Evaluation of Potential Transfer of the Pathogen Saprolegnia parasitica between Farmed Salmonids and Wild Fish
Abstract
1. Introduction
2. Results
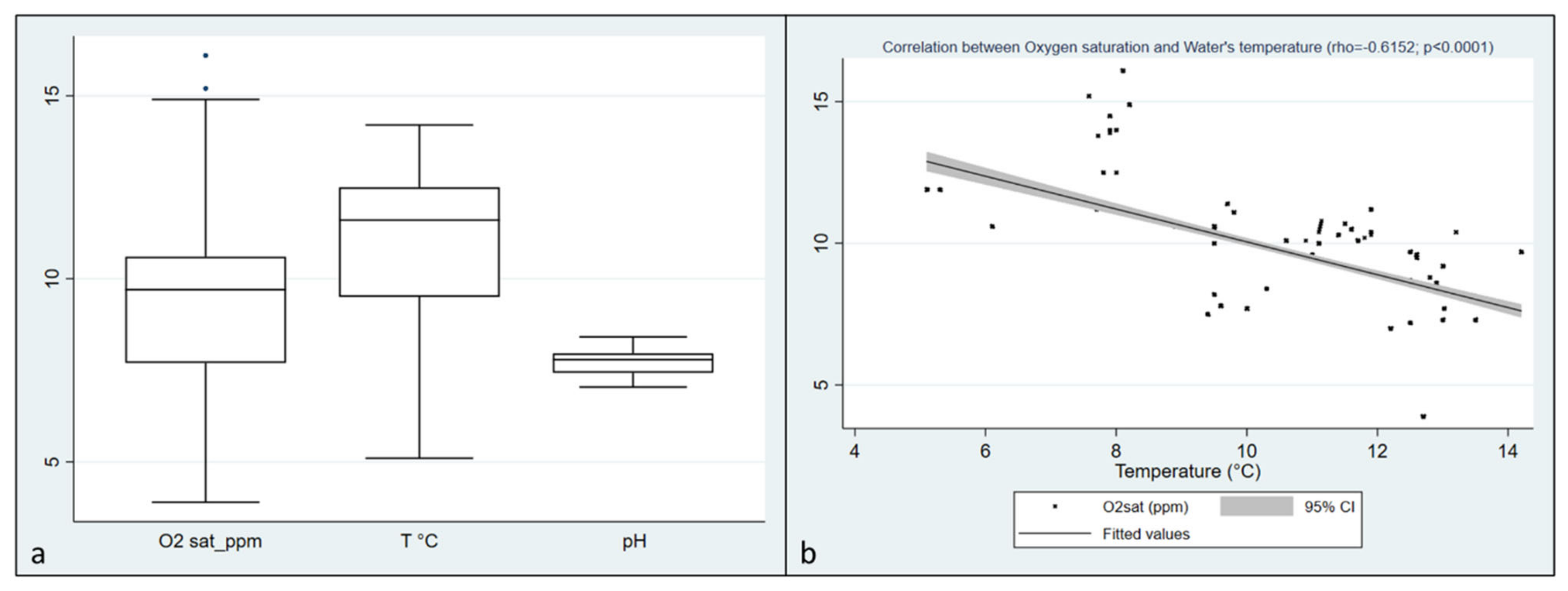
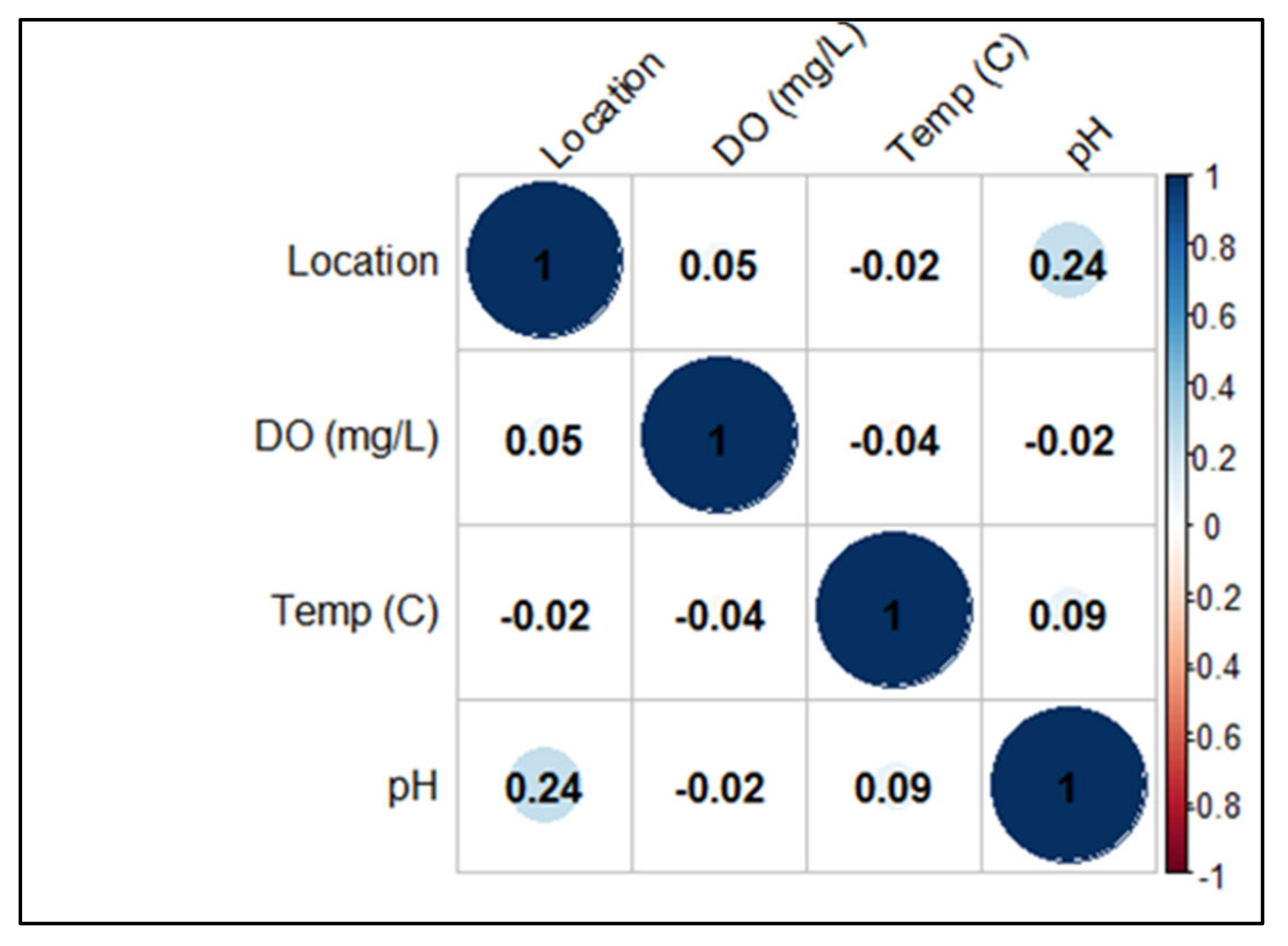
2.1. Environmental Parameters
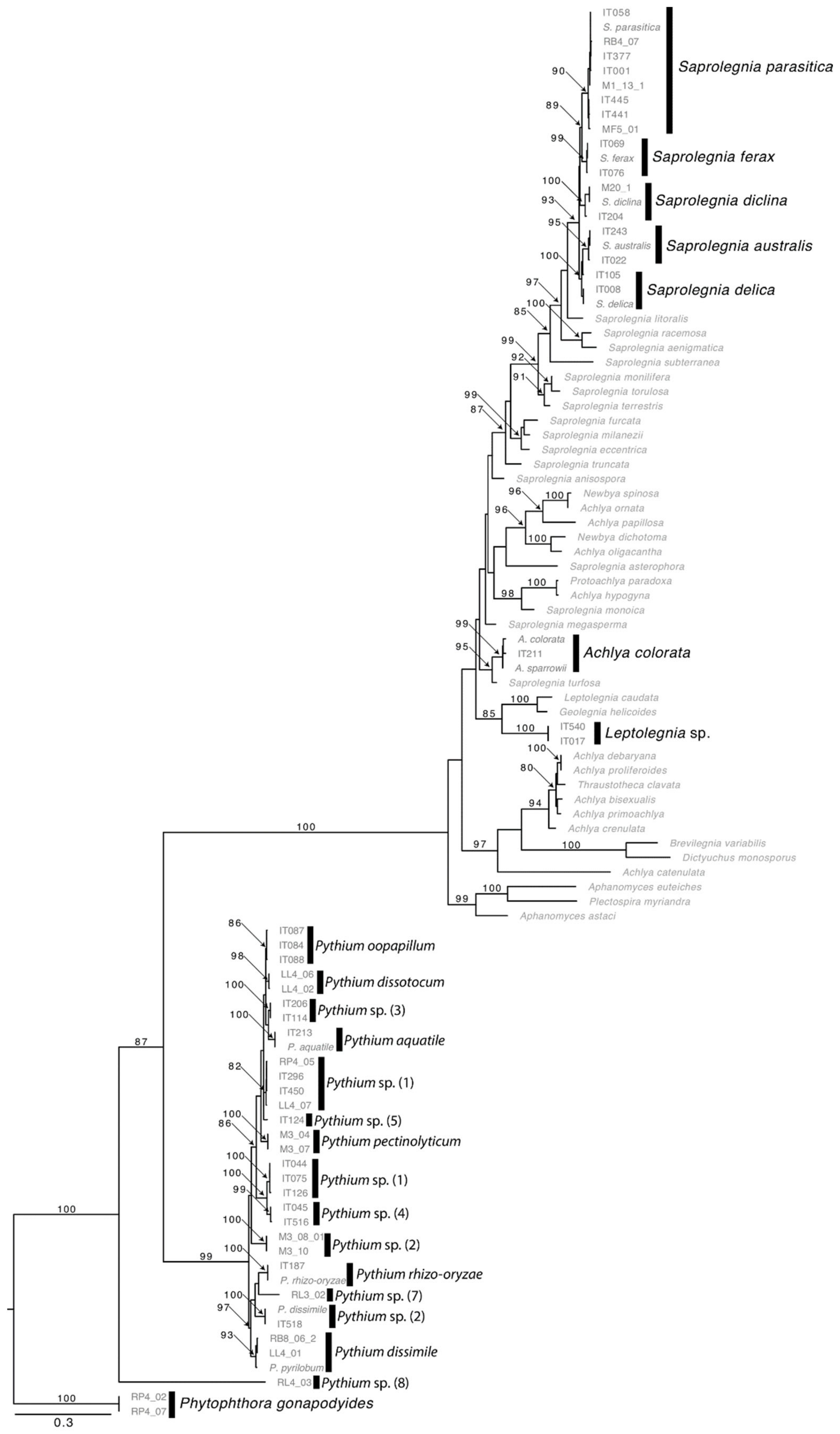
2.2. Oomycetes Diversity
2.3. Transfer of Saprolegnia parasitica between Wild and Farmed Fish and Impact of Salmonid Farms on the Spread of S. parasitica to Wild Fish
3. Discussion
3.1. Oomycetes Diversity
3.2. Transfer of Saprolegnia parasitica between Wild and Farmed Fish and Impact of Salmonid Farms on the Spread of S. parasitica to Wild Fish
4. Materials and Methods
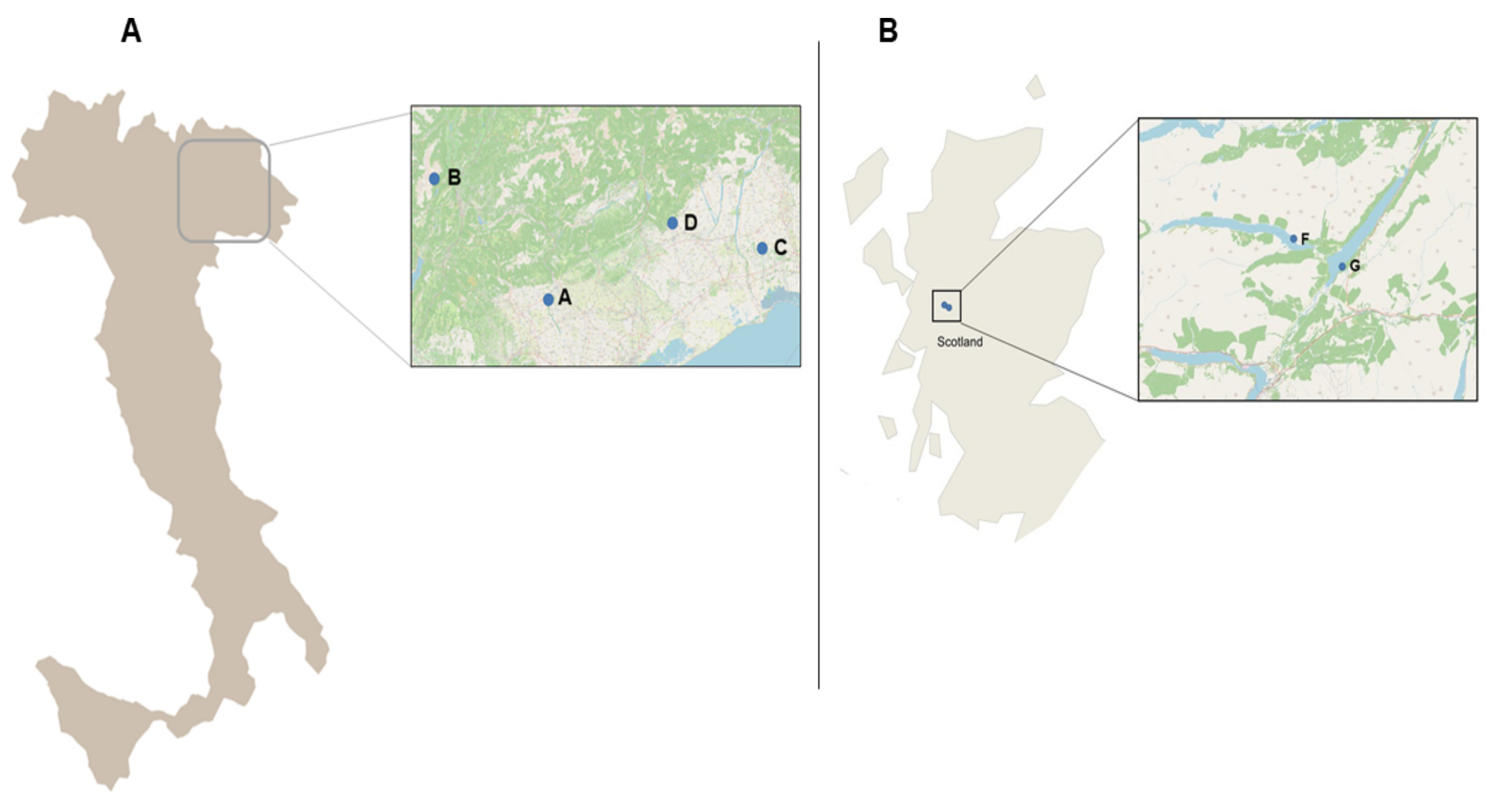
4.1. Study Areas
4.2. Collection of Fish and Water Samples
4.3. Oomycetes Isolation
4.4. DNA Extraction and PCR Conditions
4.5. Molecular Identification of Species
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johansen, L.H.; Jensen, I.; Mikkelsen, H.; Bjørn, P.A.; Jansen, P.A.; Bergh, Ø. Disease interaction and pathogens exchange between wild and farmed fish populations with special reference to Norway. Aquaculture 2011, 315, 167–186. [Google Scholar] [CrossRef]
- Krkošek, M. Host density thresholds and disease control for fisheries and aquaculture. Aquac. Environ. Int. 2010, 1, 21–32. [Google Scholar] [CrossRef]
- Krkošek, M.; Lewis, M.A.; Volpe, J.P. Transmission dynamics of parasitic sea lice from farm to wild salmon. Proc. R. Soc B Biol. Sci. 2005, 272, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Krkošek, M.; Lewis, M.A.; Morton, A.; Frazer, L.N.; Volpe, J.P. Epizootics of wild fish induced by farm fish. Proc. Natl. Acad. Sci. USA 2006, 103, 15506–15510. [Google Scholar] [CrossRef] [PubMed]
