Abstract
Leptospirosis is one of the most widespread bacterial diseases caused by pathogenic Leptospira. There are broad clinical manifestations due to varied pathogenicity of Leptospira spp., which can be classified into three clusters such as pathogenic, intermediate, and saprophytic. Intermediate Leptospira spp. can either be pathogenic or non-pathogenic and they have been reported to cause mild to severe forms of leptospirosis in several studies, contributing to the disease burden. Hence, this study aimed to estimate the global prevalence of intermediate Leptospira spp. in humans using meta-analysis with region-wise stratification. The articles were searched from three databases which include PubMed, Scopus, and ScienceDirect. Seven studies were included consisting of two regions based on United Nations geo-scheme regions, among 469 records identified. Statistical analysis was performed using RevMan software. The overall prevalence estimate of intermediate Leptospira spp. in humans was 86% and the pooled prevalences were 96% and 17% for the American and Asia regions, respectively. The data also revealed that Leptospira wolffii was the most predominantly found compared to the other intermediate species identified from the included studies, which were Leptospira inadai and Leptospira broomii. The estimated prevalence data from this study could be used to develop better control and intervention strategies in combating human leptospirosis.
1. Introduction
Leptospirosis is one of the most well-known, widespread zoonotic diseases, which accounts for high morbidity and mortality particularly in the regions with humid tropical or subtropical climates and in areas with impoverished populations. Leptospirosis has been estimated to affect around 1.03 million people and causes 58,900 deaths every year [1]. Even though the reported data is significant, there is no precise estimation of the global burden of human leptospirosis as it is often overlooked due to the wide range of clinical manifestations such as fever, diarrhea, headache, vomiting, muscle aches, malaise, jaundice, renal failure, pulmonary hemorrhage, etc. [2]. Leptospirosis also mimics several other diseases, for instance, dengue fever, malaria infection, influenza infection, Hanta virus, viral flu-like illnesses, and typhoid fever [3]. Moreover, leptospirosis is usually under-reported because of the poor health surveillance especially in the under-developed and developing countries. Therefore, leptospirosis was reported as one of the bacterial neglected tropical diseases due to its high disease burden and huge impacts on the public health following the re-emergence of this disease in several parts of the world [4].
This disease is caused by the pathogenic spirochetes of the genus Leptospira. Leptospira can be mainly classified according to methods used that are either based on a serological classification system or molecular classification system [5]. Traditionally, serology-based methods identified leptospires according to their antigenic properties found on the outer membrane of the bacteria, which was associated with the structural heterogeneity of lipopolysaccharides (LPS) [6]. This method divides these bacteria into two: Leptospira interrogans and Leptospira biflexa, which contain pathogenic and non-pathogenic strains, respectively. There are 26 serogroups and more than 300 serovars Leptospira currently identified using this classification system, which were usually detected by agglutination techniques such as Microscopic Agglutination Test (MAT) and cross agglutination absorption test (CAAT) [7,8]. On the other hand, phylogenetic or genomic classification system based on DNA relatedness using DNA-DNA hybridization and 16S-rRNA-based methods have further categorized 22 Leptospira species into three different clusters, which comprised of pathogenic, intermediate, and saprophytic. There are currently 10 pathogenic Leptospira spp. (Leptospira noguchii, Leptospira kirschneri, Leptospira interrogans, Leptospira santarosai, Leptospira mayottensis, Leptospira borgpetersenii, Leptospira alexanderi, Leptospira weilii, Leptospira alstonii, and Leptospira kmetyi) that could cause the disease. Meanwhile, there are five intermediate Leptospira spp. (Leptospira broomii, Leptospira inadai, Leptospira fainei, Leptospira wolffii, and Leptospira licerasiae) that have uncertain pathogenicity but mostly cause moderate symptoms, and seven saprophytic Leptospira spp. (Leptospira meyeri, Leptospira wolbachii, Leptospira terpstrae, Leptospira vanthielii, Leptospira biflexa, Leptospira yanagawae, and Leptospira idonii) that are commonly found in water and soil and unable to infect people [9]. Although serological classification using MAT technique remains the gold standard method, the overall prediction of the infecting species is not as reliable as genomic classification. This is because molecular methods allow the identification of the species exhibiting both pathogenic and non-pathogenic serovars, the intermediate Leptospira spp.
Intermediate Leptospira spp. were reported to cause mild to severe forms of leptospirosis in humans, however, the findings about its pathogenicity status are still unclear [10]. This indicates the need for more studies on the intermediate species since they were also isolated from the clinical samples and proved to have virulence features, which have the potential to (or might) affect the burden of leptospirosis [11]. In addition, there have been many individual studies and research that successfully identified the presence of intermediate Leptospira spp. in various areas, countries, or regions. However, to date, there is no reported meta-analysis of published data that summarize its prevalence in humans on a global scale. Therefore, it is essential to conduct meta-analysis to systematically summarize the relevant individual studies in a similar field and to obtain a more precise estimation on the overall effect measure [12].
This study aimed to estimate the overall prevalence of intermediate Leptospira spp. in humans by quantitatively synthesizing the frequency of its presence in humans from different regions via meta-analysis.
2. Materials and Methods
2.1. Literature Search Strategy
Meta-analysis for this study was conducted according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) 2009 guidelines [13]. Comprehensive search related to the human leptospirosis caused by the intermediate Leptospira spp. was performed using three databases, which include PubMed, ScienceDirect, and Scopus. The combinations of the keywords “prevalence”, “presence”, “epidemiology”, “leptospirosis”, “intermediate Leptospira”, “human”, “patient”, and species of the intermediate Leptospira were included for the search terms for the relevant studies (Table 1). Boolean connectors such as “OR” and “AND” were applied to connect the terms within and between the categories, respectively. In addition, truncation and wildcard operators such as ‘*’, ‘#’, or ‘$’ were also utilized to maximize the search for the related terms of the pertinent studies. The search strategy was slightly adjusted based on the requirements of different databases. There was no restriction posed for publication dates and languages during the initial search. The reference lists of the included studies were searched manually to seek for additional relevant papers that were not selected during the initial search. The last database search was carried out on 10 January 2021.

Table 1.
Search terms and keywords for the literature search.
