Phylogenetic Groups, Pathotypes and Antimicrobial Resistance of Escherichia coli Isolated from Western Lowland Gorilla Faeces (Gorilla gorilla gorilla) of Moukalaba-Doudou National Park (MDNP)
Abstract
1. Introduction
2. Results
2.1. E. coli Found in Gorilla Faecal Samples
2.2. Prevalence of E. coli Phylogroups
2.3. Prevalence of E. coli Pathotypes
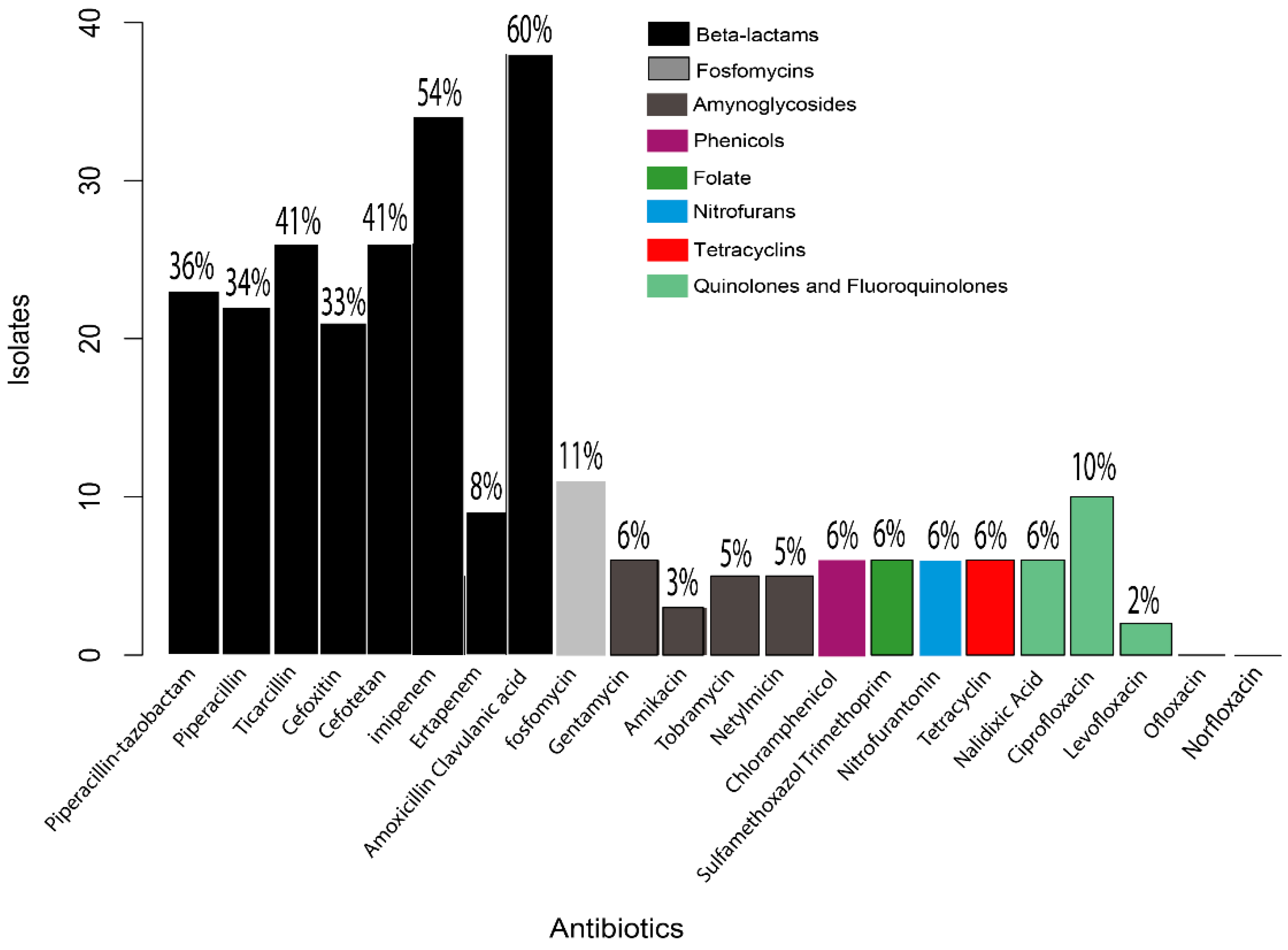
2.4. Antibiotic Susceptibility Testing
3. Discussion
3.1. Isolation of E. coli in Wild Gorillas from MDNP
3.2. Prevalence of Phylogenetic Groups
3.3. Prevalence of Pathotypes
3.4. Antibiotic Susceptibility
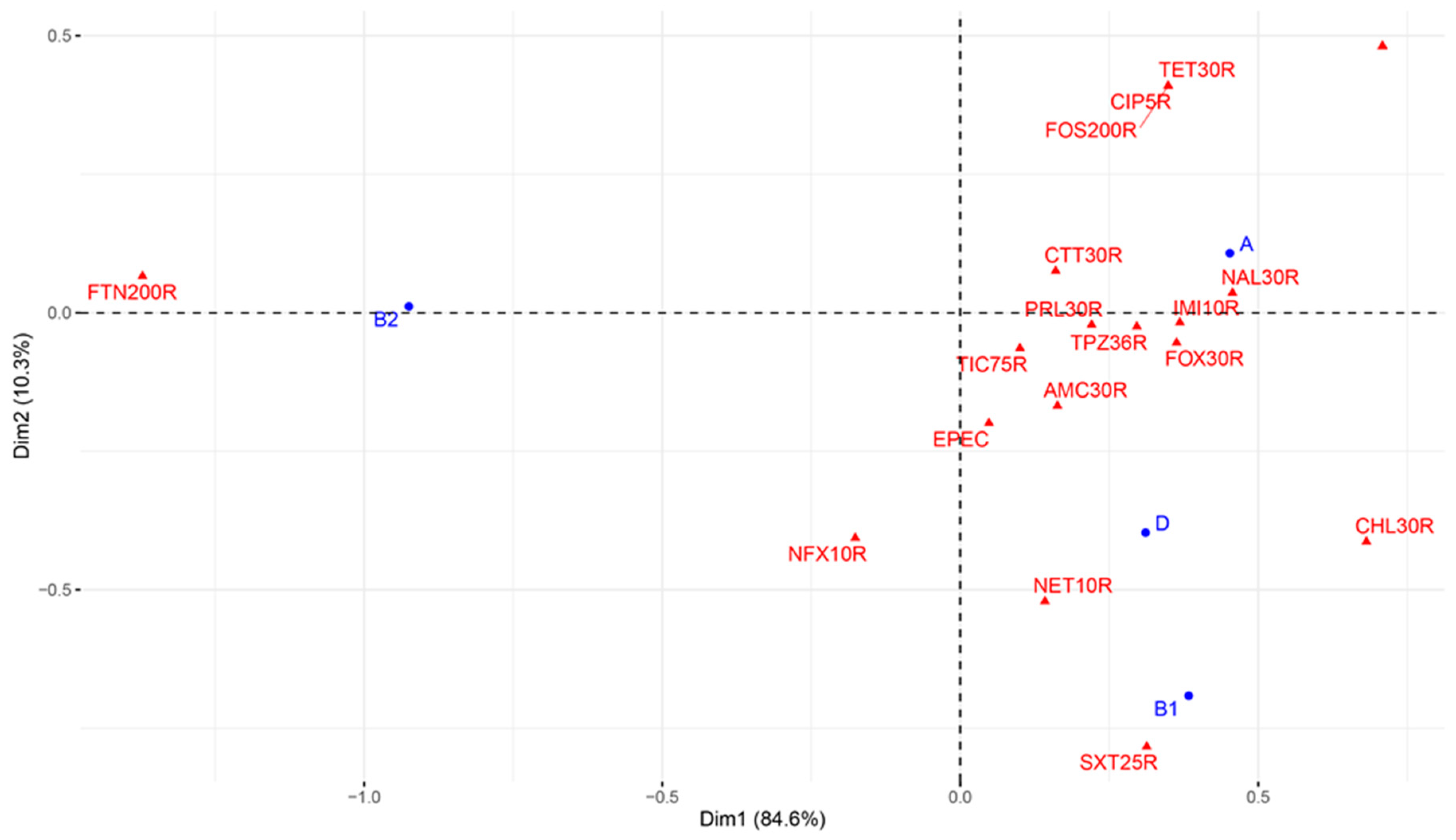
3.5. Association between Antimicrobial-Resistant DECs and Virulence in E. coli
4. Materials and Methods
- Feld research authorization
4.1. Study Area
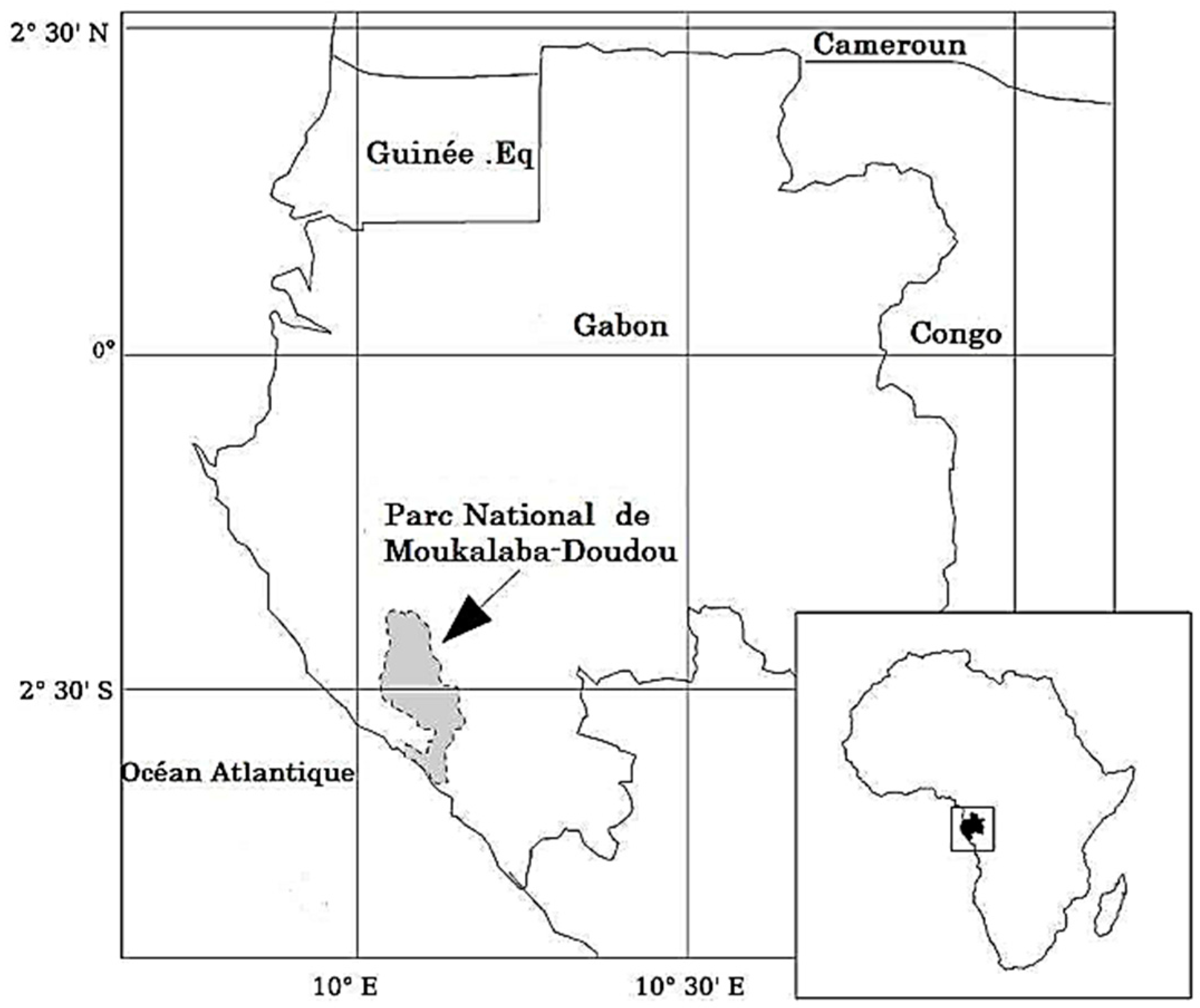
4.2. Sample Collection
4.3. Culture, Isolation and Identification of Colonies
4.4. Antimicrobial Susceptibility Testing of E. coli Isolates
4.5. Molecular Identification of E. coli Phylogenetic and Pathotype Groups
DNA Extraction
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chomel, B.B.; Belotto, A.; Meslin, F.-X. Wildlife, exotic pets, and emerging zoonoses. Emerg. Infect. Dis. 2007, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001, 78, 103–116. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, N.D.; Dunavan, C.P.; Diamond, J. Origins of major human infectious diseases. Nature 2007, 447, 279–283. [Google Scholar] [CrossRef]
- Lloyd-Smith, J.O.; Georges, D.; Pepin, K.M.; Pitzer, V.E.; Pulliam, J.R.C.; Dobson, A.P.; Hudson, P.J.; Grenfell, B.T. Epidemic dynamics at the human-animal interface. Science 2009, 326, 1362–1367. [Google Scholar] [CrossRef]
- Dobson, A.; Foufopoulos, J. Emerging infectious pathogens of wildlife. Philos. Trans. Royal. Soc. Lond. Ser. B Biol. Sci. 2001, 356, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Kahn, R.; Clouser, D.; Richt, J. Emerging infections: A tribute to the one medicine, one health concept. Zoonoses Public Health 2009, 56, 407–428. [Google Scholar] [CrossRef]
- Richt, J.; Feldmann, H. Emerging zoonoses: Recent advances and future challenges. Zoonoses Public Health 2009, 56, 257. [Google Scholar] [CrossRef]
- Kruse, H.; Kirkemo, A.-M.; Handeland, K. Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 2004, 10, 2067. [Google Scholar] [CrossRef]
- Rabinowitz, P.; Scotch, M.; Conti, L. Human and animal sentinels for shared health risks. Vet. Ital. 2009, 45, 23. [Google Scholar]
- Quammen, D. Spillover: Animal Infections and the Next Human Pandemic; WW Norton & Company: New York, NY, USA, 2012. [Google Scholar]
- Quammen, D. Ebola: The Natural and Human History of a Deadly Virus; WW Norton & Company: New York, NY, USA, 2014. [Google Scholar]
