Distribution of Theileria orientalis in Virginia Market Cattle, 2018–2020
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Blood Collection
2.2. Molecular Detection and Genotyping of T. orientalis
2.3. Statistical Analyses
2.4. Spatial Analyses
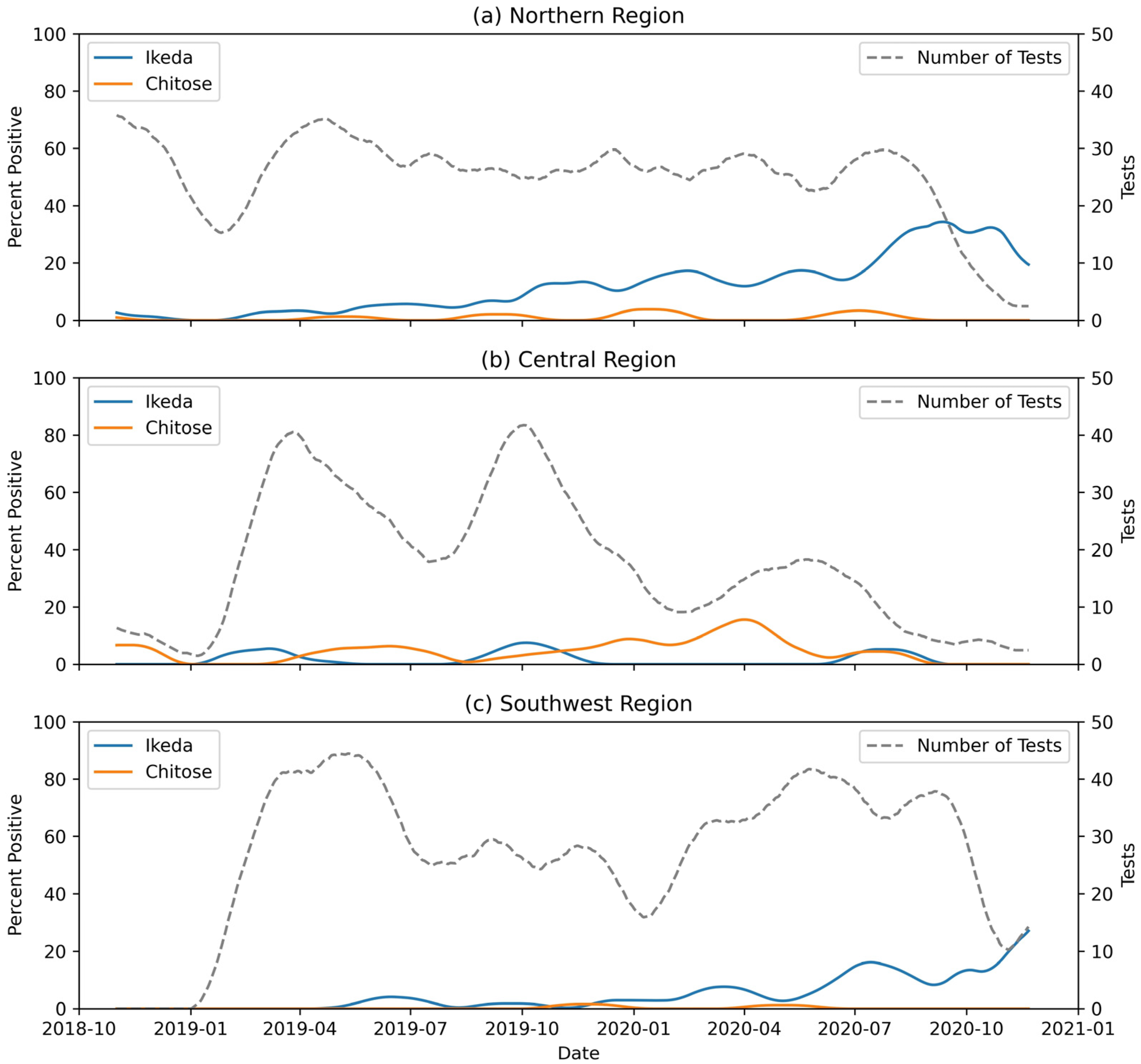
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hammer, J.F.; Emery, D.; Bogema, D.R.; Jenkins, C. Detection of Theileria orientalis genotypes in Haemaphysalis longicornis ticks from southern Australia. Parasit. Vectors 2015, 8, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, J.; Playford, M.; Hickey, K. Theileria orientalis: A review. N. Z. Vet. J. 2016, 64, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Oakes, V.J.; Yabsley, M.J.; Schwartz, D.; LeRoith, T.; Bissett, C.; Broaddus, C.; Schlater, J.L.; Todd, S.M.; Boes, K.M.; Brookhart, M.; et al. Theileria orientalis Ikeda genotype in cattle, Virginia, USA. Emerg. Infect. Dis. 2019, 25, 1653–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinkel, K.D.; Herndon, D.R.; Noh, S.M.; Lahmers, K.K.; Todd, S.M.; Ueti, M.W.; Scoles, G.A.; Mason, K.L.; Fry, L.M. A U.S. isolate of Theileria orientalis, Ikeda genotype, is transmitted to cattle by the invasive Asian longhorned tick, Haemaphysalis longicornis. Parasit. Vectors 2021, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, K.; Lawrence, B.; Hickson, R.; Hewitt, C.; Gedye, K.; Fermin, L.; Pomroy, W. Associations between Theileria orientalis Ikeda type infection and the growth rates and haematocrit of suckled beef calves in the North Island of New Zealand. N. Z. Vet. J. 2019, 67, 66–73. [Google Scholar] [CrossRef]
- Perera, P.K.; Gasser, R.B.; Firestone, S.M.; Anderson, G.A.; Malmo, J.; Davis, G.; Beggs, D.S.; Jabbar, A. Oriental theileriosis in dairy cows causes a significant milk production loss. Parasit. Vectors 2014, 7, 73. [Google Scholar] [CrossRef] [Green Version]
- Sivakumar, T.; Hayashida, K.; Sugimoto, C.; Yokoyama, N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014, 27, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Yu, D.-H.; Chae, J.-B.; Choi, K.-S.; Kim, H.-C.; Park, B.-K.; Chae, J.-S.; Park, J. Pathogenic genotype of major piroplasm surface protein associated with anemia in Theileria orientalis infection in cattle. Acta Vet. Scand. 2017, 59, 51. [Google Scholar] [CrossRef] [Green Version]