2.2. Eligibility Criteria and Study Selection
Inclusion and exclusion criteria were pre-determined using PICOS (population, intervention, comparator, outcome, study design) approach [13] (Table 2). The subjects of the studies were any individuals suspected with leptospirosis infection including those that were co-infected with the other diseases. There was no restriction imposed on age, gender, or race of the subjects. The studies were excluded if there was no leptospirosis infection and if the studies did not report the origin of the samples or patients. Any irrelevant study was also removed. As for the types of intervention, the studies must identify the presence of intermediate Leptospira spp., which were detected using any recognized diagnostic or confirmation methods. The species of the intermediate Leptospira spp. and the methods used must be specified. The outcome measures were the frequency of the samples positive for intermediate Leptospira spp. and the total number of positive samples investigated. These data were used to determine the raw prevalence outcome, which were measured in percentage (%) by dividing the number of positive samples with the total number of positive samples tested in the study. All research articles of any countries and publication year were included. However, inaccessible full-text articles, letter to editor, duplicated publications, studies using other than English language, and secondary research such as review paper, systematic reviews, and meta-analyses were removed.

Table 2.
Eligibility criteria for study selection.
All the search results from the three databases were compiled and sorted out in one Microsoft Excel spreadsheet document. The articles with redundant titles and authors were removed. The title and abstract of the remaining studies were screened by three reviewers, independently according to the inclusion and exclusion criteria. The results of the screening by the three reviewers were cross-checked, and disagreements were resolved through consensus. The qualified studies were then subjected to full-text screening to further ascertain their relevancy and studies that did not meet the criteria were excluded. The flow of study selection was done by referring to the PRISMA flow diagram [13].
2.3. Data Extraction
Data from the selected studies were extracted independently by two reviewers, which included the first author and year of publication, location where the samples were collected, number of samples positive for intermediate Leptospira spp., number of samples positive for pathogenic Leptospira spp., total sample size confirmed for leptospirosis, method used for the confirmation of leptospirosis, and the type of intermediate Leptospira spp. Data collected from both of the reviewers were compared and re-checked for better accuracy. Disagreements and inconsistencies in data extraction were resolved through consensus and discussion with the third reviewer (Z.S.).
2.4. Quality Assessment
Quality assessment of the included studies was conducted by two reviewers independently in accordance with the modified Critical Appraisal Checklist recommended by the Joanna Briggs Institute (JBI) since the use of this tool had been formally evaluated and increased in assessing prevalence studies [14,15]. Any disagreements were discussed and resolved through consensus with the third reviewer. The tool was comprised of nine questions, by which the reviewers referred to when assessing the studies. The score is either 0 or 1 for the answers of “No/Unclear” and “Yes”, respectively. Therefore, the score can range from 0 to 9.
2.5. Statistical Analysis
The extracted data were quantitatively analyzed using RevMan version 5.4 software to identify the pooled prevalence estimates of the intermediate Leptospira spp. [16]. The overall results including heterogeneity represented as I2 statistic (%) calculated for the included studies were then recorded [17]. Fixed-effect model was used in this study and the overall effect would be considered as statistically significant if the p-value was less than 0.05. Sub-group analysis based on United Nations (UN) geo-scheme regions devised by the UN Statistics Division [18] were also carried out to further reduce the heterogeneity or variations between the included studies. Apart from that, the pooled data would highlight the prevalence differences between the regions.
3. Results
3.1. Literature Search
A total of 469 records were identified from three databases, of which 112 were from ScienceDirect, 286 were from Scopus, 68 were from PubMed, and 3 from the other sources. There were 258 articles removed because they were found to be duplicated and have either the same authors, DOI, or PMID serial number after compiling the results into Microsoft Excel spreadsheet document. Further, the remaining 211 articles were reviewed by title and abstract according to the exclusion and inclusion criteria (Table 2). After excluding 189 records, the remaining 22 articles were subjected for full-text screening according to the eligibility criteria. Then, 15 records were excluded and the remaining seven studies that fit the eligibility criteria were included for meta-analysis. The flowchart of study selection and the reasons for excluding the studies were illustrated in Figure 1.
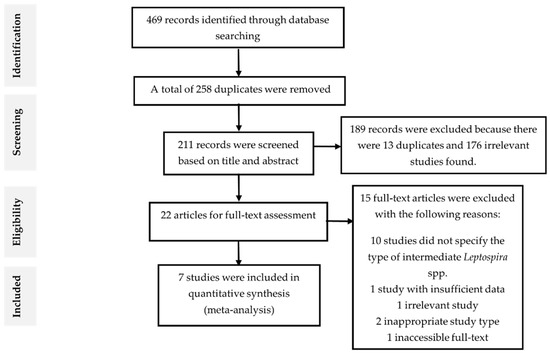
Figure 1.
Flow diagram of literature search and selection.
3.2. Characteristics of the Included Articles
There were seven studies selected for meta-analysis, which were published between 2009 to 2020 and the characteristics were detailed in Table 3. The included articles were case-control studies and most of the studies that discovered intermediate Leptospira spp. in human samples were from the Asian region and then followed by the American region. There were no eligible studies identified from the European, Oceania, and African regions. The total sample size was 403 while the total samples positive for intermediate and pathogenic Leptospira spp. were 225 and 174, respectively. Sera and blood samples collected from humans were utilized for the characterization of the leptospires using several methods such as microscopic agglutination test (MAT), IgM ELISA (enzyme-linked immunosorbent assay), PCR (polymerase chain reaction) assay, partial RNA polymerase β-subunit (rpo-β) gene sequencing, multilocus sequence typing (MLST), 16S rRNA gene sequencing, as well as partial 16S rDNA (rrs) gene sequencing. There were only three out of five intermediate Leptospira spp. found from the studies. Five studies found L. wolffii [19,20,21,22,23] and one study recorded the presence of both L. wolffii and L. inadai [10] in their study subjects. Another study reported L. wolffii and L. broomii [24]. This meta-analysis study showed that L. wolffii was the most predominant species (n = 223/225) compared to the other two species, L. inadai (n = 1/225) and L. broomii (n = 1/225).

Table 3.
Characteristics of the included studies.
3.3. Quality Assessment and Risk of Bias
Quality assessment of the selected studies using JBI appraisal checklist for the prevalence study is as shown in Table 4. The mean score of the assessment was 5 out of 9, which ranged from 3 to 9. All of the included studies are deemed to have high risk of bias since none of them were randomized.

Table 4.
Quality assessment of the included studies.