- Krkošek, M.; Ford, J.S.; Morton, A.; Lele, S.; Myers, R.A.; Lewis, M.A. Declining wild salmon populations in relation to parasites from farm salmon. Science 2007, 318, 1772–1775. [Google Scholar] [CrossRef]
- Čomić, L.; Ranković, B.; Novevska, V.; Ostojić, A. Diversity and dynamics of the fungal community in Lake Ohrid. Aquat. Biol. 2010, 9, 169–176. [Google Scholar] [CrossRef]
- Blaustein, A.R.; Wake, D.B.; Sousa, W.P. Amphibian declines: Judging stability, persistence, and susceptibility of populations to local and global extinctions. Conserv. Biol. 1994, 8, 60–71. [Google Scholar] [CrossRef]
- Kestrup, Å.M.; Thomas, S.H.; van Rensburg, K.; Ricciardi, A.; Duffy, M.A. Differential infection of exotic and native freshwater amphipods by a parasitic water mold in the St. Lawrence River. Biol. Invasions 2011, 13, 769–779. [Google Scholar] [CrossRef]
- Rezinciuc, S.; Sandoval-Sierra, J.V.; Oidtmann, B.; Diéguez-Uribeondo, J. The biology of crayfish plague pathogen Aphanomyces astaci: Current answers to most frequent questions. In Freshwater Crayfish—A Global Overview; Kawai, T., Faulkes, Z., Scholtz, G., Eds.; Taylor and Francis: Boca Raton, FL, USA, 2015; pp. 182–204. [Google Scholar]
- Van West, P. Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: New challenges for an old problem. Mycologist 2006, 20, 99–104. [Google Scholar] [CrossRef]
- Diéguez-Uribeondo, J.; Fregeneda-Grandes, J.M.; Cerenius, L.; Pérez-Iniesta, E.; Aller-Gancedo, J.M.; Tellería, M.T.; Söderhäll, K.; Martín, M.P. Re-evaluation of the enigmatic species complex Saprolegnia diclina–Saprolegnia parasitica based on morphological, physiological and molecular data. Fungal Genet. Biol. 2007, 44, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Rezinciuc, S.; Sandoval-Sierra, J.V.; Diéguez-Uribeondo, J. Molecular