The Oesophageal Squamous Cell Carcinoma Cell Line COLO-680N Fails to Support Sustained Cryptosporidium parvum Proliferation
Abstract
:1. Introduction
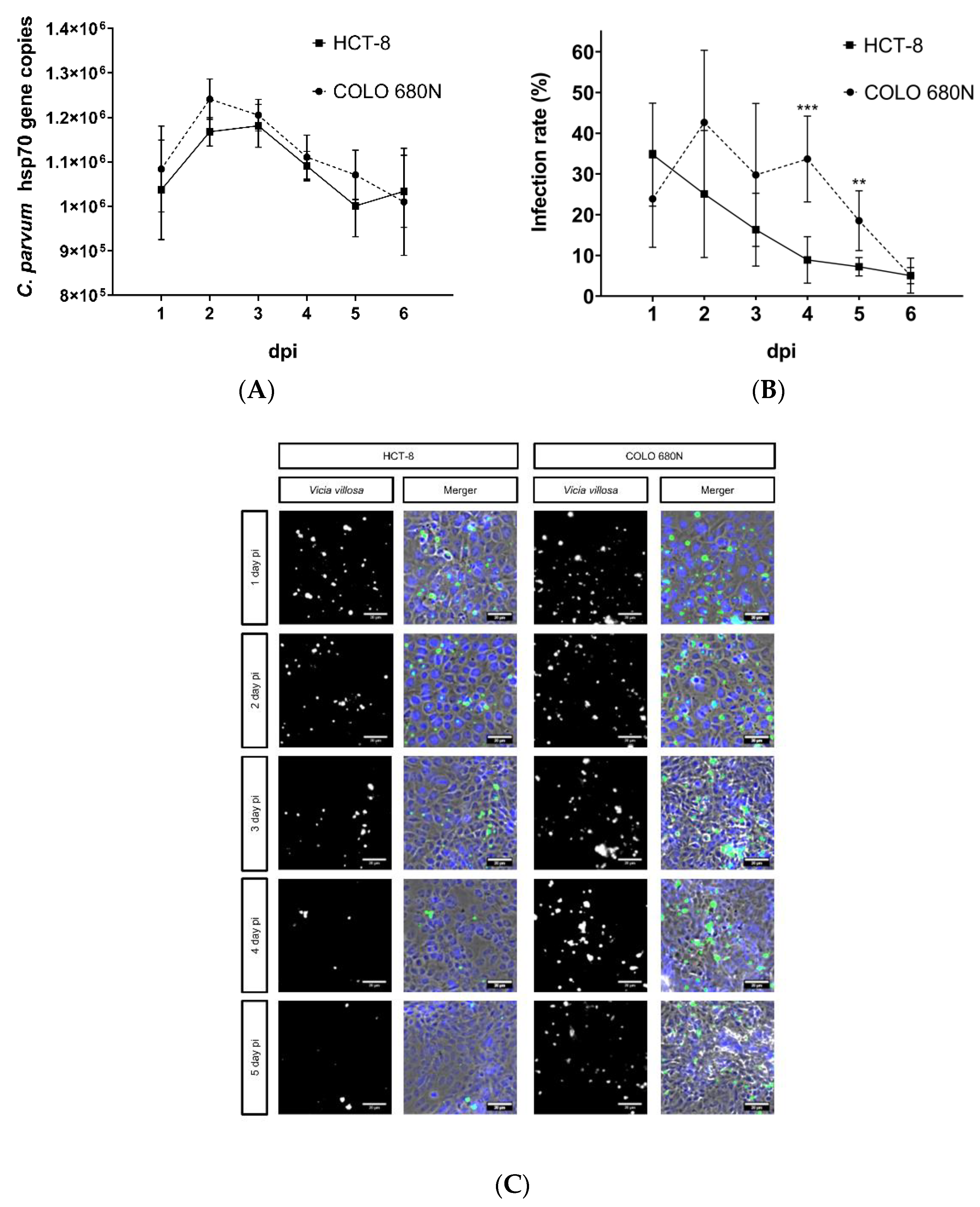
2. Results
3. Discussion
4. Materials and Methods
4.1. Host Cell Lines and Cell Culture Conditions
4.2. Parasites
4.3. Host Cell Infection
4.4. Vicia villosa Lectin-Based Detection of C. parvum
4.5. Real-Time qPCR for C. parvum Quantification
4.6. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and Aetiology of Diarrhoeal Disease in Infants and Young Children in Developing Countries (the Global Enteric Multicenter Study, GEMS): A Prospective, Case-Control Study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Hunter, P.R.; Hadfield, S.J.; Wilkinson, D.; Lake, I.R.; Harrison, F.; Chalmers, R.M. Subtypes of Cryptosporidium parvum in Humans and Disease Risk. Emerg. Infect. Dis. 2007, 13, 82–88. [Google Scholar] [CrossRef]
- O’Connor, R.M.; Shaffie, R.; Kang, G.; Ward, H.D. Cryptosporidiosis in Patients with HIV/AIDS. AIDS 2011, 25, 549–560. [Google Scholar] [CrossRef]
- Amadi, B.; Mwiya, M.; Sianongo, S.; Payne, L.; Watuka, A.; Katubulushi, M.; Kelly, P. High Dose Prolonged Treatment with Nitazoxanide Is Not Effective for Cryptosporidiosis in HIV Positive Zambian Children: A Randomised Controlled Trial. BMC Infect. Dis. 2009, 9, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Checkley, W.; White, A.C.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.-M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A Review of the Global Burden, Novel Diagnostics, Therapeutics, and Vaccine Targets for Cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Tandel, J.; English, E.D.; Sateriale, A.; Gullicksrud, J.A.; Beiting, D.P.; Sullivan, M.C.; Pinkston, B.; Striepen, B. Life Cycle Progression and Sexual Development of the Apicomplexan Parasite Cryptosporidium parvum. Nat. Microbiol. 2019, 4, 2226–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hijjawi, N.S.; Meloni, B.P.; Morgan, U.M.; Thompson, R.C.A. Complete Development and Long-Term Maintenance of Cryptosporidium parvum Human and Cattle Genotypes in Cell Culture. Int. J. Parasitol. 2001, 31, 1048–1055. [Google Scholar] [CrossRef]
- Current, W.L.; Haynes, T.B. Complete Development of Cryptosporidium in Cell Culture. Science 1984, 224, 603–605. [Google Scholar] [CrossRef]
- Karanis, P. The Truth about in Vitro Culture of Cryptosporidium Species. Parasitology 2017, 145, 1–10. [Google Scholar] [CrossRef]
- Wilke, G.; Funkhouser-Jones, L.J.; Wang, Y.; Ravindran, S.; Wang, Q.; Beatty, W.L.; Baldridge, M.T.; VanDussen, K.L.; Shen, B.; Kuhlenschmidt, M.S.; et al. A Stem-Cell-Derived Platform Enables Complete Cryptosporidium Development In Vitro and Genetic Tractability. Cell Host Microbe 2019, 26, 123–134.e8. [Google Scholar] [CrossRef] [Green Version]