3.4. Pooled Prevalence of Intermediate Leptospira spp. in Human Samples
The analysis using RevMan software provided the pooled estimates of the input data illustrated as forest plot, as well as upper and lower bounds of 95% confidence interval (CI), p-value, and heterogeneity value (I2 statistic). All the data obtained from the analysis were summarized and the prevalence estimates were bolded in Table 5.

Table 5.
Meta-analysis of the prevalence of intermediate Leptospira spp. in humans.
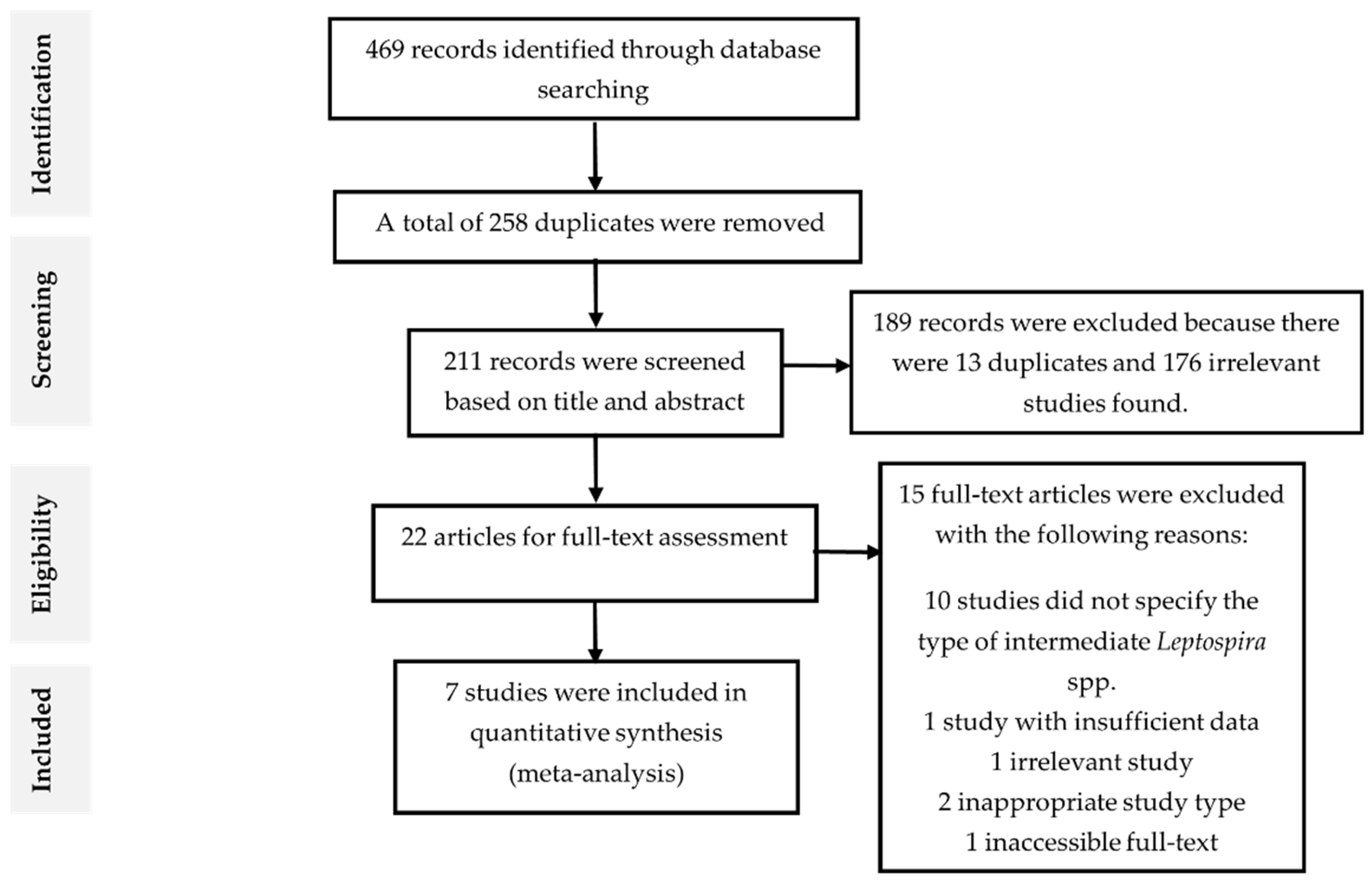
Forest plot in Figure 2 exhibited the overall prevalence estimates on the global prevalence of intermediate Leptospira spp. The diagram also showed the first author, year, and the state where the samples were collected. In addition, the proportion outcomes as well as the standard error of the mean (SEM) for the proportion of every study were calculated separately and included manually into RevMan [25]. The statistical analysis revealed that the overall pooled prevalence of intermediate Leptospira spp. in humans was 86% (95% CI: 0.85–0.88; I2 = 99%; p < 0.00001).
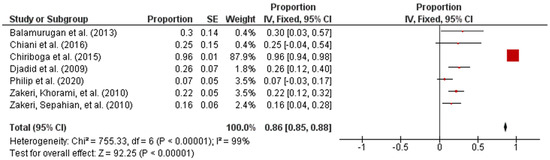
Figure 2.
Forest plot of overall prevalence of intermediate Leptospira spp. in human samples.