- Köndgen, S.; Kühl, H.; N’Goran, P.K.; Walsh, P.D.; Schenk, S.; Ernst, N.; Biek, R.; Formenty, P.; Mätz-Rensing, K.; Schweiger, B. Pandemic human vi-ruses cause decline of endangered great apes. Curr. Biol. 2008, 18, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Palacios, G.; Lowenstine, L.J.; Cranfield, M.R.; Gilardi, K.V.K.; Spelman, L.; Lukasik-Braum, M.; Kinani, J.-F.; Mudakikwa, A.; Nyirakaragire, E.; Bussetti, A.V.; et al. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerg. Infect. Dis. 2011, 17, 711. [Google Scholar] [CrossRef]
- Calvignac-Spencera, S.; Leendertza, S.A.J.; Gillespie, T.R.; Leendertza, F.H. Wild great apes as sentinels and sources of infectious disease. Clin. Microbiol. Infect. 2012, 18, 521–527. [Google Scholar] [CrossRef]
- Schaumburg, F.; Mugisha, L.; Kappeller, P.; Fichtel, C.; Köck, R.; Köndgen, S.; Becker, K.; Boesch, C.; Peters, G.; Leendertz, F.H. Evaluation of non-invasive biological samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS ONE 2013, 8, e78046. [Google Scholar] [CrossRef]
- Nowrouzian, F.L.; Wold, A.E.; Adlerberth, I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis. 2005, 191, 1078–1083. [Google Scholar] [CrossRef]
- Nowrouzian, F.L.; Wold, A.E.; Adlerberth, I. Enhanced persistence in the colonic microbiota of Escherichia coli strains be-longing to phylogenetic group B2: Role of virulence factors and adherence to colonic cells. Microbes Infect. 2006, 8, 834–840. [Google Scholar] [CrossRef]
- Johnson, J.R. Molecular epidemiology and population genetics of extraintestinal pathogenic Escherichia coli. In Population Genetics of Bacteria: A Tribute to Thomas, S. Whittam; ASM Press: Washington, DC, USA, 2011; pp. 91–107. [Google Scholar]
- Escobar-Páramo, P.; Le Menac’h, A.; Le Gall, T.; Amorin, C.; Gouriou, S.; Picard, B.; Skurnik, D.; Denamur, E. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 2006, 8, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M.; Cowling, A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: Host and geographic effects. Microbiology 2003, 149, 3575–3586. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, T.L.; Gillespie, T.R.; Rwego, I.B.; Estoff, E.L.; Chapman, C.A. Forest fragmentation as cause of bacterial transmission among nonhuman primates, humans, and livestock, Uganda. Emerg. Infect. Dis. 2008, 14, 1375. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.; Cranfield, M.; Gaffikin, L.; Kalema-Zikusoka, G.; Köndgen, S.; Leendertz, S.; Lonsdorf, E.; Muehlenbein, M.; Mugisha, L.; Nizeyi, J.B.; et al. Best Practice Guidelines for Health Monitoring and Disease Control in Great Ape Populations; Occasional Paper of the IUCN Species Survival Commission No. 56; IUCN Species Survival Commission: Gland, Switzerland, 2015; p. 56. [Google Scholar]
