Abstract
Regional differences in tick-borne disease epidemiology may be related to biological variations between vector populations. Amblyomma aureolatum (Ixodida: Ixodidae), a neotropical tick, is known from several regions in Brazil. However, only in the metropolitan area of São Paulo (SP) state are there studies that establish its role as a vector of a pathogenic rickettsia (Rickettsia rickettsii). The aim of the study was to analyze the genetic diversity, population dynamics, and rickettsia infection in A. aureolatum populations from different spotted fever scenarios in Brazil. Samples were subjected to DNA extraction, amplification and sequencing of 12S rDNA, cytochrome oxidase subunit II and D-loop mitochondrial markers for tick population analyses, and gltA, htrA, ompA, and ompB genes for rickettsia researches. Of the 7–17 tick haplotypes identified, 5–13 were exclusive to each population and 2–12 for each epidemiological scenario, as well as three haplotypes shared by all populations. Amblyomma aureolatum populations are expanding, and do not appear to be genetically structured vis-a-vis the different epidemiological scenarios studied. Rickettsia bellii (in SP) and Rickettsia felis (in Santa Catarina) were identified as infecting A. aureolatum. No relationship between tick haplotypes and rickettsia types were observed.
1. Introduction
Ticks are very important to world public health as vectors of a variety of bioagents. Brazilian spotted fever (BSF), the main tick-borne zoonotic disease in Brazil, is caused by Rickettsia rickettsii and transmitted by Amblyomma sculptum Berlese, 1888 (the star tick) and Amblyomma aureolatum (the yellow dog tick) [1]. Knowledge of BSF vectors and epidemiology, including the vector–pathogen–host interaction, is key for effective disease control strategies in the context of One Health. However, such knowledge is still incipient, especially for A. aureolatum.
Records of A. aureolatum are restricted to the neotropical region, being found in the southeastern region of South America, from Uruguay to Surinam, including records for northeastern Argentina, eastern Paraguay, French Guiana, and Brazil (largely in the states of the south and southeast regions) [2]. Atlantic Forest, with conditions of humidity and high temperature throughout the year, appears to be the natural habitat of this ixodid [3]. However, there are records of A. aureolatum in the Pampa biome, in southern Brazil, where winters reach freezing, but summers are hot and dry e.g., [4,5,6].
Immature stages of A. aureolatum feed on passerine birds and a few rodent species [2,7]. In forest areas, the main hosts for both sexes of this tick are members of the order carnivora, among them the crab-eating fox (Cerdocyon thous), and the crab-eating raccoon (Procyon cancrivorus). Thus, BSF cases associated with A. aureolatum seem to occur where human dwellings are close to forest fragments and domestic dogs have free access to it. Dogs get infested by adult A. aureolatum, then go on to carry this ixodid into their owners’ households [1]. Humans are parasitized only by adult ticks, usually by a single specimen [5], as the population density of this species is usually low [3].
Although populations of A. aureolatum are present, under similar environmental conditions (human habitations close to forest fragments and dogs infested with A. aureolatum), in several locations in Brazil, information concerning the distribution of BSF cases associated with this ixodid are restricted to the metropolitan area of São Paulo (SP) state (southeast region) [1], where the endemic sites for the disease differ from non-endemic sites due to the presence of smaller and more degraded areas of the Atlantic Forest, with low vertebrate abundance and richness [7,8]. Despite this and the fact that the highest known extent of A. aureolatum R. rickettsii vectoring occurs within the SP metropolitan region, this BSF scenario may have a wider geographic distribution, one which is currently unreported solely due to the scarcity of studies in other regions of Brazil.
Regional differences in the epidemiology of tick-borne diseases (TBD), such as BSF, may be related to pathogen behavior and biological variations between vector populations [9]. Intraspecific genetic studies of ticks can provide information on their variability, dispersal mechanisms, gene flow, population structure, and expansion, as well as TBD epidemiology and surveillance [10,11,12,13]. However, this knowledge is scarce for A. aureolatum, with just one study for SP, Brazil [14]. Accordingly, the current study aimed to analyze the genetic diversity, population dynamics, and infection by rickettsia of A. aureolatum populations from different spotted fever (SF) scenarios in Brazil.
2. Results
2.1. Molecular Identification of Ticks
Following molecular tests, 90 sequences of 12S rDNA (371 bp), 58 sequences of cytochrome oxidase subunit II (COII) (487 bp), and 110 sequences of D-loop (458 bp) were obtained. Of these markers, 12S rDNA was the only one with sequences available in GenBank for A. aureolatum, and with these the sequences of the current study gave similar values (99.42–100.00% with sequences from Paraná State, Brazil: AY277248-49).
2.2. Intrapopulational Analysis: A. aureolatum Has Unique Haplotypes per Population and Low Nucleotide Diversity
Based on the genetic variation of 12S rDNA, COII, and D-loop sequences, 7 (hI–hVII), 17 (h1–h17), and 14 (hA–hN) A. aureolatum haplotypes were identified, respectively (Tables S1–S3).
In the 371–bp data matrix of 12S rDNA, the Rio Grande do Sul (RS) population showed the highest indices for polymorphic site (5), haplotype (0.787), and nucleotide diversity (0.0046) (Table 1). Of the seven haplotypes identified, hI (51.12%) and hII (40.00%) were the most frequent, with the latter shared by all populations. Unique haplotypes were found in RS (hIII–hV) and Paraná (PR) (hVI and hVII) populations (Table S1).

Table 1.
Summary of 12S rDNA, cytochrome oxidase subunit II, and D-loop haplotype distribution, variability, genetic diversity, and demographic history measures of Amblyomma aureolatum populations analyzed from different spotted fever scenarios in Brazil.