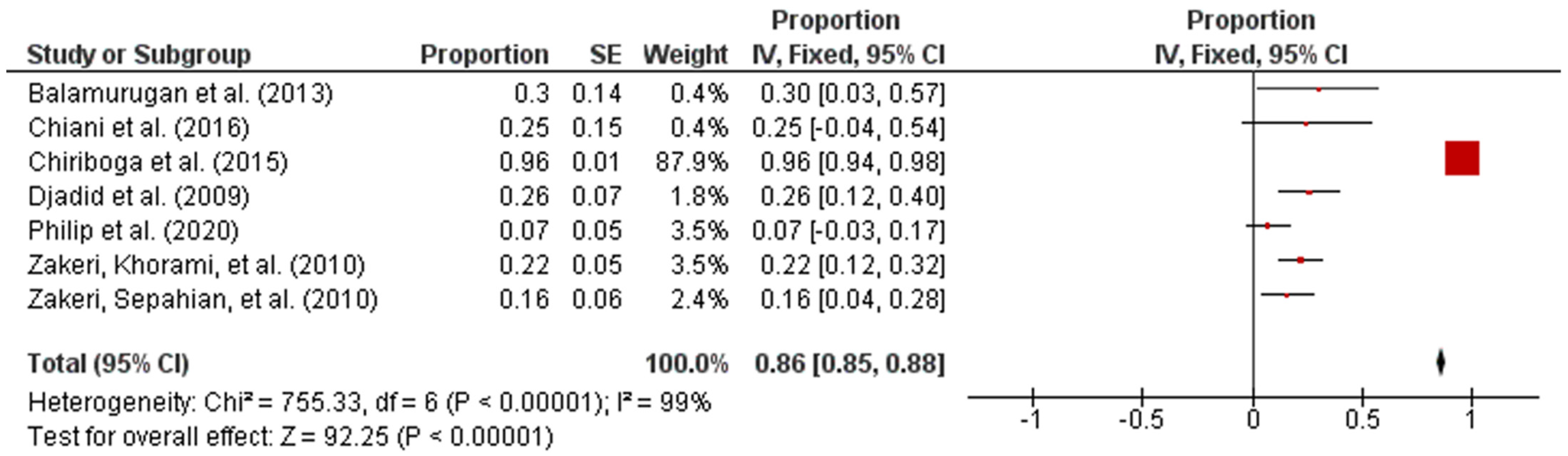
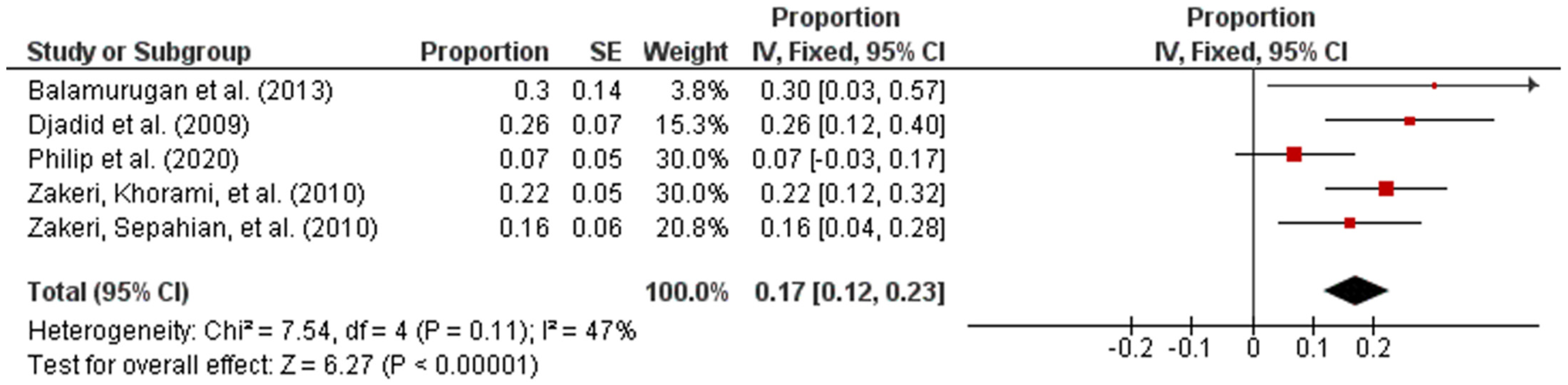
Subsequently, the studies were categorized by region in accordance with the UN geo-schemes [18]. The pooled prevalence of intermediate Leptospira spp. in human samples from the American and Asian regions were 96% (95% CI: 0.94–0.98; I2 = 96%; p < 0.00001) and 17% (95% CI: 0.12–0.23; I2 = 47%; p < 0.00001), respectively (Figure 3 and Figure 4).
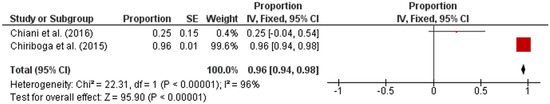
Figure 3.
Forest plot of pooled prevalence of intermediate Leptospira spp. in human samples from the American region.
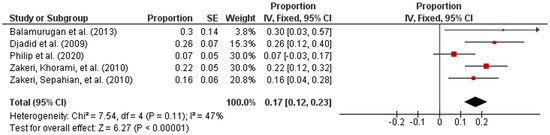
Figure 4.
Forest plot of pooled prevalence of intermediate Leptospira spp. in human samples from the Asian region.
4. Discussion
Intermediate Leptospira spp. have both pathogenic and non-pathogenic serovars, and to date, its role in human pathogenicity remains unclear and is yet to be explored. In this study, the findings from meta-analysis demonstrated that the presence of intermediate Leptospira spp. should be considered when making decisions for disease control and prevention, particularly in the regions where leptospirosis is endemic.
To our knowledge, this is the first meta-analysis that summarizes the prevalence of intermediate Leptospira spp. in humans worldwide. Meta-analysis of the included studies illuminates that the overall prevalence of the intermediate Leptospira spp. is high and significant at 86% (95% CI: 0.85–0.88; I2 = 99%; p < 0.00001). It indicates that these species are indeed contributing to the burden of the disease. Out of two UN regions identified from the included studies, the region of the Americas had the highest prevalence of intermediate Leptospira spp., which was at 96% (95% CI: 0.94–0.98; I2 = 96%; p < 0.00001) as compared to the Asian region, which was at 17% (95% CI: 0.12–0.23; I2 = 47%; p < 0.00001) (Table 5). The sub-group analysis suggests that the prevalence of intermediate Leptospira spp. in both regions involved was significant. However, the studies found from the European, Oceania, and African regions were excluded because they did not meet the inclusion criteria of this meta-analysis study such that the studies did not specify the intermediate Leptospira spp., non-English articles, and some of the studies were irrelevant for this analysis. The data from the included studies also demonstrated that there were only three out of five intermediate Leptospira spp. identified, which were L. wolffii, L. inadai, and L. broomii. The other two intermediate species, L. licerasiae and L. fainei were not found in the selected studies because both species were reported in studies related to non-human samples, which were excluded.