identification of a bronopol tolerant strain of Saprolegnia australis causing egg and fry mortality in farmed brown trout, Salmo trutta. Fungal Biol. 2014, 118, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Sierra, J.V.; Martín, M.P.; Diéguez-Uribeondo, J. Species identification in the genus Saprolegnia (Oomycetes): Defining DNA-based molecular operational taxonomic units. Fungal Biol. 2014, 118, 559–578. [Google Scholar] [CrossRef]
- Hatai, K.; Hoshiai, G. Mass mortality in cultured coho salmon (Oncorhynchus kisutch) due to Saprolegnia parasitica coker. J. Wildl. Dis. 1992, 28, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Bly, J.E.; Lawson, L.A.; Dale, D.J.; Szalai, A.J.; Durburow, R.M.; Clem, L.W. Winter saprolegniosis in channel catfish. Dis. Aquat. Organ. 1992, 13, 155–164. [Google Scholar] [CrossRef]
- Richards, R.H.; Pickering, A.D. Frequency and distribution patterns of Saprolegnia infection in wild and hatchery-reared brown trout Salmo trutta L. and char Salvelinus alpinus (L.). J. Fish Dis. 1978, 1, 69–82. [Google Scholar] [CrossRef]
- White, D.A. Ecology of an annual Saprolegnia sp. (Phycomycete) outbreak in wild brown trout. Verh. Int. Ver. Theor. Angew. Limnol. Verh. 1975, 19, 2456–2460. [Google Scholar]
- Álvarez, F.; Villena, A.; Zapata, A.; Razquin, B. Histopathology of the thymus in Saprolegnia-infected wild brown trout, Salmo trutta L. Vet. Immunol. Immunopathol. 1995, 47, 163–172. [Google Scholar] [CrossRef]
- Diéguez-Uribeondo, J.; Cerenius, L.; Söderhäll, K. Physiological characterization of Saprolegnia parasitica isolates from brown trout. Aquaculture 1996, 140, 247–257. [Google Scholar] [CrossRef]