- Walsh, P.D.; Abernethy, K.A.; Bermejo, M.; Beyers, R.; De Wachter, P.; Akou, M.E.; Huijbregts, B.; Mambounga, D.I.; Toham, A.K.; Kilbourn, A.M.; et al. Catastrophic ape decline in western equatorial Africa. Nature 2003, 422, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.M.; Rouquet, P.; Formenty, P.; Souquière, S.; Kilbourne, A.; Froment, J.-M.; Bermejo, M.; Smit, S.; Karesh, W.; Rollin, P.E. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science 2004, 303, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Genton, C.; Cristescu, R.; Gatti, S.; Levréro, F.; Bigot, E.; Caillaud, D.; Pierre, J.-S.; Ménard, N. Recovery potential of a western lowland gorilla population following a major Ebola outbreak: Results from a ten year study. PLoS ONE 2012, 7, e37106. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, M.G. CTX-M-type β-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef]
- Lawick-Goodall, J.V. The Shadow of Man; Houghton Mifflin Company: Boston, MA, USA, 1971. [Google Scholar]
- Wallis, J. Prevention of disease transmission in primate conservation. Ann. N. Y. Acad. Sci. 2000, 916, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Ferber, D. Human diseases threaten great apes. Science 2000, 289, 1277–1278. [Google Scholar] [CrossRef] [PubMed]
- Woodford, M.H.; Butynski, T.M.; Karesh, W.B. Habituating the great apes: The disease risks. Oryx 2002, 36, 153–160. [Google Scholar] [CrossRef]
- Kaur, T.; Singh, J.; Tong, S.; Humphrey, C.; Clevenger, D.; Tan, W.; Szekely, B.; Wang, Y.; Li, Y.; Muse, E.A.; et al. Descriptive epidemiology of fatal respiratory outbreaks and detection of a human-related metapneumovirus in wild chimpanzees (Pan troglodytes) at Mahale Mountains National Park, Western Tanzania. Am. J. Primatol. 2008, 70, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Deem, S.L. Conservation Medicine: A Solution-Based Approach for Saving Nonhuman Primates. In Ethnoprimatology. Developments in Primatology: Progress and Prospects; Waller, M.T., Ed.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Estrada, A.; Garber, P.A.; Rylands, A.B.; Roos, C.; Fernandez-Duque, E.; Di Fiore, A.; Nekaris, K.A.-I.; Nijman, V.; Heymann, E.W. Impending extinction crisis of the world’s primates: Why primates matter. Sci. Adv. 2017, 3, e1600946. [Google Scholar] [CrossRef]
- Gillespie, T.R.; Nunn, C.L.; Leendertz, F.H. Integrative approaches to the study of primate infectious disease: Implications for biodiversity conservation and global health. Am. J. Phys. Anthr. 2008, 137, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, T.L.; Gillespie, T.R.; Rwego, I.B.; Estoff, E.L.; Chapman, C.A. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol. Conserv. 2007, 135, 511–517. [Google Scholar] [CrossRef]
- Rwego, I.B.; Isabirye-Basuta, G.; Gillespie, T.R.; Goldberg, T.L. Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conserv. Biol. 2008, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Yamagiwa, J. Creating a Future in which Human Beings and Wild Animals Can Live in Harmony in the Tropical Forests of Africa. Japan Science and Technology Agency. 2006. Available online: https://www.jst.go.jp/global/english/kadai/h2006_gabon.html (accessed on 7 November 2013).