The higher prevalence of intermediate Leptospira spp. in the American region than the Asian region indicates that most of the countries in the region of the Americas have more access to health care facilities and better health surveillance system with accurate diagnostic and confirmation methods, which were able to detect and identify the species that infected the patients. This was supported by a report by Schneider et al. (2011) [26], in which they pointed out that the surveillance and control strategies for leptospirosis were developed in many countries in the region of the Americas. Moreover, the improved method in detecting leptospiral DNA has also enabled the identification of intermediate clusters from patients with febrile symptoms in this region [10]. For instance, the utilization of the amplified leptospiral 16S rrs gene and sequencing instead of the common PCR protocols that amplify genes present only in the pathogenic species. Furthermore, there are approximately 10 million people that are affected by natural disasters such as floods (35%) and storms (41%) in the American region every year, and there have been several studies that reported the outbreaks of leptospirosis associated with these events from different countries in Central and South America [27,28]. The results from local studies performed in Central America showed that leptospirosis cases were prevalent among the communities residing in the rural areas, which depend mostly on the animals such as bovine and porcine for their income and for daily protein intake [29]. All these factors might have increased the chances of the communities being exposed to the intermediate Leptospira spp. Apart from that, the data also revealed that L. wolffii, L. inadai, and L. broomii were found in the American region (Table 3), which signified that varieties of the intermediate species contributed to the increased burden of leptospirosis in this region.
On the other hand, the prevalence of the intermediate Leptospira spp. in the Asian countries, specifically in the South-Eastern Asian and Southern Asian countries, may not be high when compared to the prevalence in the American region, nonetheless it was also statistically significant. The significant presence of the intermediate Leptospira spp. in this region, particularly in the Southern Asian countries such as in India and Iran, may be due to the poor access to safe water supplies, poor hygiene, as well as inadequate sanitation. Even though the latest estimates in 2019 showed the improvement in the access to the water supply in India, the water safety and security planning for several districts in India was still lacking and less than 50% of the population has access to safe water supply [30]. Other than that, Zakeri et al. [23] mentioned that 18.5% of the examined cases in northern Iran had collected drinking water from wells and 52% of them had been infected with leptospirosis. The same study also revealed that L. wolffii was one of the species isolated from the samples tested. This suggests that unsafe water sources played a role in the transmission of the disease, which may be attributed to the indirect exposure to the intermediate Leptospira spp. found in the contaminated water, influencing the incidence of leptospirosis in this region.
In addition, the prevalence of intermediate Leptospira spp. in the Asian region was considerably low even though leptospirosis was endemic and causing sporadic outbreaks in most of the South-East Asian developing countries, especially those with humid subtropical and tropical climates such as Malaysia [1,31,32]. This may possibly be due to the diagnostic capabilities of the disease, of which the tools used were less sensitive in detecting the species infecting the patients. Even though culture and microscopic agglutination tests (MAT) are the gold standard methods for laboratory diagnostic testing and the most widely used in this region, they require experts in handling the live pathogens; hence, these methods were offered by only a few hospitals and laboratories in several countries in the Asian region [31]. Additionally, it was said to have little value in predicting the infecting serogroup of the patients since the screening of the serum samples is mostly based on 25 reference serovars, representing only a fraction of over 200 serovars found globally [31,33,34,35,36]. There were several alternative methods to MAT in detecting the acute infection such as enzyme-linked immunosorbent assay (ELISA), IgM dipstick, lateral flow assay, and latex agglutination test, nonetheless, these assays have low sensitivity especially during the acute phase [37,38,39,40]. The accuracies of these techniques are also poor in some areas where leptospirosis is endemic [41,42]. According to Gamage et al. [33], the available laboratory facilities were still poor and inadequate specifically in certain South-East Asian countries, and as reported by WHO in 2009, India, Indonesia, Thailand, and Sri Lanka were the only WHO Member States that have fully or partially implemented laboratory facilities for the diagnosis of leptospirosis [43]. Besides, as most of the South-East Asian countries were the major importers for the agricultural products such as Malaysia, Philippines, and Indonesia [44], the significant prevalence in this region may be contributed to by the occupational factors such as the contact with intermediate Leptospira spp. in the contaminated water and soil through farming. This was corroborated with the findings of intermediate Leptospira spp. being isolated from the environmental and water samples in several countries in this region [19,45,46].
The data collected from the seven included studies showed that L. wolffii was the most predominant species (n = 223/225) as compared to the other intermediate species (L. inadai; 1/225; L. broomii: 1/225). L. wolffii was first isolated from an individual with suspected leptospirosis in Thailand [47], and was also found in all of the included studies, which were in India [19], Argentina [24], Ecuador [10], Iran [20,22,23], and Malaysia [21], which may suggest that L. wolffii was the dominant intermediate Leptospira circulating in most of the areas. The majority of samples were collected from the patients with acute, febrile illness with other common symptoms for the suspected leptospirosis such as fever, myalgia, chills, rigors, gastrointestinal problems, as well as a more serious symptom like jaundice [10,20,21,22,23]. In one study, several patients with fever, jaundice, hematuria, icteric discoloration with hepatomegaly, as well as weakness on the left side were confirmed to be infected with L. wolffii [19]. In other similar studies conducted in Argentina and Malaysia, L. wolffii was isolated from patients with fatal cases, particularly respiratory syndrome [21,24]. In addition, as L. wolffii was categorized as pathogenic Leptospira using nested PCR-RFLP due to the absence of ApoI restriction sites, further sequencing analysis of the samples was required, by which they showed that 26% of the tested DNA belonged to L. wolffii [20]. All the tested samples from this study were collected from symptomatic patients manifesting fever with headache, body aches related to jaundice for several days, and headache with myalgia, which all required hospitalization. Furthermore, L. wolffii was also isolated from environments and animals such as cattle, rats, pigs, sheep, and dog in several previous reports [10,22,48]. This indicated that L. wolffii was prevalent in various environmental and animal reservoirs and that it played a significant role in the transmission cycle of leptospirosis. Therefore, all this evidence suggested that they had the highest pathogenicity compared to other intermediates. L. broomii, on the other hand, was identified from one of the human samples in Argentina [24], while L. inadai was isolated from one of the human samples in Ecuador [10]. Chiani et al. [24] reported that L. broomii was identified from the patient with no signs of severe leptospirosis. This indicates that L. broomii likely caused milder disease, which thus explained the smaller number of L. broomii being identified from human samples than L. wolffii. Meanwhile, L. inadai was mainly isolated from animal samples such as cattle, rats, dogs, and pigs [7], suggesting its predominance in the animal reservoirs rather than in humans. It is also noteworthy that one of the included studies found that 96% of leptospiral DNA isolated from human serum belonged to the intermediate species including L. wolffii and L. inadai, rather than pathogenic cluster strains [10].