Analysis of 487 nucleotides of the A. aureolatum COII gene identified two (Rio de Janeiro (RJ)) to 10 (RS) polymorphic sites that defined the 17 haplotypes described in A. aureolatum populations studied in Brazil (Table 1 and Table S2). Of these, the most frequent was h1 (31.04%), found in PR and Santa Catarina (SC), while h15 was shared by all populations (27.60%). Other haplotypes shared between populations were h2 (SC and São Paulo (SP)), h4 (PR and SC), and h14 (PR, RS, and SP). All populations had unique, low-frequency haplotypes: PR (h11–h13), SC (h3), RJ (h5), RS (h8–h10, h16, and h17), and SP (h6 and h7) (Table S2), where wide haplotype diversity (0.911–0.533) and low nucleotide diversity (0.0048–0.0015) were observed (Table 1).
Of the 14 A. aureolatum D-loop haplotypes, hA was the most frequent (70.01%) and identified in all populations. Other haplotypes shared among populations were hB (PR, SC, RJ, and SP), hC (PR and SC), and hF (PR and RJ). Unique haplotypes were found in all populations: hD (SC), hE (RJ), hG–hJ (SP), hK–hL (RS), and hM–hN (PR); with the exception of hK, all of the other haplotypes showed low frequency (Table S3). The 458 bp fragments aligned from D-loop revealed three (SC and RJ) to six (PR and SP) polymorphic sites, resulting in the low nucleotide (0.0007 in SC; 0.0042 in RS) and haplotype diversity (0.331 in SC; 0.601 in PR) detected in A. aureolatum populations (Table 1).
For all of the analyzed markers, in general, Group I populations (PR, SC, RJ, and RS) had higher diversity index values and more haplotypes than the population from Group II (SP) (Table 1 and Tables S1–S3).
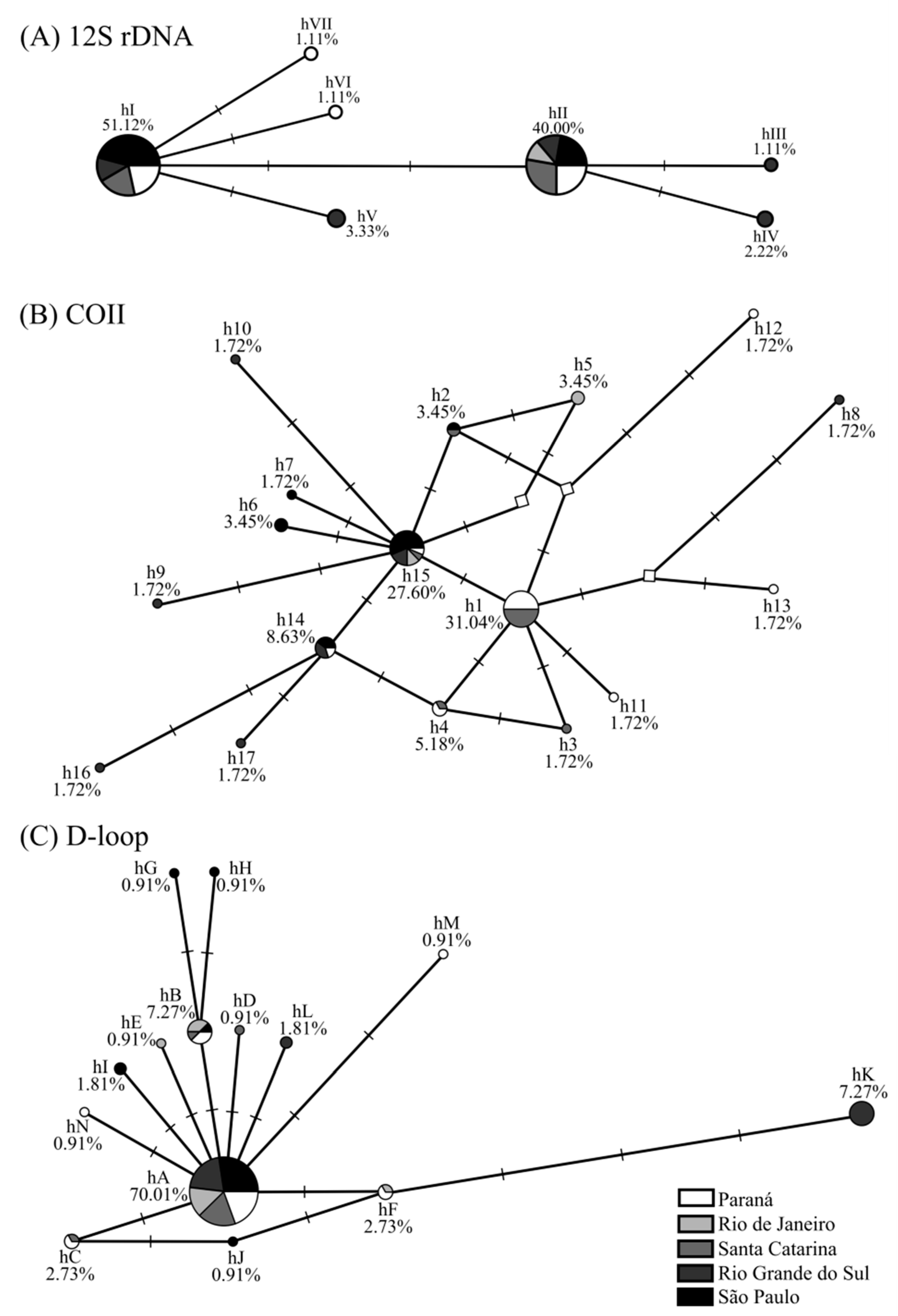
The topology of 12S rDNA, COII, and D-loop median-joining networks was similar: the presence of central haplotypes (hI and hII; h1 and h15; and hA, respectively) had a high frequency and was shared by several populations. Among these haplotypes, separated by 1–4 mutations, are low-frequency haplotypes unique to particular populations. The star-shaped network, mainly for COII and D-loop, was due to the low nucleotide diversity observed in the genes of A. aureolatum populations (0.0000–0.0048) (Table 1). No haplogroups were found (Figure 1).