- Mancini, M.; Rodriguez, C.; Barberis, C.; Astoreca, A.; Brol, G. Registro de saprolegniasis en ejemplares silvestres de pejerrey Odontesthes bonariensis (Pisces: Atherinopsidae) de Argentina. Braz. J. Vet. Med. 2010, 32, 46–50. [Google Scholar]
- De la Bastide, P.Y.; Leung, W.L.; Hintz, W.E. Species composition of the genus Saprolegnia in fin fish aquaculture environments, as determined by nucleotide sequence analysis of the nuclear rDNA ITS regions. Fungal. Biol. 2015, 119, 27–43. [Google Scholar] [CrossRef]
- Pavić, A.; Miljanović, D.; Grbin, Š.L.; Vladušić, T.; Galuppi, R.; Tedesco, P.; Bielen, A. Identification and molecular characterization of oomycete isolates from trout farms in Croatia, and their upstream and downstream water environments. Aquaculture 2021. [Google Scholar] [CrossRef]
- Sandoval-Sierra, J.V.; Diéguez-Uribeondo, J. A comprehensive protocol for improving the description of Saprolegniales (Oomycota): Two practical examples (Saprolegnia aenigmatica sp. nov. and Saprolegnia racemosa sp. nov.). PLoS ONE 2015, 10, e0132999. [Google Scholar]
- Fernández-Benéitez, M.J.; Ortiz-Santaliestra, M.E.; Lizana, M.; Diéguez-Uribeondo, J. Saprolegnia diclina: Another species responsible for the emergent disease ‘Saprolegnia infections’ in amphibians. FEMS Microbiol. Lett. 2008, 279, 23–29. [Google Scholar] [CrossRef]
- Kiesecker, J.M.; Blaustein, A.R.; Belden, L.K. Complex causes of amphibian population declines. Nature 2001, 410, 681. [Google Scholar] [CrossRef] [PubMed]
- Montalva, C.; dos Santos, K.; Collier, K.; Rocha, L.F.; Fernandes, É.K.; Castrillo, L.A.; Luz, C.; Humber, R.A. First report of Leptolegnia chapmanii (Peronosporomycetes: Saprolegniales) affecting mosquitoes in central Brazil. J. Invertebr. Pathol. 2016, 136, 109–116. [Google Scholar] [CrossRef]