Despite the increasing studies concerning leptospirosis disease in the African region, there were no eligible studies that reported the intermediate Leptospira spp. in this region, which may be due to the inadequate sampling and poor access to the diagnostic facilities, leading to the under-reporting of leptospirosis. Hence, there is insufficient information regarding the species that infected the patients from the African region [49]. Besides, there were also no eligible studies that reported the presence of intermediate Leptospira spp. in the Oceania region, albeit the previous systematic review had revealed that the incidence of leptospirosis was notably high (150.68 cases per 100,000 per year), especially in the temperate parts of the region such as Australia and New Zealand [50,51]. However, there were too little information and investigations regarding the specific species that infected the patients from this region. Other than that, there were also no eligible studies that reported the presence of intermediate Leptospira spp. in the European region. This was because leptospirosis was not a common disease with 0.2 confirmed cases per 100,000 population, which is considered a low rate in European countries in comparison with other regions [52].
There were several limitations of this study, which include the possibility of missing some of the relevant studies during the literature search procedure. Also, the information from the other countries and regions was lacking. This prevented us from making a thorough analysis of the prevalence data needed for species-wise stratification, reducing the accuracy of the obtained data. This also limited our study from analyzing the pathogenicity status of the intermediate Leptospira spp. Furthermore, the high risk of bias of all the studies included and high I2 statistic for the overall prevalence (99%) might influence the accuracy of prevalence estimates in this analysis. Differences in the design, complexity, geographical regions, environments, and study settings of the included studies contributed to the high heterogeneity in this analysis. However, the chances of getting correct estimates are higher using a random-effect model with increasing heterogeneity, than using fixed-effect model [53]. Thus, optimizing the effect model based on the heterogeneity instead of outright rejecting the result due to the prominent heterogeneity is preferred [54]. Other than that, publication bias assessment using a funnel plot was not carried out since there were only seven included studies in this study. Higgins et al. [17] stated that the power of the tests would be too low to distinguish the chance from the real asymmetry if the studies are fewer than 10. Additionally, Debray et al. [55] also reported that the power for the tests of the funnel plot asymmetry usually remained less than 50% even when there were ≥ 50 studies available for meta-analysis. Therefore, it is best to use a funnel plot only when there is a minimum of 10 studies. Lastly, based upon the advanced literature survey, we found only a few articles that addressed the presence of intermediate Leptospira spp. and identified the exact type of species in human samples.
5. Conclusions
In conclusion, this is the first meta-analysis on the prevalence of intermediate Leptospira spp. from human samples worldwide. The overall prevalence estimate of intermediate Leptospira spp. was high and statistically significant, and the pooled prevalence estimates based on the UN regions showed the highest prevalence of intermediate Leptospira spp. in the American region followed by the Asian region. The data from the included studies also demonstrated that L. wolffii was the most predominant species found in the human samples as compared to L. inadai and L. broomii. All the findings suggest that intermediate Leptospira spp. played an important role in the transmission of human leptospirosis. This calls for more investigations using molecular analysis, as it would give accurate species identification, which can be used to break the chain of leptospirosis and reduce the disease burden. Also, further studies on the effect of the species on the clinical outcome of the patients are required to gain better understanding on the pathogenicity status and capacity of intermediate Leptospira spp.
Author Contributions
Conceptualization, K.T., N.J. and A.N.A.R.; methodology, A.N.A.R., K.T. and N.J.; validation, K.T. and N.J.; formal analysis, K.T., N.J., A.N.A.R., N.H.H.H. and Z.S.; investigation, A.N.A.R., N.H.H.H. and Z.S.; resources, A.N.A.R., N.H.H.H., K.T. and N.J.; data curation, A.N.A.R., N.H.H.H., K.T., N.J. and Z.S.; writing—original draft preparation, A.N.A.R.; writing—review and editing, K.T. and N.J.; supervision, K.T. and N.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors would like to thank Cindy Stern, from Faculty of Health and Medical Sciences, University of Adelaide, Australia for her advice on using JBI appraisal tool for quality assessment. We would also like to thank the reviewers for very helpful and constructive feedbacks and suggestions for improving the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/leptospirosis/symptoms/index.html (accessed on 20 March 2021).
- Wang, S.; Stobart Gallagher, M.A.; Dunn, N. Leptospirosis; StatPearls Publishing: Treasure Island, FL, USA, 2020; Available online: http://europepmc.org/books/NBK441858 (accessed on 20 March 2021).
- Hotez, P.J.; Aksoy, S.; Brindley, P.J.; Kamhawi, S. What constitutes a neglected tropical disease? PLoS Negl. Trop. Dis. 2020, 14, e0008001. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef]
- Cerqueira, G.M.; Picardeau, M. A century of Leptospira strain typing. Infect. Genet. Evol. 2009, 9, 760–768. [Google Scholar] [CrossRef]
- Hartskeel, R.A.; Smythe, L.D. The role of leptospirosis reference laboratories. Curr. Top. Microbiol. Immunol. 2015, 387, 273–288. [Google Scholar] [CrossRef]
- Nagraik, R.; Kaushal, A.; Gupta, S.; Sharma, A.; Kumar, D. Leptospirosis: A systematic review. J. Microbiol. Biotech. Food Sci. 2020, 9, 1099–1109. [Google Scholar] [CrossRef]
- Chiriboga, J.; Barragan, V.; Arroyo, G.; Sosa, A.; Birdsell, D.N.; España, K.; Mora, A.; Espín, E.; Mejía, M.E.; Morales, M.; et al. High prevalence of intermediate Leptospira spp. DNA in febrile humans from urban and rural Ecuador. Emerg. Infect. Dis. 2015, 21, 2141. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, M.; Koizumi, N.; Hayakawa, K.; Kanagawa, S.; Ohmagari, N.; Kato, Y. Imported Leptospira licerasiae infection in traveler returning to Japan from Brazil. Emerg. Infect. Dis. 2017, 23. [Google Scholar] [CrossRef]
- Haidich, A.B. Meta-analysis in medical research. Hippokratia 2010, 14, 29–37. [Google Scholar] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef]
- Munn, Z.; MClinSc, S.M.; Lisy, K.; Riitano, D.; Tufanaru, C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int. J. Evid. Based Health 2015, 13, 147–153. [Google Scholar] [CrossRef]
- Borges Migliavaca, C.; Stein, C.; Colpani, V.; Barker, T.H.; Munn, Z.; Falavigna, M. How are systematic reviews of prevalence conducted? A methodological study. BMC Med. Res. Methodol. 2020, 20, 96. [Google Scholar] [CrossRef]
- Review Manager; Version 5.4; Nordic Cochrane Centre, The Cochrane Collaboration: Copenhagen, Denmark, 2020.