- Matsuura, N.; Takenoshita, Y.; Yamagiwa, J. Eco-anthropologie et primatologie pour la conservation de la biodiversité: Un projet collaboratif dans le Parc National de Moukalaba-Doudou, Gabon. Rev. Primatol. 2013. [CrossRef]
- Alvarez, D.A.K.A.; Völlm, J. Maladies zoonotiques partagées par les gorilles et les humains. Gorilla J. 2004, 29, 17–18. (In French). [Google Scholar]
- Nguema, P.P.M.; Okubo, T.; Tsuchida, S.; Fujita, S.; Yamagiwa, J.; Tamura, Y.; Ushida, K. Isolation of multiple drug-resistant enteric bacteria from feces of wild Western Lowland Gorilla (Gorilla gorilla gorilla) in Gabon. J. Vet. Med. Sci. 2015, 77, 619–623. [Google Scholar] [CrossRef]
- Nguema, P.P.M.; Tsuchida, S.; Ushida, K. Bacteria culturing and isolation under field conditions of Moukalaba-Doudou National Park, Gabon, and preliminary survey on bacteria carrying antibiotic resistance genes. Tropics 2015, 23, 165–174. [Google Scholar] [CrossRef]
- Nguema, P.P.M.; Onanga, R.; Atome, G.R.N.; Mbeang, J.C.O.; Mabika, A.M.; Yaro, M.; Lounnas, M.; Dumont, Y.; Zohra, Z.F.; Godreuil, S.; et al. Characterization of ESBL-producing enterobacteria from fruit bats in an unprotected area of Makokou, Gabon. Microorganisms 2020, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.A.; Godreuil, S.; Bodenham, R.; Ratiarison, S.; Devos, C.; Petretto, M.-O.; Raymond, M.; Escobar-Páramoa, P. No evidence for transmission of antibiotic-resistant Escherichia coli strains from humans to wild western lowland gorillas in Lopé National Park, Gabon. Appl. Environ. Microbiol. 2012, 78, 4281–4287. [Google Scholar] [CrossRef]
- Nguema, P.P.M.; Onanga, R.; Atome, G.R.N.; Tewa, J.J.; Mabika, A.M.; Nzambe, J.U.M.; Mbeang, J.C.O.; Essono, P.Y.B.; Bretagnolle, F.; Godreuil, S. High level of intrinsic phenotypic antimicrobial resistance in enterobacteria from terrestrial wildlife in Gabonese national parks. PLoS ONE 2021, 16, e0257994. [Google Scholar]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H.; et al. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS ONE 2012, 7, e53039. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Shaik, S.; Ranjan, A.; Nandanwar, N.; Tiwari, S.K.; Majid, M.; Baddam, R.; Qureshi, I.A.; Semmler, T.; Wieler, L.H.; et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 2017, 8, 2120. [Google Scholar] [CrossRef] [PubMed]
- Wieler, L.H.; Ewers, C.; Guenther, S.; Walther, B.; Lübke-Becker, A. Methicillin-resistant staphylococci (MRS) and extended-spectrum beta-lactamases (ESBL)-producing Enterobacteriaceae in companion animals: Nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. Int. J. Med. Microbiol. 2011, 301, 635–641. [Google Scholar] [PubMed]
- Mazurek-Popczyk, J.; Pisarska, J.; Bok, E.; Baldy-Chudzik, K. Antibacterial activity of bacteriocinogenic commensal Escherichia coli against zoonotic strains resistant and sensitive to antibiotics. Antibiotics 2020, 9, 411. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M.; Clermont, O.; Tolley, H.; Denamur, E. Assigning Escherichia coli strains to phylogenetic groups: Multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 2008, 10, 2484–2496. [Google Scholar] [CrossRef]
- Koneman, E.W.; Allen, S.D.; Janda, W.; Schreckenberger, P.; Winn, W. Diagnostic Microbiology. The Nonfermentative Gram-Negative Bacilli; Lippincott-Raven Publishers: Philadelphia, PA, USA, 1997; pp. 253–320. [Google Scholar]
- Baneth, G.; Aroch, I.; Tal, N.; Harrus, S. Hepatozoon species infection in domestic cats: A retrospective study. Vet. Parasitol. 1998, 79, 123–133. [Google Scholar] [CrossRef]
- Zogg, A.L.; Zurfluh, K.; Schmitt, S.; Nüesch-Inderbinen, M.; Stephan, R. Antimicrobial resistance, multilocus sequence types and virulence profiles of ESBL producing and non-ESBL producing uropathogenic Escherichia coli isolated from cats and dogs in Switzerland. Vet. Microbiol. 2018, 216, 79–84. [Google Scholar] [CrossRef]
- Onanga, R.; Nguema, P.P.M.; Atome, G.R.N.; Mabika, A.M.; Ngoubangoye, B.; Tonda, W.L.K.; Mbeang, J.C.O.; Lebibi, J. Prevalence of Extended-Spectrum β-Lactamases in E. coli of Rats in the Region North East of Gabon. Vet. Med. Int. 2020, 2020, 5163493. [Google Scholar] [CrossRef]
- Renaud, F.; Vittecoq, M. Faune Sauvage et Antibiorésistances. In La Revue d’Humanité et Biodiversité; Humanité et Biodiversité: Paris, France, 2015; p. 84. [Google Scholar]
- Nys, G. Revue Bibliographique des Antibiorésistances Portées par les Entérobactéries Isolées de Reptiles et Evaluation du Profil de Résistance d’Escherichia coli Isolées de Reptiles en Belgique. Master’s Thesis, Université de Liège, Liège, Belgium, 2021. [Google Scholar]
- Aberkane, S. Dépistage et Caractérisation de Bactéries Multirésistantes aux Antibiotiques au Sein d’un Réservoir Aviaire Méditerranéen. Ph.D. Thesis, Université de Montpellier, Montpellier, France, 2017. [Google Scholar]