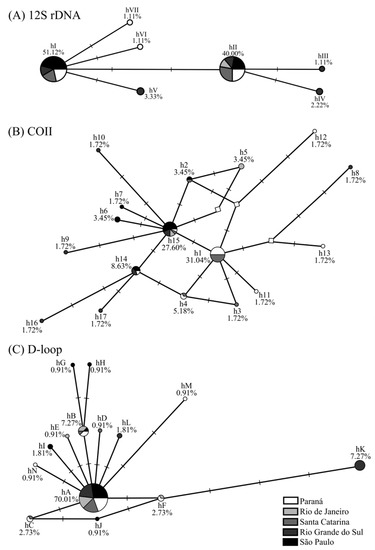
Figure 1.
(A) Haplotype network for 12S rDNA, (B) cytochrome oxidase subunit II (COII), and (C) D-loop markers from Amblyomma aureolatum collected under different spotted fever scenarios in Brazil and their relative frequencies. Networks were generated with the median-joining method, using maximum parsimony post-processing. The circle size corresponds to the frequency of each haplotype sequence, each dash on the line (─) represents one mutational step, and unsampled/intermediate haplotypes are shown by a white diamond (◊).
2.3. Population Differentiation, Structuring, and Demographic Dynamics in A. aureolatum from Brazilian Spotted Fever Scenarios
With the exception of those from PR and SC, all populations showed significant genetic differentiation values for at least one marker in the pairwise FST analyzes (Table 2). Additionally, SF scenario-grouped A. aureolatum populations showed significant FST values for 12S rDNA and COII genes, suggesting that there was genetic divergence between populations of A. aureolatum that acted (SP) or not (PR, SC, RJ, and RS) as vectors of R. rickettsii (Table 3). However, Mantel test results indicated no significant correlation between genetic and geographic distances (12S rDNA: r = 0.1862, p = 0.276; COII: r = 0.1165, p = 0.298; D-loop: r = 0.1934, p = 0.268).

Table 2.
Pairwise comparisons for genetic differentiation (FST) estimated for all Amblyomma aureolatum populations sampled from different spotted fever scenarios in Brazil.

Table 3.
Pairwise comparisons for genetic differentiation (FST) estimated between groups of Amblyomma aureolatum populations from different spotted fever scenarios in Brazil.
AMOVA analysis indicated that the majority of total molecular variation in A. aureolatum haplotypes occurred within populations (91.32% in 12S rDNA, 70.01% in COII, and 90.84% in D-loop) (Table 4). These results, plus the lack of significance in both the Φ statistic among groups (Table 4) and the Mantel tests, may indicate that no A. aureolatum population structure correlated with the current SF epidemiological scenarios.

Table 4.
Analysis of molecular variance (AMOVA) for the analyzed genetic sequences of the 12S rDNA, cytochrome oxidase subunit II, and D-loop markers of groups and studied Brazilian populations of Amblyomma aureolatum.
Statistically significant values were obtained for the neutrality test of COII and/or D-loop markers in the PR, SC, RS, and SP A. aureolatum populations (Table 1). These results may be indicative of recent population expansion under both epidemiological scenarios studied.
2.4. Identification of Detected Rickettsiae
A total of five A. aureolatum samples collected from dogs in epidemiological scenario 1 (ES 1) (SC) (4749A) and epidemiological scenario 2 (ES 2) (SP) (6249H, I1, K, and R) amplified fragments of gltA and htrA genes from rickettsia (Table S4). BLAST analysis revealed 100% nucleotide identity rates with Rickettsia felis in samples from ES 1 and with Rickettsia bellii in samples from ES 2. In the phylogenetic reconstruction, these bacteria clustered with the same rickettsia species to which they showed similarity (Figure S1).
The majority of A. aureolatum samples identified with rickettsia in ES 1 and ES 2 belonged to the haplotypes both most frequent and shared by several populations (12S rDNA-hI, hII; COII-h1, h14, h15; and D-loop hA, hB). However, the same haplotype was detected with different rickettsia species, and the same rickettsia species was found in different A. aureolatum haplotypes (Table S4).
3. Discussion
This is the first study to provide a better understanding of SF epidemiology by integrating A. aureolatum population genetics data using mitochondrial genes and rickettsia infection. The number of identified haplotypes (7–17) was relatively small when compared to other Amblyomma species that are involved in the SF scenario in Brazil [12,13] (Tables S1–S3). This may be due to the low population density of A. aureolatum [3] and the high rate of endogamy previously reported for populations of this tick [14]. Of the analyzed markers, COII was the most sensitive for polymorphism detection, while 12S rDNA proved to be the most conserved in the A. aureolatum populations analyzed (Table 1 and Table S1–S3).
The A. aureolatum populations from RJ, RS, and PR had intermediate/non-sampled haplotypes (Figure 1B), suggesting that diversity in these populations may be greater than reported. The results of tick population genetics are strongly influenced by the sampling analyzed [10]. Accordingly, the greater number of exclusive haplotypes and higher diversity index values obtained in ES 1 populations compared to ES 2 (Table 1) may be associated with the larger area, collection period, populations/individuals, and different host species sampled (Tables S4 and S5). In contrast, individuals analyzed for ES 2 were all from the same location (Santo André/SP), and were all collected during the same period from dogs (Tables S4 and S5).
In general, A. aureolatum haplotypes differ from each other by few mutations (Figure 1). The presence of unique and low-frequency haplotypes to individual populations (Tables S1–S3) when added to the star-shaped network (COII and D-loop) (Figure 1B,C), as well as the high values of haplotype diversity, but low nucleotide diversity, and significant values in the neutrality tests for populations in ES 1 (SC, PR, and RS) and ES 2 (SP) (Table 1), may be indicative of a rapid and recent population expansion [15]. Climate and anthropogenic changes can influence tick demographic dynamics, which may in turn impact public health via increases in vector populations and, possibly, TBD bioagents [10]. This population expansion scenario has been observed previously for other tick species, such as A. sculptum in the Brazilian Cerrado biome [12]. The demographic pattern observed for A. aureolatum may be associated with Atlantic Forest fragmentation, which consequently leads to reductions in both the abundance and species richness of vertebrate hosts. This contrasts with the high density of domestic dogs, animals that can access the forest, then return to home infested with A. aureolatum to cause tick expansion on a local scale [7,8]. Additionally, the rapid population growth of this ixodid deserves attention since, under laboratory conditions, A. aureolatum adults infected with R. rickettsii can transmit this bacterium in about 10 min following initial skin attachment to a second host [16].