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. Cochrane Handbook for Systeatic Reviews of Interventions. Version 6.0 (Updated July 2019). Available online: http://www.training.cochrane.org/handbook (accessed on 15 June 2021).
- United Nations. Geographic Regions. Available online: http://unstats.un.org/unsd/methods/m49/m49regin.htm (accessed on 15 April 2021).
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.A.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. SpringerPlus 2013, 2, 1–9. [Google Scholar] [CrossRef]
- Djadid, N.D.; Ganji, Z.F.; Gouya, M.M.; Rezvani, M.; Zakeri, S. A simple and rapid nested polymerase chain reaction-restriction fragment length polymorphism technique for differentiation of pathogenic and nonpathogenic Leptospira spp. Diagn. Microbiol. Infect. Dis. 2009, 63, 251–256. [Google Scholar] [CrossRef]
- Philip, N.; Affendy, N.B.; Ramli, S.N.A.; Arif, M.; Raja, P.; Nagandran, E.; Renganathan, P.; Taib, N.M.; Masri, S.N.; Yuhana, M.Y.; et al. Leptospira interrogans and Leptospira kirschneri are the dominant Leptospira species causing human leptospirosis in central Malaysia. PLoS Negl. Trop. Dis. 2020, 14, e0008197. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, S.; Khorami, N.; Ganji, Z.F.; Sepahian, N.; Malmasi, A.-A.; Gouya, M.M.; Djadid, N.D. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect. Genet. Evol. 2010, 10, 273–277. [Google Scholar] [CrossRef]
- Zakeri, S.; Sepahian, N.; Afsharpad, M.; Esfandiari, B.; Ziapour, P.; Djadid, N.D. Molecular epidemiology of Leptospirosis in northern Iran by nested polymerase chain reaction/restriction fragment length polymorphism and sequencing methods. Am. J. Trop. Med. Hyg. 2010, 82, 899–903. [Google Scholar] [CrossRef]
- Chiani, Y.; Jacob, P.; Varni, V.; Landolt, N.; Schmeling, M.F.; Pujato, N.; Caimi, K.; Vanasco, B. Isolation and clinical sample typing of human leptospirosis cases in Argentina. Infect. Genet. Evol. 2016, 37, 245–251. [Google Scholar] [CrossRef]
- Research Gate. Available online: https://www.researchgate.net/post/How-to-calculate-pooled-prevalence-using-RevMan (accessed on 1 March 2021).
- Schneider, M.C.; Aguilera, X.P.; Smith, R.M.; Moynihan, M.J.; Silva, J.B.D., Jr.; Aldighieri, S.; Almiron, M. Importance of the animal/human interface in potential public health emergencies of international concern in the Americas. Pan Am. J. Public Health 2011, 29, 371–379. [Google Scholar] [CrossRef]
- Schneider, M.C.; Jancloes, M.; Buss, D.F.; Aldighieri, S.; Bertherat, E.; Najera, P.; Galan, D.I.; Durski, K.; Espinal, M.A. Leptospirosis: A silent epidemic disease. Int. J. Environ. Res. Public Health 2013, 10, 7229–7234. [Google Scholar] [CrossRef]
- Schneider, M.C.; Tirado, M.C.; Rereddy, S.; Dugas, R.; Borda, M.I.; Peralta, E.A.; Aldighieri, S.; Cosivi, O. Natural disasters and communicable diseases in the Americas: Contribution of veterinary public health. Vet. Ital. 2012, 48, 193–218. [Google Scholar]
- Pan American Health Organization. Leptospirosis in the Americas Region from an Outbreak Perspective. Available online: https://www.paho.org/en/node/49202 (accessed on 2 April 2021).
- United Nations International Children’s Emergency Fund. Available online: https://www.unicef.org/india/what-we-do/water-sanitation-hygiene (accessed on 16 April 2021).
- Garba, B.; Bahaman, A.R.; Bejo, S.K.; Zakaria, Z.; Mutalib, A.R.; Bande, F. Major epidemiological factors associated with leptospirosis in Malaysia. Acta Trop. 2018, 178, 242–247. [Google Scholar] [CrossRef]
- Victoriano, A.F.B.; Smythe, L.D.; Gloriani-Barzaga, N.; Cavinta, L.L.; Kasai, T.; Limpakarnjanarat, K.; Ong, B.L.; Gongal, G.; Hall, J.; Coulombe, C.A.; et al. Leptospirosis in the Asia Pacific region. BMC Infect. Dis. 2009, 9, 147. [Google Scholar] [CrossRef]
- Gamage, C.D.; Tamashiro, H.; Ohnishi, M.; Koizumi, N. Epidemiology, surveillance and laboratory diagnosis of leptospirosis in the WHO South-East Asia region. In Zoonosis; InTech: Rijeka, Croatia, 2012; pp. 213–226. [Google Scholar] [CrossRef]
- Levett, P.N. Usefulness of serologic analysis as a predictor of the infecting serovar in patients with severe leptospirosis. Clin. Infect. Dis. 2003, 36, 447–452. [Google Scholar] [CrossRef]
- Katz, A.R.; Effler, P.V.; Ansdell, V.E. Comparison of serology and isolates for the identification of infecting leptospiral serogroups in Hawaii, 1979–1998. Trop. Med. Int. Health 2003, 8, 639–642. [Google Scholar] [CrossRef]
- Smythe, L.D.; Wuthiekanun, V.; Chierakul, W.; Suputtamongkol, Y.; Tiengrim, S.; Dohnt, M.F.; Symonds, M.L.; Slack, A.T.; Apiwattanaporn, A.; Chueasuwanchai, S.; et al. The microscopic agglutination test (MAT) is an unreliable predictor of infecting Leptospira serovar in Thailand. Am. J. Trop. Med. Hyg. 2009, 81, 695–697. [Google Scholar] [CrossRef]
- Smits, H.L.; Eapen, C.K.; Sugathan, S.; Kuriakose, M.; Gasem, M.H.; Yersin, C.; Sasaki, D.; Pujianto, B.; Vestering, M.; Abdoel, T.H.; et al. Lateral-flow assay for rapid serodiagnosis of human leptospirosis. Clin. Diagn. Lab. Immunol. 2001, 8, 166–169. [Google Scholar] [CrossRef]