- Waititu, K.K. Molecular Characterization and Antimicrobial Susceptibility Patterns of Escherichia coli in Captive and Wild Olive Baboon (Papio anubis) Gut. Master’s Thesis, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya, 2020. [Google Scholar]
- Kolappaswamy, K.; Nazareno, J.; Porter, W.P.; Klein, H.J. Outbreak of pathogenic Escherichia coli in an outdoor-housed non-human primate colony. J. Med. Primatol. 2014, 43, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gouali, M.; Weill, F.-X. Les Escherichia coli entérohémorragiques: Des entérobactéries d’actualité. Presse Méd. 2013, 42, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Le Bouguénec, C. Diagnostic des différents pathovars d’Escherichia coli responsables de diarrhées chez l’homme. Rev. Francoph. Lab. 1999, 314, 33–37. [Google Scholar]
- Szalo, I.M.; Goffaux, F.; Pirson, V.; Piérard, D.; Ball, H.; Mainila, J. Presence in bovine enteropathogenic (EPEC) and enterohaemorrhagic (EHEC) Escherichia coli of genes encoding for putative adhesins of human EHEC strains. Res. Microbiol. 2002, 153, 653–658. [Google Scholar] [CrossRef]
- Hu, J.; Torres, A.G. Enteropathogenic Escherichia coli: Foe or innocent bystander? Clin. Microbiol. Infect. 2015, 21, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Frankel, G. Enteropathogenic Escherichia Coli: Unravelling Pathogenesis. FEMS Microbiol. Rev. 2005, 29, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Milton, A.A.P.; Agarwal, R.K.; Priya, G.B.; Aravind, M.; Athira, C.K.; Rose, L.; Saminathan, M.; Sharma, A.K.; Kumar, A. Captive wildlife from India as carriers of Shiga toxin-producing, Enteropathogenic and Enterotoxigenic Escherichia coli. J. Vet. Med. Sci. 2018, 81, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.G.; Lin, K.-C.; Newman, J.; Schauer, D.; MacKey, J.; Lackner, A.A.; Carville, A. Identification of Enteropathogenic Escherichia coli in Simian Immunodeficiency Virus-Infected Infant and Adult Rhesus Macaques. J. Clin. Microbiol. 2001, 39, 971–976. [Google Scholar] [CrossRef]
- Mansfield, K.G.; Lin, K.-C.; Xia, D.; Newman, J.V.; Schauer, D.B.; Mackey, J.; Lackner, A.A.; Carville, A. Enteropathogenic Escherichia coli and Ulcerative Colitis in Cotton-Top Tamarins (Saguinus Oedipus). J. Infect. Dis. 2001, 184, 803–807. [Google Scholar] [CrossRef]
- Carvalho, V.d.; Irino, K.; Onuma, D.; Castro, A.F. Random amplification of polymorphic DNA reveals clonal relationships among enteropathogenic Escherichia coli isolated from non-human primates and humans. Braz. J. Med. Biol. Res. 2007, 40, 237–241. [Google Scholar] [CrossRef]
- Clayton, J.B.; Danzeisen, J.L.; Trent, A.M.; Emurphy, T.; Johnson, T.J. Longitudinal characterization of Escherichia coli in healthy captive non-human primates. Front. Vet. Sci. 2014, 1, 24. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Munns, K.; Alexander, T.; Entz, T.; Mirzaagha, P.; Yanke, L.J.; Mulvey, M.; Topp, E.; McAllister, T. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 2008, 74, 6178–6186. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.G.; Bueris, V.; Porangaba, T.M.; Navarro-Garcia, F.; Elias, W.P. Auto transporter protein-encoding genes of diarrheagenic Escherichia coli are found in both typical and atypical enteropathogenic E. coli strains. Appl. Environ. Microbiol. 2012, 79, 4114. [Google Scholar] [CrossRef]
- Carvalho, V.M.; Gyles, C.L.; Ziebell, K.; Ribeiro, M.A.; Catão-Dias, J.L.; Sinhorini, I.L.; Otman, J.; Keller, R.; Trabulsi, L.R.; de Castro, A.F.P. Characterization of monkey enteropathogenic Escherichia coli (EPEC) and human typical and atypical EPEC serotype isolates from neotropical nonhuman primates. J. Clin. Microbiol. 2003, 41, 1225–1234. [Google Scholar] [CrossRef]
- Chen, F.G. Enteropathogenic Escherichia coli: Unravelling pathogenesis. FEMS Microbiol. Rev. 2005, 29, 83–98. [Google Scholar] [CrossRef]
- Drzewiecka, D. Significance and roles of Proteus spp. bacteria in natural environments. Microb. Ecol. 2016, 72, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Mwova, J.P. Antimicrobial Resistance Genes Harboured in Enterococci Isolated from the Faeces of Captive Baboons. Ph.D. Thesis, University of Nairobi, Nairobi, Kenya, 2016. [Google Scholar]
- Bachiri, T.; Bakour, S.; Ladjouzi, R.; Thongpan, L.; Rolain, J.M.; Touati, A. High rates of CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae in wild boars and Barbary macaques in Algeria. J. Glob. Antimicrob. Resist. 2017, 8, 35–40. [Google Scholar] [CrossRef]
- Dolejska, M.; Cizek, A.; Literak, I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 2007, 103, 11–19. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Lanza, V.F.; Rodríguez-Beltrán, J.; Galán, J.C.; Millán, A.S.; Cantón, R.; Coque, T.M. Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 2021, 34, e0005019. [Google Scholar] [CrossRef]
- Baron, S.A.; Mediannikov, O.; Abdallah, R.; Yimagou, E.K.; Medkour, H.; Dubourg, G.; Elamire, Y.; Afouda, P.; Ngom, I.I.; Angelakis, E. Multidrug-Resistant Klebsiella Pneumoniae Clones from Wild Chimpanzees and Termites in Senegal. Antimicrob. Agents Chemother. 2021, 65, AAC0255720. [Google Scholar] [CrossRef]
