int. 2021, 83, 102374. [Google Scholar] [CrossRef]


| Year | Ra | Rb | Rm |
|---|---|---|---|
| 2004 | 0.000 | 0.001 | 3 |
| 2013–2015 | 0.058 | 3.2 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naw, H.; Kang, J.-M.; Moe, M.; Lee, J.; Lê, H.G.; Võ, T.C.; Mya, Y.Y.; Myint, M.K.; Htun, Z.T.; Kim, T.-S.; et al. Temporal Changes in the Genetic Diversity of Plasmodium vivax Merozoite Surface Protein-1 in Myanmar. Pathogens 2021, 10, 916. https://doi.org/10.3390/pathogens10080916
Naw H, Kang J-M, Moe M, Lee J, Lê HG, Võ TC, Mya YY, Myint MK, Htun ZT, Kim T-S, et al. Temporal Changes in the Genetic Diversity of Plasmodium vivax Merozoite Surface Protein-1 in Myanmar. Pathogens. 2021; 10(8):916. https://doi.org/10.3390/pathogens10080916
Chicago/Turabian StyleNaw, Haung, Jung-Mi Kang, Mya Moe, Jinyoung Lee, Hương Giang Lê, Tuấn Cường Võ, Yi Yi Mya, Moe Kyaw Myint, Zaw Than Htun, Tong-Soo Kim, and et al. 2021. "Temporal Changes in the Genetic Diversity of Plasmodium vivax Merozoite Surface Protein-1 in Myanmar" Pathogens 10, no. 8: 916. https://doi.org/10.3390/pathogens10080916