The presence of exclusive haplotypes for the ES 1 and ES 2 populations (Figure 1; Tables S1–S3) and the significant values in the pairwise (except for SC and PR) (Table 2) and FST group analyses (Table 3) together suggest genetic differentiation between A. aureolatum populations involved in different epidemiological scenarios analyzed. However, the following may indicate the presence of the gene flow and absence of A. aureolatum population structure between the different SF scenarios studied: the presence of haplotypes shared by all populations in all evaluated markers (Figure 1; Tables S1–S3); no haplogroup in the network (Figure 1); the AMOVA (most of the molecular variance occurred within studied populations); and the lack of significance in the Φ statistic results between groups (Table 4). In addition, the Mantel test confirms that genetic differentiation is not associated with geographical distances for the studied populations. The absence of population structure with low and non-significant pairwise FST values was also observed when comparing A. aureolatum from endemic and non-endemic BSF areas in SP [14]. Consequently, it is possible that other factors are responsible for the genetic differentiation observed in the analyzed populations that are not associated with the susceptibility of this ixodid to infection and vectoring of R. rickettsii.
Movements by A. aureolatum hosts could potentially influence the lack of structure observed between the tick populations, since this arthropod alone does not have a large dispersion capacity. Some bird groups that are infested by immature A. aureolatum may be the main agent responsible for maintaining the gene flow between different geographic populations of this ixodid, as canids (the main hosts for adult stage) and small rodents (hosts for larva and nymph) are likely to have a lower dispersion potential [14], mainly with the increase of Atlantic Forest fragmentation. Consequently, it is possible that structural and functional ecological connections between tick hosts could sustain gene flow between populations of this arthropod, explaining the observed patterns. The absence of a population structure associated with the movement of hosts has already been suggested for A. sculptum in Brazil by [11,12], who considered that low parasite specificity, availability of preferred hosts in degraded areas, and the human transportation of infested hosts (e.g., horses) were the main agents responsible for the maintenance of the gene flow between A. sculptum populations.
Although A. aureolatum and A. ovale have some similar ecological and morphological characteristics, mainly in areas where they occur in sympathy, the results of population genetics obtained for the two species were different. While A. aureolatum showed an absence of a population structure [14] associated with population growth, A. ovale populations were structured, possibly associated with ecological factors and geographical distances between the studied populations [13]. These differences are not unexpected, as ticks are a highly diverse group of ectoparasites, and each species may have specific characteristics relating to life history strategies and/or evolutionary pressures that can result in distinct genetic signatures [10].
Estimates of genetic structure and dispersal in ticks may serve as a surrogate for dispersal estimates of tick-borne pathogens [10]. However, no relationship was observed between A. aureolatum genetic patterns and the different SF scenarios in Brazil. Thus, other factors, such as ecology and environment, may influence the action of A. aureolatum as a R. rickettsii vector in certain areas and not in others. However, the absence of population structure associated with anthropic actions that have been causing environmental changes, such as the fragmentation of the Atlantic Forest, as observed for the endemic area for SF associated with A. aureolatum in SP [8], may cause this tick over time to become an important vector of R. rickettsii in other degraded areas of the Atlantic Forest throughout the country. Thus, it can generate new outbreaks of SF cases with an impact on public health. Although A. aureolatum and A. sculptum are vectors of R. rickettsii, the first one is more susceptible to this infection [17].
Detection of R. bellii during environmental surveillance for SF occurred in an area with previous records of BSF cases in SP (Figure S1, Table S4). There are previous reports of this bacterium in A. aureolatum in the Brazilian states of SP, RJ, and Espírito Santo (e.g., [7,18,19]). Even though the pathogenicity of this rickettsia to humans is unknown, it can play an important role in the epidemiology of rickettsial diseases by inhibiting the maintenance of other pathogenic rickettsia species in the tick [20,21]. Consequently, the detection of R. bellii in the metropolitan region of SP is important, since, in that area, A. aureolatum has been identified as the main vector of R. rickettsii [1].
Rickettsia felis is the bioagent of flea-borne spotted fever (FBSF), an emerging disease. However, basic knowledge of its biology and FBSF epidemiology remains incipient in Brazil. The presence of a confirmed FBSF case in the country [22] highlights the need for research related to this bacterium. There is a previous report of R. felis in A. aureolatum in SP [7]. However, this is the first detection of this rickettsia in this tick for SC. This infection may be due to a simultaneous infestation of the dog by both A. aureolatum- and R. felis-infected fleas (Ctenocephalides spp.), which are admittedly vectors of this rickettsia. However, we cannot rule out the possibility of A. aureolatum participation in the R. felis cycle. Although this tick acts as an important vector of R. rickettsii in the metropolitan region of SP [1], it may be that in other areas of the country this ixodid has a relevant role in the cycle of other rickettsia. In southern Brazil, A. aureolatum is abundant [2,23], although there is, so far, no confirmation of infection by R. rickettsii [1]. The detection of R. felis, a bioagent associated with mild SF cases, in A. aureolatum from SC, a region with confirmed cases of but no deaths from SF during SF case investigation, may indicate the participation of this tick in different eco-epidemiological scenarios of the disease in Brazil.
Thus, it is possible that the role of A. aureolatum as a potential vector for rickettsia in southern Brazil, where this ixodid has already been detected to be infected with the Rickettsia parkeri strain Atlantic rainforest [24], Rickettsia parkeri sensu stricto [4,6], and Rickettsia asembonensis [6], is different from what is known to-date for the metropolitan region of SP [1]. Further studies are needed to assess whether A. aureolatum is also capable of transmitting R. felis. In addition, evaluating the implications associated with this rickettsia in the country, as well as the participation of A. aureolatum, should be investigated.