- Bamunusinghage, N.P.D.; Neelawala, R.G.; Magedara, H.P.; Ekanayaka, N.W.; Kalupahana, R.S.; Silva-Fletcher, A.; Kottawatta, S.A. Antimicrobial Resistance Patterns of Fecal Escherichia coli in Wildlife, Urban Wildlife, and Livestock in the Eastern Region of Sri Lanka, and Differences between Carnivores, Omnivores, and Herbivores. J. Wildl. Dis. 2022, 58, 380–383. [Google Scholar] [CrossRef]
- Turchi, B.; Dec, M.; Bertelloni, F.; Winiarczyk, S.; Gnat, S.; Bresciani, F.; Viviani, F.; Cerri, D.; Fratini, F. Antibiotic Susceptibility And Virulence Factors In Escherichia coli From Sympatric Wildlife Of The Apuan Alps Regional Park (Tuscany, Italy). Microb. Drug Resist. 2019, 25, 772–780. [Google Scholar] [CrossRef]
- Albrechtova, K.; Papousek, I.; de Nys, H.; Pauly, M.; Anoh, E.; Mossoun, A.; Dolejska, M.; Masarikova, M.; Metzger, S.; Couacy-Hymann, E. Low rates of antimicrobial-resistant Enterobacteriaceae in wildlife in Taï National Park, Côte d’Ivoire, surrounded by villages with high prevalence of multiresistant ESBL-producing Escherichia coli in people and domestic animals. PLoS ONE 2014, 9, e113548. [Google Scholar] [CrossRef] [PubMed]
- Janatova, M.; Albrechtova, K.; Petrzelkova, K.J.; Dolejska, M.; Papousek, I.; Masarikova, M.; Cizek, A.; Todd, A.; Shutt, K.; Kalousova, B.; et al. Antimicrobial-resistant Enterobacteriaceae from humans and wildlife in Dzanga-Sangha protected area, Central African Republic. Vet. Microbiol. 2014, 171, 422–431. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment-occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef] [PubMed]
- Verlicchi, P.; Al Aukidy, M.; Galletti, A.; Petrovic, M.; Barceló, D. Hospital effluent: Investigation of the concentrations and distribution of pharmaceuticals and environmental risk assessment. Sci. Total Environ. 2012, 430, 109–118. [Google Scholar] [CrossRef]
- Cardoza, L.A.; Knapp, C.W.; Larive, C.K.; Belden, J.B.; Lydy, M.; Graham, D.W. Factors affecting the fate of ciprofloxacin in aquatic field systems. Water Air Soil. Poll. 2005, 161, 383–398. [Google Scholar] [CrossRef]
- Hanna, N.; Sun, P.; Sun, Q.; Li, X.; Yang, X.; Ji, X.; Zou, H.; Ottoson, J.; Nilsson, L.E.; Berglund, B.; et al. Presence of antibiotic residues in various environmental compartments of Shandong province in eastern China: Its potential for resistance development and ecological and human risk. Environ. Int. 2018, 114, 131–142. [Google Scholar] [CrossRef]
- Reis, E.O.; Foureaux, A.F.S.; Rodrigues, J.S.; Moreira, V.R.; Lebron, Y.A.R.; Santos, L.V.S.; Amaral, M.C.S.; Langea, L.C. Occurrence, removal and seasonal variation of pharmaceuticals in Brasilian drinking water treatment plants. Environ. Pollut. 2019, 250, 773–781. [Google Scholar] [CrossRef]
- Marcusson, L.L.; Frimodt-Møller, N.; Hughes, D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009, 5, e1000541. [Google Scholar] [CrossRef]
- Baker, S.; Duy, P.T.; Nga, T.V.T.; Dung, T.T.N.; Phat, V.V.; Chau, T.T.; Turner, A.K.; Farrar, J.; Boni, M.F. Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. eLife 2013, 2, e01229. [Google Scholar] [CrossRef]
- Machuca, J.; Briales, A.; Labrador, G.; Díaz-de-Alba, P.; López-Rojas, R.; Docobo-Pérez, F.; Martínez-Martínez, L.; Rodríguez-Baño, J.; Pachón, M.E.; Pascual, Á. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J. Antimicrob. Chemother. 2014, 69, 3203–3215. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Sokurenko, E.V.; Tchesnokova, V. Intensity and Mechanisms of Fluoroquinolone Resistance within the H 30 and H 30Rx Subclones of Escherichia coli Sequence Type 131 Compared with Other Fluoroquinolone-Resistant, E. coli. Antimicrob. Agents Chemother. 2015, 59, 4471–4480. [Google Scholar] [CrossRef]
- Fuzi, M.; Szabo, D.; Csercsik, R. Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front. Microbiol. 2017, 8, 2261. [Google Scholar] [CrossRef] [PubMed]
- Rolland, R.M.; Hausfater, G.; Marshall, B. Antibiotic-resistant bacteria in wild primates: Increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 1985, 49, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, D.; Ruimy, R.; Andremont, A.; Amorin, C.; Rouquet, P.; Picard, B.; Denamur, E. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 1215–1219. [Google Scholar] [CrossRef]
- Literák, I.; Vanko, R.; Dolejská, M.; Čížek, A.; Karpíšková, R. Resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett. Appl. Microbiol. 2007, 45, 616–621. [Google Scholar] [CrossRef]
- Cohen, M.L. Epidemiology of drug resistance: Implications for a post-antimicrobial era. Science 1992, 257, 1050–1055. [Google Scholar] [CrossRef]
- Weiss, D.; Wallace, R.M.; Rwego, I.B.; Gillespie, T.R.; Chapman, C.A.; Singer, R.S.; Goldberg, T.L. Antibiotic-resistant Escherichia coli and class 1 integrons in humans, domestic animals, and wild primates in Rural Uganda. Appl. Environ. Microbiol. 2018, 84, e01632-18. [Google Scholar] [CrossRef]