- Effler, P.V.; Bogard, A.K.; Domen, H.Y.; Katz, A.R.; Higa, H.Y.; Sasaki, D.M. Evaluation of eight rapid screening tests for acute leptospirosis in Hawaii. J. Clin. Microbiol. 2002, 40, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Hull-Jackson, C.; Glass, M.B.; Ari, M.D.; Bragg, S.L.; Branch, S.L.; Whittington, C.U.; Edwards, C.N.; Levett, P.N. Evaluation of a commercial latex agglutination assay for serological diagnosis of leptospirosis. J. Clin. Microbiol. 2006, 44, 1853–1855. [Google Scholar] [CrossRef]
- McBride, A.J.; Santos, B.L.; Queiroz, A.; Santos, A.C.; Hartskeerl, R.A.; Reis, M.G.; Ko, A.I. Evaluation of four whole-cell Leptospira-based serological tests for diagnosis of urban leptospirosis. Clin. Vaccine Immunol. 2007, 14, 1245–1248. [Google Scholar] [CrossRef]
- Blacksell, S.D.; Smythe, L.; Phetsouvanh, R.; Dohnt, M.; Hartskeerl, R.; Symonds, M.; Slack, A.; Vongsouvath, M.; Davong, V.; Lattana, O.; et al. Limited diagnostic capacities of two commercial assays for the detection of Leptospira immunoglobulin M antibodies in Laos. Clin. Vaccine Immunol. 2006, 13, 1166–1169. [Google Scholar] [CrossRef]
- Myint, K.S.; Gibbons, R.V.; Murray, C.K.; Rungsimanphaiboon, K.; Supornpun, W.; Sithiprasasna, R.; Gray, M.R.; Pimgate, C.; Mammen, M.P., Jr.; Hospenthal, D.R. Leptospirosis in Kamphaeng Phet, Thailand. Am. J. Trop. Med. Hyg. 2007, 76, 135–138. [Google Scholar] [CrossRef]
- World Health Organization. Informal Expert Consultation on Surveillance, Diagnosis and Risk Reduction of Leptospirosis; World Health Organization Regional Office for South-East Asia: Chennai, India, 2009; Available online: http://www.searo.who.int/LinkFiles/Communicable_Diseases_Surveillance_and_response_SEA-CD-217.pdf (accessed on 2 April 2021).
- United States Department of Agriculture. Available online: https://www.fas.usda.gov/data/trade-opportunities-southeast-asia-indonesia-malaysia-and-philippines#:~:text=Indonesia%2C%20Malaysia%2C%20and%20the%20Philippines%2C%20are%20among%20the%20region’s,to%20Southeast%20Asia%20in%202017 (accessed on 29 March 2021).
- Narkkul, U.; Thaipadungpanit, J.; Srilohasin, P.; Srilohasin, P.; Singkhaimuk, P.; Thongdee, M.; Chaiwattanarungruengpaisan, S.; Krairojananan, P.; Pan-Ngum, W. Optimization of culture protocols to isolate Leptospira spp. from environmental water, field investigation, and identification of factors associated with the presence of Leptospira spp. from the environment. Trop. Med. Infect. Dis. 2020, 5, 94. [Google Scholar] [CrossRef]
- Pui, C.F.; Bilung, L.M.; Apun, K.; Su’ut, L. Diversity of Leptospira spp. in Rats and Environment from Urban Areas of Sarawak, Malaysia. J. Trop. Med. 2017. [Google Scholar] [CrossRef]
- Slack, A.T.; Kalambaheti, T.; Symonds, M.L.; Dohnt, M.F.; Galloway, R.L.; Steigerwalt, A.G.; Chaicumpa, W.; Bunyaraksyotin, G.; Craig, S.; Harrower, B.J.; et al. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int. J. Syst. Evol. Microbiol. 2008, 58, 2305–2308. [Google Scholar] [CrossRef]
- Mohd Ali, M.R.; Mohamad Safiee, A.W.; Yusof, N.Y.; Fauzi, M.H.; Yean, C.Y.; Ismail, N. Isolation of Leptospira kmetyi from residential areas of patients with leptospirosis in Kelantan, Malaysia. J. Infect. Public Health 2018, 11, 578–580. [Google Scholar] [CrossRef]
- Allan, K.J.; Biggs, H.M.; Halliday, J.E.; Kazwala, R.R.; Maro, V.P.; Cleaveland, S.; Crump, J.A. Epidemiology of leptospirosis in Africa: A systematic review of a neglected zoonosis and a paradigm for ‘One Health’ in Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003899. [Google Scholar] [CrossRef]
- Guernier, V.; Allan, K.J.; Goarant, C. Advances and challenges in barcoding pathogenic and environmental Leptospira. J. Parasitol. 2018, 145, 595–607. [Google Scholar] [CrossRef]
- Berlioz-Arthaud, A.; Kiedrzynski, T.; Singh, N.; Yvon, J.F.; Roualen, G.; Coudert, C.; Uluiviti, V. Multicentre survey of incidence and public health impact of leptospirosis in the Western Pacific. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 714–721. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Report Leptospirosis-Annual Epidemiological Report 2016 (2014 Data). Available online: https://www.ecdc.europa.eu/en/publications-data/leptospirosis-annual-epidemiological-report-2016-2014-data (accessed on 1 March 2021).
- Melsen, W.G.; Bootsma, M.C.J.; Rovers, M.M.; Bonten, M.J.M. The effects of clinical and statistical heterogeneity on the predictive values of results from meta-analyses. Clin. Microbiol. Inf. 2014, 20, 123–129. [Google Scholar] [CrossRef]
- Ryan, R. Cochrane Consumers and Communication Group: Meta-Analysis. Cochrane Consumers and Communication Review Group. Available online: https://cccrg.cochrane.org/sites/cccrg.cochrane.org/files/public/uploads/meta-analysis_revised_december_1st_1_2016.pdf (accessed on 15 June 2021).
- Debray, T.P.; Moons, K.G.; Riley, R.D. Detecting small-study effects and funnel plot asymmetry in meta-analysis of survival data: A comparison of new and existing tests. Res. Synth. Methods 2018, 9, 41–50. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).