- Van der Plaats-Niterink, A.J. Monograph of the Genus Pythium; Centraalbureau voor Schimmelcultures: Baarn, NL, USA, 1981; Volume 21. [Google Scholar]
- Czeczuga, B.; Kozłowska, M.; Godlewska, A. Zoosporic aquatic fungi growing on dead specimens of 29 freshwater crustacean species. Limnologica 2002, 32, 180–193. [Google Scholar] [CrossRef][Green Version]
- Pickering, A.D.; Willoughby, L.G. Saprolegnia infections of salmonid fish. In Microbial Diseases of Fish; Roberts, R.J., Ed.; Academic Press Inc.: London, UK, 1982; pp. 271–297. [Google Scholar]
- Toor, H.S.; Sehgal, H.; Sehdev, R.S. A case study of acute fish diseases in tanks loaded with high levels of organic manures. Aquaculture 1983, 35, 277–282. [Google Scholar] [CrossRef]
- Carballo, M.; Muñoz, M.J. Effect of sublethal concentrations of four chemicals on susceptibility of juvenile rainbow trout (Oncorhynchus mykiss) to saprolegniosis. Appl. Environ. Microbiol. 1991, 57, 1813–1816. [Google Scholar] [CrossRef]
- Seymour, R.L. The genus Saprolegnia. Nova Hedwig. 1970, 19, 1–124. [Google Scholar]
- Alderman, D.J.; Polglase, J.L. A comparative investigation of the effects of fungicides on Saprolegnia parasitica and Aphanomyces astaci. Trans. Br. Mycol. Soc. 1984, 83, 313–318. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform. 2008, 9, 212. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Kim, H.Y. Statistical notes for clinical researchers: Chi-squared test and Fisher’s exact test. Restor. Dent. Endod. 2017, 42, 152–155. [Google Scholar] [CrossRef] [PubMed]



| Environmental Parameters (Min–Max) | n° Samples Collected | |||||||
|---|---|---|---|---|---|---|---|---|
| Farm ID (Farmed Species/Water Supply | T (°C) | pH | O2 (mg/L) | Upstream | Farm | Downstream | Tot | |