- Foster-Nyarko, E.; Alikhan, N.-F.; Ravi, A.; Thilliez, G.; Thomson, N.M.; Baker, D.; Kay, G.; Cramer, J.D.; O’Grady, J.; Antonio, M.; et al. Genomic diversity of Escherichia coli isolates from non-human primates in the Gambia. Microb. Genom. 2020, 6, mgen000428. [Google Scholar] [CrossRef] [PubMed]
- Beatty, A.; Scott, K.; Tsai, P. Achieving Sustainable Global Capacity for Surveillance and Response to Emerging Diseases of Zoonotic Origin: Workshop Summary; National Academies Press: Washington, DC, USA, 2008. [Google Scholar]
- He, T.; Wei, R.; Zhang, L.; Sun, L.; Pang, M.; Wang, R.; Wang, Y. Characterization of Ndm-5-Positive Extensively Resistant Escherichia coli Isolates from Dairy Cows. Vet. Microbiol. 2017, 207, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Levy, K.; Trueba, G.; Cevallos, W.; Trostle, J.; Foxman, B.; Marrs, C.F.; Eisenberg, J.N.S. Effects of selection pressure and genetic association on the relationship between antibiotic resistance and virulence in Escherichia coli. Antimicrob. Agents Chemother. 2015, 59, 6733–6740. [Google Scholar] [CrossRef] [PubMed]
- Vega-Manriquez, X.D.; Ubiarco-López, A.; Verdugo-Rodríguez, A.; Hernández-Chiñas, U.; Navarro-Ocaña, A.; Ahumada-Cota, R.E.; Ramírez-Badillo, D.; de León, N.H.-D.; Eslava, C.A. Pet dogs potential transmitters of pathogenic Escherichia coli with resistance to antimicrobials. Arch. Microbiol. 2020, 202, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Walk, S.T.; Alm, E.W.; Calhoun, L.M.; Mladonicky, J.M.; Whittam, T.S. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 2007, 9, 2274–2288. [Google Scholar] [CrossRef]
- Gordon, D.M. The Influence of Ecological Factors on the Distribution and the Genetic Structure of Escherichia coli. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef] [PubMed]
- Pérez, C.; Gómez-Duarte, O.G.; Arias, M.L. Diarrheagenic Escherichia coli in children from Costa Rica. Am. J. Trop. Med. Hyg. 2010, 83, 292. [Google Scholar] [CrossRef] [PubMed]
- Adib, N.; Ghanbarpour, R.; Solatzadeh, H.; Alizade, H. Antibiotic resistance profile and virulence genes of uropathogenic Escherichia coli isolates in relation to phylogeny. Trop. Biomed. 2014, 31, 17–25. [Google Scholar] [PubMed]
- Vande Weghe, J. Les Parcs Nationaux du Gabon: Moukalaba Doudou; Agence Nationale des Parcs Nationaux: Libreville, Gabon, 2012; p. 296. [Google Scholar]
- Takenoshita, Y.; Ando, C.; Yamagiwa, J. Fruit phenology of the great ape habitat in the Moukalaba-Doudou National Park, Gabon. Afr. Study Monogr. 2008, 39, 23–39. [Google Scholar]
- Takenoshita, Y.; Yamagiwa, J. Estimating gorilla abundance by dung count in the northern part of Moukalaba-Doudou National Park, Gabon. Afr. Study Monogr. 2008, 39, 41–54. [Google Scholar]
- Nguelet, F.L.M.; Koumba, C.R.Z.; Mavoungou, J.F.; Nzengue, E.; Akomo-Okoue, E.F.; Nakashima, Y.; Hongo, S.; Ella, G.W.E.; Koumba, L.B.M.; M’batchi, B. Étude de la relation entre l’abondance des grands mammifères frugivores et celle des fruits dans le Parc National de Moukalaba-Doudou, Gabon. Int. J. Biol. Chem. Sci. 2016, 10, 1969–1982. [Google Scholar] [CrossRef][Green Version]
- Oluduro, A.O. Antibiotic-Resistant Commensal Escherichia coli in Faecal Droplets from Bats and Poultry in Nigeria. Vet. Ital. 2012, 48, 297–308. [Google Scholar] [PubMed]
- Jaja, I.F.; Green, E.; Muchenje, V. Aerobic mesophilic, coliform, Escherichia coli, and Staphylococcus aureus counts of raw meat from the formal and informal meat sectors in South Africa. Int. J. Environ. Res. Public Health 2018, 15, 819. [Google Scholar] [CrossRef] [PubMed]
- Jaja, I.F.; Jaja, C.-J.I.; Chigor, N.V.; Anyanwu, M.U.; Maduabuchi, E.K.; Oguttu, J.W.; Green, E. Antimicrobial resistance phenotype of Staphylococcus aureus and Escherichia coli isolates obtained from meat in the formal and informal sectors in South Africa. BioMed Res. Int. 2020, 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. M100: Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute. M100: Performance Standards for Antimicrobial Susceptibility Testing, 20th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Ibrahim, M.; Bilal, N.; Hamid, M. Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan. Afr. Health Sci. 2012, 12, 368–375. [Google Scholar] [CrossRef]
- Sjöling, Å.; Sadeghipoorjahromi, L.; Novak, D.; Tobias, J. Detection of major diarrheagenic bacterial pathogens by multiplex PCR panels. Microbiol. Res. 2015, 172, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.Y.C.; Lee, S.K.Y.; Law, T.W.F.; Law, S.H.W.; Wu, R.S.S. Detection of major diarrheagenic bacterial pathogens by multiplex PCR panels. Water Res. 2002, 36, 2802–2812. [Google Scholar] [CrossRef]
- Rodas, C.; Mamani, R.; Blanco, J.; Blanco, J.E.; Wiklund, G.; Svennerholm, A.-M.; Sjöling, Å.; Iniguez, V. Enterotoxins, colonization factors, serotypes and antimicrobial resistance of enterotoxigenic Escherichia coli (ETEC) strains isolated from hospitalized children with diarrhea in Bolivia. Braz. J. Infect. Dis. 2011, 15, 132–137. [Google Scholar] [PubMed]