Although A. aureolatum is an efficient R. rickettsii vector [1,17], the absence of this infection in the analyzed samples, even from BSF endemic area in SP, does not allow us to exclude the existence of this bacterium and/or another rickettsia species infecting the tick populations within the studied locations. Low infection rates (1–10%) of R. rickettsii are reported in A. aureolatum in the wild, possibly related with the deleterious effect that this bioagent has on this tick [7,18].
The lack of a relationship between tick haplotypes and rickettsia detected in A. aureolatum may be associated with the lack of genetic structuring among the analyzed populations of this arthropod. A previous study also detected weak differentiation and a lack of structure between A. aureolatum populations in endemic and non-endemic BSF areas in SP, which did not support the hypothesis that analyzed genetic characteristics of A. aureolatum populations would be responsible for the presence or absence of R. rickettsii in these areas of SP [14].
For a better understanding of A. aureolatum participation in SF epidemiological scenarios in Brazil, it may be necessary to analyze other genetic regions of this tick that are under strong selective pressure related to forms of natural resistance to infection by R. rickettsii and/or other rickettsia [14]. In addition, ecological and environmental factors can influence the presence or absence of rickettsia and their amplifying hosts [7]. Atlantic Forest fragmentation is correlated with a high risk of BSF associated with A. aureolatum in SP [8], since deforestation can result in habitat loss and create environmental conditions unfavorable to the maintenance of populations of large carnivores, and this often makes domestic dogs the only available host for the adult stage of this ixodid. With access to the forest and without competition from wild carnivores, dogs can roam freely in the vegetation, increasing the chance of A. aureolatum parasitism. Upon returning parasitized to human habitations, these animals increase the risk of human infection with rickettsia [8]. Future studies that seek a broader understanding of the epidemic and enzootic cycles of SF involving A. aureolatum should analyze samples of the different stages of this ixodid from birds, small rodents, and wild canids. For future phylogenetic and population analyzes of the tick, other markers can be used, such as the 16S rDNA that is commonly used for the group, which enable a greater understanding of the inter- and intraspecific relationships of ixodids in the different epidemiological scenarios of SF.
The absence of structure among analyzed A. aureolatum populations associated with the expansion of this vector in PR, SC, RS, and SP requires public health attention, since population growth of this ixodid, a competent vector for R. rickettsii, could potentially cause enhanced pathogen dispersion [10], thus generating new foci of SF. The integrated use of tick population genetics data, the presence of pathogens, transcriptome and microbiome studies, and ecological analysis can be useful tools for a broader understanding of the processes surrounding, and perhaps regulating, TBDs in Brazil.
4. Materials and Methods
4.1. Sampling and Taxonomy
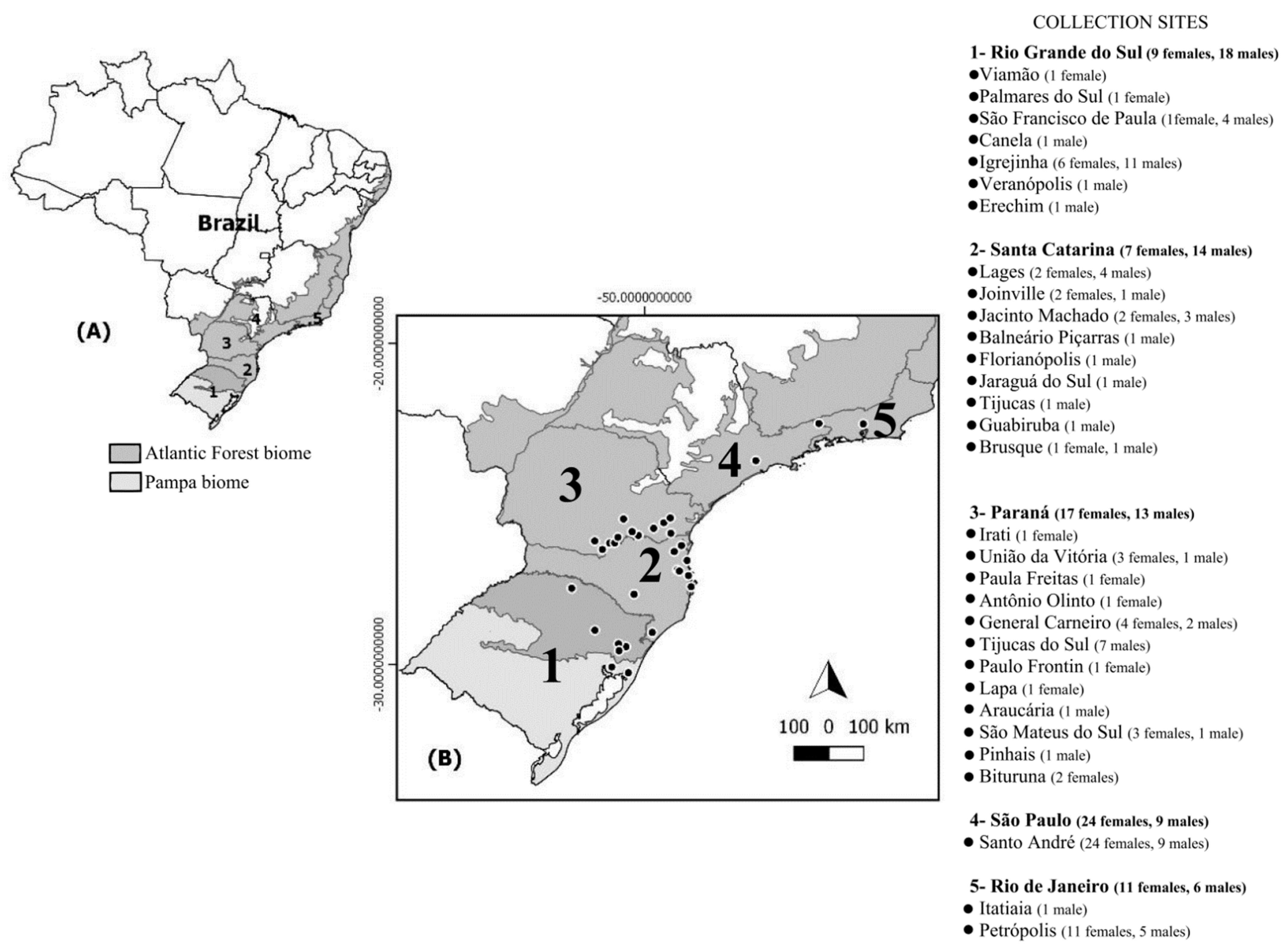
In total, 128 (68 females and 60 males) A. aureolatum were collected from vertebrate hosts (Canis familiaris (102), Felis catus (9), Homo sapiens (9), Bos taurus (1), and Nasua nasua (1)) and the environment (six human habitations), between 2012 to 2017 during SF-focused investigations and environmental surveillance in Brazil in localities with altitudes varying from 2–1648 m by teams from five state departments of health (Tables S4 and S5). The samples came from states where there is no evidence of A. aureolatum involvement in the BSF epidemic cycle (PR, SC, RS, and RJ; ES 1) and from an area where this ixodid demonstrably participates in the BSF cycle as the vector of R. rickettsii (metropolitan region of SP; ES 2) (Figure 2). Obtained A. aureolatum were stored in isopropyl alcohol, then sent to the Laboratory of the National Reference of Rickettsial Vectors, where they were morphologically identified [23] and analyzed individually with molecular biology techniques. The states and epidemiological scenarios of tick collection were used to determine populations in genetic analyzes.