| A (1/3) | 5.2–9.8 | 7.25–8.41 | 10–16.8 | fish baits | 20 34 | 30 32 | 16 38 | 66 105 |
| B (1/3) | 11.7–13 | 7.05–7.45 | 3.9–10.1 | fish baits | 1 38 | 30 45 | 42 30 | 73 113 |
| C (2/4) | 9.5–13.2 | 7.04–8.21 | 8.2–11.2 | fish baits | 15 39 | 25 27 | 20 35 | 60 101 |
| D (2/3) | 9.4–14.2 | 7.45–7.85 | 7.3–9.7 | fish baits | 10 39 | 20 30 | 10 24 | 40 93 |
| Environmental Parameters (Min–Max) | n° Samples Collected | |||||||
|---|---|---|---|---|---|---|---|---|
| upstream | farm | downstream | tot | |||||
| Farm ID | T (°C) | pH | O2 (mg/L) | |||||
| F | 6.1–16.7 | 4.5–7.9 | 91–93 | fish baits | 0 240 | 200 240 | 0 240 | 200 720 |
| G | 6.5–14.7 | 6.5–7.6 | 90–94 | fish baits | 0 240 | 200 240 | 0 240 | 200 720 |
| Farm ID | n° Isolates Identified | Species Identified | ||||||
|---|---|---|---|---|---|---|---|---|
| Upstream | Farm | Downstream | Tot | Upstream | Farm | Downstream | ||
| A | fish | 4 | 35 | 6 | 45 | 2 S. parasitica 1 Pythium sp. 1 S delica | 31 S. parasitica 4 S delica | 6 Leptolegnia sp. |
| baits | 35 | 32 | 38 | 105 | 1 S. australis 11 S. parasitica 13 S. delica 9 Pythium sp. 1 S. ferax | 24 S. delica 6 S. australis 1 S. ferax 1 S. parasitica | 13 S. parasitica 11 S. delica 7 Pythium sp. 4 S. australis 1 S. ferax 2 S. hypogyna | |
| B | fish | 0 | 2 | 5 | 7 | 0 | 2 Pythium sp. | 3 Pythium sp. 1 S. parasitica 1 S.ferax |
| baits | 38 | 45 | 30 | 113 | 19 Pythium sp. 19 S. ferax | 29 S. ferax 6 Pythium sp. 10 S. delica | 18 S. ferax 4 S. australis 5 Pythium sp. 3 S. delica | |
| C | fish | 4 | 12 | 10 | 26 | 4 S. parasitica | 12 S. parasitica | 9 S. parasitica 1 S. australis |