- Morada, M.; Lee, S.; Gunther-Cummins, L.; Weiss, L.M.; Widmer, G.; Tzipori, S.; Yarlett, N. Continuous Culture of Cryptosporidium parvum Using Hollow Fiber Technology. Int. J. Parasitol. 2016, 46, 21–29. [Google Scholar] [CrossRef]
- Alcantara Warren, C.; Destura, R.V.; Sevilleja, J.E.A.D.; Barroso, L.F.; Carvalho, H.; Barrett, L.J.; O’Brien, A.D.; Guerrant, R.L. Detection of Epithelial-Cell Injury, and Quantification of Infection, in the HCT-8 Organoid Model of Cryptosporidiosis. J. Infect. Dis. 2008, 198, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.N.; Jossé, L.; Brown, I.; Blakeman, B.; Povey, J.; Yiangou, L.; Price, M.; Cinatl, J.; Xue, W.-F.; Michaelis, M.; et al. A Cell Culture Platform for Cryptosporidium That Enables Long-Term Cultivation and New Tools for the Systematic Investigation of Its Biology. Int. J. Parasitol. 2018, 48, 197–201. [Google Scholar] [CrossRef]
- Varughese, E.A.; Bennett-Stamper, C.L.; Wymer, L.J.; Yadav, J.S. A New in Vitro Model Using Small Intestinal Epithelial Cells to Enhance Infection of Cryptosporidium parvum. J. Microbiol. Methods 2014, 106, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Shahiduzzaman, M.; Dyachenko, V.; Obwaller, A.; Unglaube, S.; Daugschies, A. Combination of Cell Culture and Quantitative PCR for Screening of Drugs against Cryptosporidium parvum. Vet. Parasitol. 2009, 162, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Broglia, A.; Reckinger, S.; Cacció, S.M.; Nöckler, K. Distribution of Cryptosporidium parvum Subtypes in Calves in Germany. Vet. Parasitol. 2008, 154, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Holzhausen, I.; Lendner, M.; Göhring, F.; Steinhöfel, I.; Daugschies, A. Distribution of Cryptosporidium parvum Gp60 Subtypes in Calf Herds of Saxony, Germany. Parasitol. Res. 2019, 118, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Mauzy, M.J.; Enomoto, S.; Lancto, C.A.; Abrahamsen, M.S.; Rutherford, M.S. The Cryptosporidium parvum Transcriptome during In Vitro Development. PLoS ONE 2012, 7, e31715. [Google Scholar] [CrossRef] [Green Version]
- Borowski, H.; Thompson, R.C.A.; Armstrong, T.; Clode, P.L. Morphological Characterization of Cryptosporidium parvum Life-Cycle Stages in an in Vitro Model System. Parasitology 2010, 137, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bessoff, K.; Sateriale, A.; Lee, K.K.; Huston, C.D. Drug Repurposing Screen Reveals FDA-Approved Inhibitors of Human HMG-CoA Reductase and Isoprenoid Synthesis That Block Cryptosporidium parvum Growth. Antimicrob. Agents Chemother. 2013, 57, 1804–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clode, P.L.; Koh, W.H.; Thompson, R.C.A. Life without a Host Cell: What Is Cryptosporidium? Trends Parasitol. 2015, 31, 614–624. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Vanopdenbosch, E.; Ortega-Mora, L.M.; Abbassi, H.; Peeters, J.E. A Review of the Importance of Cryptosporidiosis in Farm Animals. Int. J. Parasitol. 1999, 29, 1269–1287. [Google Scholar] [CrossRef]