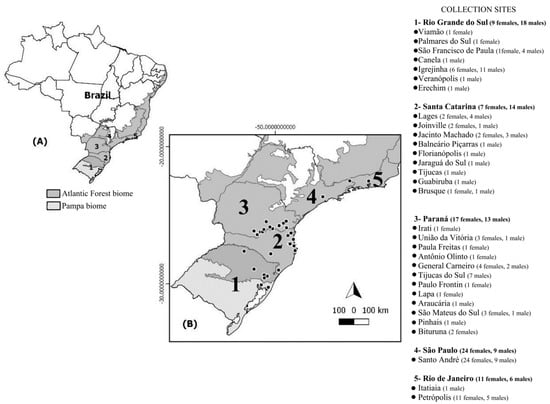
Figure 2.
(A) Map of Brazil with indication of studied states (1–5) and respective biomes; (B) number of individuals sampled at each collection sites (•) for the 128 Amblyomma aureolatum specimens analyzed from Brazil.
4.2. DNA Extraction, Amplification, and Sequencing
In all samples, DNA was extracted using the saturated sodium chloride solution protocol [25]. For A. aureolatum population analysis, a set of mitochondrial gene fragments (12S rDNA [26], D-loop [27], and COII [11]) was amplified by a polymerase chain reaction (PCR) following previously established protocols [11].
Rickettsia-infected ticks were detected by PCR screening for the rickettsial gltA (CS2-78/CS2-323) [28], ompA (Rr l90.70p/Rr l90.602n) [29], ompB (120.M59/120.807) [30], and htrA (nested PCR, first-round primers 17k-5/17k-3) [31]; (second round primers 17kD1/17kD2) [32] genes. In these amplifications, DNA-free reactions was used as negative control, with R. rickettsii DNA as positive control.
PCR products were visualized by electrophoresis in 2% agarose gel stained with ethidium bromide and purified using the Wizard®SV Gel and PCR Clean-Up System kit (Promega Corp., Madison, WI, USA), following manufacturer protocols. DNA was sequenced in both directions, applying the same PCR primers and the BigDye Terminator™ version 3.1 Cycle Sequencing® Kit (Applied Biosystems, Foster City, CA, USA) on an automated ABI 3730xl DNA analyser (Applied Biosystems, Foster City, CA, USA,). Sequences from tick haplotypes and from identified rickettsia were deposited in GenBank (accession numbers MF175654-8, MF175680-91, MF175738-47, MF175769-72, MF175786-8, MT578021-31) (Table S4).
4.3. Sequence Editing, Identification, and Alignment
The obtained sequences were assembled and edited with CHROMASPRO, version 1.5 (Technelysium Pty Ltd., Tewantin, QLD, Australia), and subjected to BLASTn analyses (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 16 February 2021) to infer closest similarities with other organisms available in GenBank. Multiple alignments were conducted using the ClustalW algorithm, and then checked manually. Protein coding genes were translated into amino acids where no stop codon was observed.
4.4. Amblyomma aureolatum Population Genetics Analysis
Haplotype definition (considered distinct when differing by at least one base pair), polymorphic sites (s), number of haplotypes (n), both haplotype (h) and nucleotide diversity (nd), and the average number of nucleotide differences (k) were calculated in ARLEQUIN software, version 3.5.2.2 [33].
Analysis of molecular variance (AMOVA) [34] and pairwise FST structuring values were used to estimate molecular genetic differentiation between A. aureolatum populations. Calculations of Φ statistics indices were based on AMOVA values. Mantel tests analyzed the association between genetic (using FST values) and geographic distances (from Google Earth). Additionally, FST and AMOVA tests were determined hierarchical supra-population levels (Group I and II) to check if A. aureolatum genetic differentiation and population structure varied between SF scenarios, where: Group I-samples from ES 1 (PR, SC, RJ and RS) and Group II-samples from ES 2 (metropolitan area of SP). These analyzes were performed using ARLEQUIN, with 1000 repeats each.
The Tajima’s D [15], Fu’s Fs [35] and R2 [36] neutrality tests were performed to investigate the hypothesis of neutral mutation and population balance. To estimate the indices, ARLEQUIN software was used for Tajima’s D and Fu’s Fs, and DNASP v. 6.12.03 [37] for the R2 test, with 1000 permutations in each case.
A haplotype network was constructed for each marker using NETWORK 10.0.0.0 software (Fluxus Technology Ltd., Cambridge, MA, U.K.), combined with a median joining algorithm [38].