- Vidal, R.; Vidal, M.; Lagos, R.; Levine, M.; Prado, V. Multiplex PCR for diagnosis of enteric infections associated with diarrheagenic Escherichia coli. J. Clin. Microbiol. 2004, 42, 1787–1789. [Google Scholar] [CrossRef]
| Subject Studied | Sample | chuA | yjaA | TSPE4.C2 | Phylogroup Assignation | Frequency (%) |
|---|---|---|---|---|---|---|
| Gorilla | Faecal | - | - | - | A (n = 44) | 69 |
| Gorilla | Faecal | - | + | - | A (n = 0) | 0 |
| Gorilla | Faecal | - | - | + | B1 (n = 6) | 10 |
| Gorilla | Faecal | + | + | - | B2 (n = 0) | 0 |
| Gorilla | Faecal | + | + | + | B2 (n = 13) | 20 |
| Gorilla | Faecal | + | - | - | D (n = 1) | 1 |
| Gorilla | Faecal | + | - | + | D (n = 0) | 0 |
| Samples | Pathotype Group | Frequency (%) |
|---|---|---|
| Faecal | EPEC (n = 17) | 85 |
| Faecal | EPEC/EHEC (n = 3) | 15 |
| Isolate | Animal | ATB | Class | Resistance Phenotype Profile | Phylogroup | Pathotype | MARI | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E19 | Gorilla | 5 | 1 | TIC | PIP | TZP | FOX | AMC | D | EPEC | 0.23 | ||||||||
| E24 | Gorilla | 5 | 1 | TIC | PIP | TZP | AMC | CTT | B2 | 0.23 | |||||||||
| E33 | Gorilla | 5 | 2 | PIP | TZP | AMC | NAL | CTT | A | 0.23 | |||||||||
| E32 | Gorilla | 6 | 3 | PIP | TZP | AMC | TET | NAL | CTT | A | 0.28 | ||||||||
| E40 | Gorilla | 6 | 3 | PIP | TZP | IPM | AMC | FOF | TOB | A | EPEC | 0.28 | |||||||
| E41 | Gorilla | 6 | 3 | PIP | TZP | IPM | TET | NAL | CTT | A | 0.28 | ||||||||
| E1 | Gorilla | 7 | 2 | TIC | PIP | FOX | IPM | ETP | FOF | CTT | A | 0.33 | |||||||
| E8 | Gorilla | 7 | 3 | TIC | TPZ | PIP | IPM | ETP | FOF | CIP | A | 0.33 | |||||||
| E28 | Gorilla | 7 | 1 | TIC | PIP | TZP | FOX | IPM | AMC | CTT | B2 | 0.33 | |||||||
| E45 | Gorilla | 8 | 2 | TIC | PIP | TZP | FOX | IPM | AMC | NAL | CTT | B1 | 0.38 | ||||||
| E57 | Gorilla | 8 | 2 | TIC | FOX | IPM | AMC | GEN | AMK | TOB | NET | A | 0.38 | ||||||
| E59 | Gorilla | 8 | 2 | TIC | FOX | IPM | AMC | GEN | AMK | TOB | NET | A | EPEC | 0.38 | |||||
| E64 | Gorilla | 8 | 6 | FOX | IPM | TET | CHL | NAL | SXT | NIT | CTT | A | EPEC | 0.38 | |||||
| E4 | Gorilla | 9 | 5 | TZP | IPM | ETP | FOF | GEN | TOB | CIP | NIT | LEV | A | 0.42 | |||||
| E7 | Gorilla | 9 | 3 | TZP | IPM | ETP | AMC | FOF | GEN | AMK | NIT | LEV | A | 0.42 | |||||
| E37 | Gorilla | 9 | 3 | TIC | PIP | TZP | FOX | IPM | AMC | NAL | SXT | CTT | B1 | 0.42 | |||||
| E50 | Gorilla | 9 | 3 | TIC | PIP | TZP | FOX | IPM | AMC | NET | SXT | CTT | B1 | 0.42 | |||||
| E49 | Gorilla | 11 | 6 | TIC | TZP | FOX | IPM | AMC | TET | CHL | NAL | SXT | NIT | CTT | A | 0.52 | |||
| E63 | Gorilla | 12 | 7 | TIC | PIP | TZP | FOX | IPM | AMC | NET | TET | CHL | NAL | CIP | NIT | B2 | EPEC | 0.57 | |
| E10 | Gorilla | 13 | 6 | TIC | TZP | PIP | FOX | IPM | ETP | AMC | FOF | TOB | CHL | NAL | CIP | CTT | A | 0.61 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oyaba Yinda, L.E.D.; Onanga, R.; Mbehang Nguema, P.P.; Akomo-Okoue, E.F.; Nsi Akoue, G.; Longo Pendy, N.M.; Otsaghe Ekore, D.; Lendamba, R.W.; Mabika-Mabika, A.; Mbeang, J.C.O.; et al. Phylogenetic Groups, Pathotypes and Antimicrobial Resistance of Escherichia coli Isolated from Western Lowland Gorilla Faeces (Gorilla gorilla gorilla) of Moukalaba-Doudou National Park (MDNP). Pathogens 2022, 11, 1082. https://doi.org/10.3390/pathogens11101082
Oyaba Yinda LED, Onanga R, Mbehang Nguema PP, Akomo-Okoue EF, Nsi Akoue G, Longo Pendy NM, Otsaghe Ekore D, Lendamba RW, Mabika-Mabika A, Mbeang JCO, et al. Phylogenetic Groups, Pathotypes and Antimicrobial Resistance of Escherichia coli Isolated from Western Lowland Gorilla Faeces (Gorilla gorilla gorilla) of Moukalaba-Doudou National Park (MDNP). Pathogens. 2022; 11(10):1082. https://doi.org/10.3390/pathogens11101082
Chicago/Turabian StyleOyaba Yinda, Leresche Even Doneilly, Richard Onanga, Pierre Philippe Mbehang Nguema, Etienne François Akomo-Okoue, Gontran Nsi Akoue, Neil Michel Longo Pendy, Desire Otsaghe Ekore, Roméo Wenceslas Lendamba, Arsène Mabika-Mabika, Jean Constant Obague Mbeang, and et al. 2022. "Phylogenetic Groups, Pathotypes and Antimicrobial Resistance of Escherichia coli Isolated from Western Lowland Gorilla Faeces (Gorilla gorilla gorilla) of Moukalaba-Doudou National Park (MDNP)" Pathogens 11, no. 10: 1082. https://doi.org/10.3390/pathogens11101082
APA StyleOyaba Yinda, L. E. D., Onanga, R., Mbehang Nguema, P. P., Akomo-Okoue, E. F., Nsi Akoue, G., Longo Pendy, N. M., Otsaghe Ekore, D., Lendamba, R. W., Mabika-Mabika, A., Mbeang, J. C. O., Poungou, N., Ibrahim, Mavoungou, J. F., & Godreuil, S. (2022). Phylogenetic Groups, Pathotypes and Antimicrobial Resistance of Escherichia coli Isolated from Western Lowland Gorilla Faeces (Gorilla gorilla gorilla) of Moukalaba-Doudou National Park (MDNP). Pathogens, 11(10), 1082. https://doi.org/10.3390/pathogens11101082