- Woodmansee, D.; Pohlenz, J.F.L. Development of Cryptosporidium sp. in a Human Rectal Tumor Cell Line. Proc. Fourth Int. Symp. Neonatal Diarrhea 1983, 1983, 306–319. [Google Scholar]
- Upton, S.J.; Tilley, M.; Brillhart, D.B. Comparative Development of Cryptosporidium parvum (Apicomplexa) in 11 Continuous Host Cell Lines. FEMS Microbiol. Lett. 1994, 118, 233–236. [Google Scholar] [CrossRef]
- Cai, X.; Woods, K.M.; Upton, S.J.; Zhu, G. Application of Quantitative Real-Time Reverse Transcription-PCR in Assessing Drug Efficacy against the Intracellular Pathogen Cryptosporidium parvum In Vitro. Antimicrob. Agents Chemother. 2005, 49, 4437–4442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashim, A.; Mulcahy, G.; Bourke, B.; Clyne, M. Interaction of Cryptosporidium hominis and Cryptosporidium parvum with Primary Human and Bovine Intestinal Cells. Infect. Immun. 2006, 74, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Girouard, D.; Gallant, J.; Akiyoshi, D.E.; Nunnari, J.; Tzipori, S. Failure to Propagate Cryptosporidium spp. in Cell-Free Culture. J. Parasitol. 2006, 92, 399–400. [Google Scholar] [CrossRef] [PubMed]
- Vélez, J.; Lange, M.K.; Zieger, P.; Yoon, I.; Failing, K.; Bauer, C. Long-Term Use of Yeast Fermentation Products in Comparison to Halofuginone for the Control of Cryptosporidiosis in Neonatal Calves. Vet. Parasitol. 2019, 269, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Chopra, D.P.; Dombkowski, A.A.; Stemmer, P.M.; Parker, G.C. Intestinal Epithelial Cells In Vitro. Stem Cells Dev. 2010, 19, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [Green Version]
- Shanmugasundram, A.; Gonzalez-Galarza, F.F.; Wastling, J.M.; Vasieva, O.; Jones, A.R. Library of Apicomplexan Metabolic Pathways: A Manually Curated Database for Metabolic Pathways of Apicomplexan Parasites. Nucleic Acids Res. 2013, 41, D706–D713. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Roellig, D.M.; Guo, Y.; Li, N.; Frace, M.A.; Tang, K.; Zhang, L.; Feng, Y.; Xiao, L. Evolution of Mitosome Metabolism and Invasion-Related Proteins in Cryptosporidium. BMC Genom. 2016, 17, 1006. [Google Scholar] [CrossRef] [Green Version]
- Ng, J.S.Y.; Ryan, U.; Trengove, R.D.; Maker, G.L. Development of an Untargeted Metabolomics Method for the Analysis of Human Faecal Samples Using Cryptosporidium-Infected Samples. Mol. Biochem. Parasitol. 2012, 185, 145–150. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, H.; Guo, F.; Sun, M.; Zhu, G. A Unique Hexokinase in Cryptosporidium parvum, an Apicomplexan Pathogen Lacking the Krebs Cycle and Oxidative Phosphorylation. Protist 2014, 165, 701–714. [Google Scholar] [CrossRef] [Green Version]
- Gray, G.M. Carbohydrate Digestion and Absorption. Available online: https://www.nejm.org/doi/pdf/10.1056/NEJM197506052922308 (accessed on 13 September 2020).