4.5. Phylogenetic Relationships of Detected Rickettsiae
A concatenated (gltA + htrA, 1383 bp) Maximum-Likelihood phylogeny was inferred using PhyML software Version 3.0 [39], with evolutionary model GTR+G indicated by MEGA 6.0, through BIC. Alignments were concatenated with Seaview software [40]. Rickettsiae sequences generated in this study were used in the phylogenetic reconstruction, plus those available in GenBank. The reliability of the tree topology was estimated with an approximate likelihood ratio test (aLRT) with 1000 replicas.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10091146/s1, Figure S1: Concatenated phylogeny of rickettsia gltA and htrA genes fragment (1013 + 370 bp) detected in Amblyomma aureolatum in Brazil, inferred by Maximum Likelihood analysis with evolution model GTR+G. Numbers on the branches represent support values (70% cut-off), Table S1: Distribution and frequency of 12S rDNA haplotypes identified in the populations of Amblyomma aureolatum analyzed from Brazil, Table S2: Distribution and frequency of cytochrome oxidase subunit II haplotypes identified in the populations of Amblyomma aureolatum analyzed from Brazil, Table S3: Distribution and frequency of D-loop haplotypes identified in the populations of Amblyomma aureolatum analyzed from Brazil, Table S4: Sample data, haplotype and associated GenBank accession numbers for Amblyomma aureolatum 12S rDNA, cytochrome oxidase subunit II (COII), D-loop and rickettsia sequences analyzed from Brazil, Table S5: Collection data for Amblyomma aureolatum samples analyzed from Brazil.
Author Contributions
Conceptualization, K.B. and G.S.G.; methodology, K.B., M.A., S.V.d.O. and G.S.G.; formal analysis, K.B.; investigation, K.B., S.V.d.O. and G.S.G.; resources, M.A., S.V.d.O. and G.S.G.; data curation, K.B.; writing—original draft preparation, K.B.; writing—review and editing, K.B., M.A., S.V.d.O. and G.S.G.; supervision, G.S.G.; funding acquisition, M.A., S.V.d.O. and G.S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundo Nacional de Saúde/Minitério da Saúde, grant number TC 334/2013, SIAFI 677.202-FNS/FIOCRUZ, Processo Fiocruz no 25000.204745/2013-08.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available within the article, in Supplementary Materials and in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accessed on 11 March 2021, accession numbers: MF175654-8, MF175680-91, MF175738-47, MF175769-72, MF175786-8, MT578021-31.
Acknowledgments
The authors would like to thank the Paraná, Santa Catarina, Rio de Janeiro, Rio Grande do Sul Departments of Health and Robson Oliveira Lopes (Manager of Zoonoses Control at Santo André/SP Sanitary Surveillance) for the tick collections and for sending samples; the Genomic Platform DNA Sequencing (PDTIS, Oswaldo Cruz Foundation) for support with sample sequencing; and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for scholarship. Adrian Barnett helped with the English of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Szabó, M.P.J.; Pinter, A.; Labruna, M.B. Ecology, biology and distribution of spotted fever tick vectors in Brazil. Front. Cell. Infect. Microbiol. 2013, 27, 27. [Google Scholar] [CrossRef] [Green Version]
- Guglielmone, A.A.; Estrada-Peña, A.; Mangold, A.J.; Barros-Battesti, D.M.; Labruna, M.B.; Martins, J.R.; Venzal, J.M.; Arzua, M.; Keirans, J.E. Amblyomma aureolatum (Pallas, 1772) and Amblyomma ovale Kock, 1844: Hosts, distribution and 16S rDNA sequences. Vet. Parasitol. 2003, 113, 273–288. [Google Scholar] [CrossRef]
- Pinter, A.; Dias, R.A.; Gennari, S.M.; Labruna, M.B. Study of the seasonal dynamics, life cycle, and host specificity of Amblyomma aureolatum (Acari: Ixodidae). J. Med. Entomol. 2004, 41, 324–332. [Google Scholar] [CrossRef]
- Dall’Agnol, B.; Souza, U.A.; Weck, B.; Trigo, T.C.; Jardim, M.M.A.; Costa, F.B.; Labruna, M.B.; Peters, F.B.; Favarini, M.O.; Mazim, F.D.; et al. Rickettsia parkeri in free-ranging wild canids from Brazilian Pampa. Transboundary Emerg. Dis. 2018, 65, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Reck, J.; Souza, U.; Souza, G.; Kieling, E.; Dall’Agnol, B.; Webster, A.; Michel, T.; Doyle, R.; Martins, T.F.; Labruna, M.B.; et al. Records of ticks on humans in Rio Grande do Sul state, Brazil. Ticks Tick Borne Dis. 2018, 9, 1296–1301. [Google Scholar] [CrossRef]
- Souza, U.A.; Webster, A.; Dall’Agnol, B.; Peters, F.B.; Favarini, M.O.; Schott, D.; Zitelli, L.C.; Mazim, F.D.; Kasper, C.B.; Ott, R.; et al. Ticks, mites, fleas, and vector-borne pathogens in free-ranging neotropical wild felids from southern Brazil. Ticks Tick Borne Dis. 2021, 12, 101706. [Google Scholar] [CrossRef] [PubMed]
- Ogrzewalska, M.; Saraiva, D.G.; Moraes-Filho, J.; Martins, T.F.; Costa, F.B.; Pinter, A.; Labruna, M.B. Epidemiology of Brazilian spotted fever in the Atlantic Forest, state of São Paulo, Brazil. Parasitology 2012, 139, 1283–1300. [Google Scholar] [CrossRef]
- Scinachi, C.A.; Takeda, G.A.; Mucci, L.F.; Pinter, A. Association of the occurrence of Brazilian spotted fever and Atlantic rain forest fragmentation in the São Paulo metropolitan region, Brazil. Acta Trop. 2017, 166, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Gray, J. Biology of Ixodes species ticks in relation to tick-borne zoonoses. Wien Klin Wochenschr. 2002, 114, 473–478. [Google Scholar] [PubMed]