| baits | 39 | 27 | 35 | 101 | 23 S. ferax 3 S. parasitica 6 Pythium sp. 5 S. australis 2 S. delica | 6 S. parasitica 11 S. ferax 7 S. australis 2 S. delica 1 Pythium sp. | 22 S. australis 7 S. parasitica 4 Pythium sp. 1 S. delica 1 S. ferax | |
| D | fish | 3 | 32 | 7 | 42 | 2 S. parasitica 1 Pythium sp. | 30 S. parasitica 2 S. ferax | 6 S. parasitica 1 S. australis |
| baits | 39 | 30 | 24 | 93 | 11 S. australis 7 S. parasitica 13 S. ferax 6 Pythium sp. 1 Saprolegnia sp. 1 Achlya colorata | 11 S. australis 10 S. ferax 5 S. delica 4 S. parasitica | 7 S. parasitica 7 S. australis 3 S. ferax 6 S. delica 1 Pythium sp. | |
| Farm ID | n° Isolates Identified | Species Identified | ||||||
|---|---|---|---|---|---|---|---|---|
| Upstream | Farm | Downstream | Tot | Upstream | Farm | Downstream | ||
| F | fish | 0 | 61 | 0 | 61 | 0 | 2 P. dissotocum 2 P. flevoense 2 P. monospermum 1 P. pachycaule 5 S. australis 6 S. diclina 41 S. parasitica 2 Saprolegnia sp1 | 0 |
| baits | 14 | 8 | 18 | 40 | 2 Phy. gonapodyides | 3 P. dissotocum | 6 P. dissotocum | |
| 7 P. dissotocum | 2 P. pachycaule | 1 P. pachycaule | ||||||
| 2 P. pachycaule | 2 P. pyrilobum | 7 P. pyrilobum | ||||||
| 2 P. pyrilobum | 1 S. parasitica | 2 Pythium sp03 | ||||||
| 1 S. parasitica | 1 Pythium sp05 | |||||||
| 1 S. parasitica | ||||||||
| G | fish | 0 | 67 | 0 | 67 | 0 | 17 P. dissotocum 1 P. flevoense 1 P. monospermum 1 P. pachycaule 7 P. pyrilobum 1 Pythium sp01 2 Pythium sp03 1 Pythium sp05 1 Pythium sp08 8 S. australis 1 S. delica 4 S. diclina 2 S. parahypogyna 20 S. parasitica. | 0 |
| baits | 18 | 9 | 14 | 41 | 6 P. dissotocum | 5 P. dissotocum 1 P. pachycaule 3 P. pyrilobum | 10 P. dissotocum 1 S. diclina 1 S. delica 2 Pythium spp. | |
| 1 P. pachycaule | ||||||||
| 7 P. pyrilobum | ||||||||
| 2 Pythium sp03 | ||||||||
| 1 Pythium sp05 | ||||||||