- Zheng, L.; Kelly, C.J.; Colgan, S.P. Physiologic Hypoxia and Oxygen Homeostasis in the Healthy Intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 2015, 309, C350–C360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vélez, J.; Velasquez, Z.; Silva, L.M.R.; Gärtner, U.; Failing, K.; Daugschies, A.; Mazurek, S.; Hermosilla, C.; Taubert, A. Metabolic Signatures of Cryptosporidium parvum-Infected HCT-8 Cells and Impact of Selected Metabolic Inhibitors on C. parvum Infection under Physioxia and Hyperoxia. Biology 2021, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.; Sconza, S.; Lorenz, I.; Otranto, G.; Rademacher, G.; Famigli-Bergamini, P.; Klee, W. D-Lactic Acidosis in Calves as a Consequence of Experimentally Induced Ruminal Acidosis. J. Vet. Med. Ser. A 2004, 51, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, I.; Gentile, A. D-Lactic Acidosis in Neonatal Ruminants. Vet. Clin. North Am. Food Anim. Pract. 2014, 30, 317–331. [Google Scholar] [CrossRef]
- Vélez, J.; Silva, L.M.; Gärtner, U.; Daugschies, A.; Mazurek, S.; Hermosilla, C.; Taubert, A. First Metabolic Insights into Ex Vivo Cryptosporidium parvum-Infected Bovine Small Intestinal Explants Studied under Physioxic Conditions. Biology 2021, 10, 963. [Google Scholar] [CrossRef]
- Ludington, J.G.; Ward, H.D. The Cryptosporidium parvum C-Type Lectin CpClec Mediates Infection of Intestinal Epithelial Cells via Interactions with Sulfated Proteoglycans. Infect. Immun. 2016, 84, 1593–1602. [Google Scholar] [CrossRef] [Green Version]

| Infection Protocol | Excystation Medium | Excystation Time | Infection Medium | Infection Time * | Agitation | Reference |
|---|---|---|---|---|---|---|
| I | 100 μL of 0.01% trypsin and 400 μL of 0.5% sodium hypochlorite | 1 h | RPMI 1640 | 24 h | intermittent vortexing (every ~10 min) | Miller et al., 2018 |
| II | RPMI 1640 NaTC (0.4%) with pre-bleached oocysts | 3 h | RPMI 1640 + NaTC (0.4%) | 3 h | - | Shahiduzzaman et al., 2009 |
| III | acidified (pH of 2.0) and non-acidified 1x HBSS | 20 min | RPMI 1640 | 3 h | During the whole excystation time ** | Varughese et al., 2014 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vélez, J.; Silva, L.M.R.; Kamena, F.; Daugschies, A.; Mazurek, S.; Taubert, A.; Hermosilla, C. The Oesophageal Squamous Cell Carcinoma Cell Line COLO-680N Fails to Support Sustained Cryptosporidium parvum Proliferation. Pathogens 2022, 11, 49. https://doi.org/10.3390/pathogens11010049
Vélez J, Silva LMR, Kamena F, Daugschies A, Mazurek S, Taubert A, Hermosilla C. The Oesophageal Squamous Cell Carcinoma Cell Line COLO-680N Fails to Support Sustained Cryptosporidium parvum Proliferation. Pathogens. 2022; 11(1):49. https://doi.org/10.3390/pathogens11010049
Chicago/Turabian StyleVélez, Juan, Liliana M. R. Silva, Faustin Kamena, Arwid Daugschies, Sybille Mazurek, Anja Taubert, and Carlos Hermosilla. 2022. "The Oesophageal Squamous Cell Carcinoma Cell Line COLO-680N Fails to Support Sustained Cryptosporidium parvum Proliferation" Pathogens 11, no. 1: 49. https://doi.org/10.3390/pathogens11010049
APA StyleVélez, J., Silva, L. M. R., Kamena, F., Daugschies, A., Mazurek, S., Taubert, A., & Hermosilla, C. (2022). The Oesophageal Squamous Cell Carcinoma Cell Line COLO-680N Fails to Support Sustained Cryptosporidium parvum Proliferation. Pathogens, 11(1), 49. https://doi.org/10.3390/pathogens11010049