- Araya-Anchetta, A.; Busch, J.D.; Scoles, G.A.; Wagner, D.M. Thirty years of tick population genetics: A comprehensive review. Infect. Genet. Evol. 2015, 29, 164–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitencourth, K.; Voloch, C.M.; Serra-Freire, N.M.; Machado-Ferreira, E.; Amorim, M.; Gazêta, G.S. Analysis of Amblyomma sculptum haplotypes in an area endemic for Brazilian spotted fever. Med. Vet. Entomol. 2016, 30, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Bitencourth, K.; Amorim, M.; Oliveira, S.V.; Caetano, R.L.; Voloch, C.M.; Gazêta, G.S. Amblyomma sculptum: Genetic diversity and rickettsias in the Brazilian Cerrado biome. Med. Vet. Entomol. 2017, 31, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Bitencourth, K.; Amorim, M.; Oliveira, S.V.; Voloch, C.M.; Gazêta, G.S. Genetic diversity, population structure and rickettsias in Amblyomma ovale in areas of epidemiological interest for spotted fever in Brazil. Med. Vet. Entomol. 2019, 33, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Ogrzewalska, M.; Schwarcz, K.; Bajay, M.M.; Bajay, S.K.; Pinheiro, J.B.; Zucchi, M.I.; Pinter, A.; Labruna, M.B. Characterization of genetic variability and population structure of the tick Amblyomma aureolatum (Acari: Ixodidae). J. Med. Entomol. 2016, 53, 843–850. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Saraiva, D.G.; Soares, H.S.; Soares, J.F.; Labruna, M.B. Feeding period required by Amblyomma aureolatum ticks for transmission of Rickettsia rickettsii to vertebrate hosts. Emerg. Infect. Dis. 2014, 20, 1504–1510. [Google Scholar] [CrossRef]
- Labruna, M.B.; Ogrzewalska, M.; Martins, T.F.; Pinter, A.; Horta, M.C. Comparative susceptibility of larval stages of Amblyomma aureolatum, Amblyomma cajennense, and Rhipicephalus sanguineus to infection by Rickettsia rickettsii. J. Med. Entomol. 2008, 45, 1156–1159. [Google Scholar] [CrossRef]
- Pinter, A.; Labruna, M.B. Isolation of Rickettsia rickettsii and Rickettsia bellii in cell culture from the tick Amblyomma aureolatum in Brazil. Ann. N. Y. Acad. Sci. 2006, 1078, 523–529. [Google Scholar] [CrossRef]
- Luz, H.R.; Faccini, J.L.H.; McIntosh, D. Molecular analyses reveal an abundant diversity of ticks and rickettsial agents associated with wild birds in two regions of primary Brazilian Atlantic Rainforest. Ticks Tick Borne Dis. 2017, 8, 657–665. [Google Scholar] [CrossRef]
- Burgdorfer, W.; Hayes, S.F.; Mavros, A.J. Non pathogenic rickettsiae in Dermacentor andersoni: A limiting factor for the distribution of Rickettsia rickettsii. In Rickettsiae and Rickettsial Diseases; Burgdorfer, W., Anacker, R.L., Eds.; Academic Press: New York, NY, USA, 1981; pp. 585–594. [Google Scholar]
- Sakai, R.K.; Costa, F.B.; Ueno, T.E.; Ramirez, D.G.; Soares, J.F.; Fonseca, A.H.; Labruna, M.B.; Barros-Battesti, D.M. Experimental infection with Rickettsia rickettsii in an Amblyomma dubitatum tick colony, naturally infected by Rickettsia bellii. Ticks Tick Borne Dis. 2014, 5, 917–923. [Google Scholar] [CrossRef]
- Raoult, D.; La Scola, B.; Enea, M.; Fournier, P.E.; Roux, V.; Fenollar, F.; Galvão, M.A.; Lamballerie, X. A flea-associated Rickettsia pathogenic for humans. Emerg. Infect. Dis. 2001, 7, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Barros-Battesti, D.M.; Arzua, M.; Bechara, G.H. Carrapatos de Importância Médico- Veterinária da Região Neotropical: Um guia Ilustrado para Identificação de Espécies; Butantan: São Paulo, Brazil, 2006; 223p. [Google Scholar]
- Medeiros, A.P.; Souza, A.P.; Moura, A.B.; Lavina, M.S.; Bellato, V.; Sartor, A.A.; Nieri-Bastos, F.A.; Richtzenhain, L.J.; Labrina, M.B. Spotted fever group Rickettsia infecting ticks (Acari: Ixodidae) in the state of Santa Catarina, Brazil. Mem. Inst. Oswaldo Cruz 2011, 106, 926–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR- based techniques. Nucleic Acids Res. 1997, 15, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Beati, L.; Keirans, J.E. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribossomal DNA gene sequences and morphological characters. J. Parasitol. 2001, 87, 32–48. [Google Scholar] [CrossRef]
- Burkman, E.J. Genetic structure of Amblyomma cajennense (Acari: Ixodidae) populations based on mitochondrial gene sequences. Master’s Thesis, Georgia Southern University, Statesboro, GA, USA, 2009. [Google Scholar]
- Labruna, M.B.; Whitworth, T.; Horta, M.C.; Bouyer, D.H.; McBride, J.W.; Pinter, A.; Popov, V.; Gennari, S.; Walker, D.H. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 2004, 42, 90–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regnery, R.L.; Spruill, C.L.; Plikaytis, B.D. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 1991, 173, 1576–1589. [Google Scholar] [CrossRef] [Green Version]
- Roux, V.; Raoult, D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (OmpB). Int. J. Syst. Evol. Microbiol. 2000, 50, 1449–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labruna, M.B.; McBride, J.W.; Bouyer, D.H.; Camargo, L.M.A.; Camargo, E.P.; Walker, D.H. Molecular evidence for a spotted fever group Rickettsia species in the tick Amblyomma longirostre in Brazil. J. Med. Entomol. 2004, 41, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Webb, L.; Mitchell, C.; Malloy, D.C.; Dasch, G.A.; Azad, A.F. Detection of murine typhus infection in fleas by using the polymerase chain reaction. J. Clin. Microbiol. 1990, 28, 530–534. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Onsins, S.E.; Rozas, J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Del Barrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).