| 1 S. parasitica | ||||||||
| Categories | OR | p-Value | 95% CI | |
|---|---|---|---|---|
| fish * | in-farm | baseline | - | - |
| upstream | 6.39 | <0.001 | 2.92–13.95 | |
| downstream | 5.97 | <0.001 | 3.31–10.78 | |
| baits | in-farm | baseline | - | - |
| upstream | 1.8 | 0.132 | 0.26–1.19 | |
| downstream | 3.02 | 0.004 | 1.43–6.39 | |
| T (°C) | - | 1.13 | 0.001 | 1.05–1.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tedesco, P.; Saraiva, M.; Sandoval-Sierra, J.V.; Fioravanti, M.L.; Morandi, B.; Dieguez-Uribeondo, J.; van West, P.; Galuppi, R. Evaluation of Potential Transfer of the Pathogen Saprolegnia parasitica between Farmed Salmonids and Wild Fish. Pathogens 2021, 10, 926. https://doi.org/10.3390/pathogens10080926
Tedesco P, Saraiva M, Sandoval-Sierra JV, Fioravanti ML, Morandi B, Dieguez-Uribeondo J, van West P, Galuppi R. Evaluation of Potential Transfer of the Pathogen Saprolegnia parasitica between Farmed Salmonids and Wild Fish. Pathogens. 2021; 10(8):926. https://doi.org/10.3390/pathogens10080926
Chicago/Turabian StyleTedesco, Perla, Marcia Saraiva, Jose Vladimir Sandoval-Sierra, Maria Letizia Fioravanti, Benedetto Morandi, Javier Dieguez-Uribeondo, Pieter van West, and Roberta Galuppi. 2021. "Evaluation of Potential Transfer of the Pathogen Saprolegnia parasitica between Farmed Salmonids and Wild Fish" Pathogens 10, no. 8: 926. https://doi.org/10.3390/pathogens10080926
APA StyleTedesco, P., Saraiva, M., Sandoval-Sierra, J. V., Fioravanti, M. L., Morandi, B., Dieguez-Uribeondo, J., van West, P., & Galuppi, R. (2021). Evaluation of Potential Transfer of the Pathogen Saprolegnia parasitica between Farmed Salmonids and Wild Fish. Pathogens, 10(8), 926. https://doi.org/10.3390/pathogens10080926