- Bogema, D.R.; Micallef, M.L.; Liu, M.; Padula, M.P.; Djordjevic, S.P.; Darling, A.E.; Jenkins, C. Analysis of Theileria orientalis draft genome sequences reveals potential species-level divergence of the Ikeda, Chitose and Buffeli genotypes. BMC Genom. 2018, 19, 298. [Google Scholar] [CrossRef]
- Chae, J.; Allsopp, B.A.; Waghela, S.D.; Park, J.; Kakuda, T.; Sugimoto, C.; Allsopp, M.T.E.P.; Gale Wagner, G.; Holman, P.J. A study of the systematics of Theileria spp. based upon small-subunit ribosomal RNA gene sequences. Parasitol. Res. 1999, 85, 877–883. [Google Scholar] [CrossRef]
- Cossio-Bayugar, R.; Pillars, R.; Schlater, J.; Holman, P.J. Theileria buffeli infection of a Michigan cow confirmed by small subunit ribosomal RNA gene analysis. Vet. Parasitol. 2002, 105, 105–110. [Google Scholar] [CrossRef]
- Stockham, S.L.; Kjemtrup, A.M.; Conrad, P.A.; Schmidt, D.A.; Scott, M.A.; Robinson, T.W.; Tyler, J.W.; Johnson, G.C.; Carson, C.A.; Cuddihee, P. Theileriosis in a Missouri Beef Herd Caused by Theileria buffeli. Vet. Pathol. 2000, 37, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Gebrekidan, H.; Gasser, R.B.; Stevenson, M.A.; Jabbar, A. multiplexed tandem PCR (MT-PCR) assay using the major piroplasm surface protein gene for the diagnosis of Theileria orientalis infection in cattle. J. Clin. Microbiol. 2018, 56, e01661-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.T.; White, S.; Shaw, D.; Egizi, A.; Lahmers, K.; Ruder, M.G.; Yabsley, M.J. Theileria orientalis Ikeda in host-seeking Haemaphysalis longicornis in Virginia, U.S.A. Ticks Tick. Borne. Dis. 2020, 11, 101450. [Google Scholar] [CrossRef] [PubMed]
- Rainey, T.; Occi, J.L.; Robbins, R.G.; Egizi, A. Discovery of Haemaphysalis longicornis (Ixodida: Ixodidae) parasitizing a sheep in New Jersey, United States. J. Med. Entomol. 2018, 55, 757–759. [Google Scholar] [CrossRef] [PubMed]
- Beard, C.B.; Occi, J.; Bonilla, D.L.; Egizi, A.M.; Fonseca, D.M.; Mertins, J.W.; Backenson, B.P.; Bajwa, W.I.; Barbarin, A.M.; Bertone, M.A.; et al. Multistate infestation with the exotic disease–vector tick Haemaphysalis longicornis—United States, August 2017–September 2018. MMWR. Morb. Mortal. Wkly. Rep. 2018, 67, 1310–1313. [Google Scholar] [CrossRef] [Green Version]
- Heath, A. Biology, ecology and distribution of the tick, Haemaphysalis longicornis Neumann (Acari: Ixodidae) in New Zealand. N. Z. Vet. J. 2016, 64, 10–20. [Google Scholar] [CrossRef]
- Fujisaki, K.; Kawazu, S.; Kamio, T. The taxonomy of the bovine Theileria spp. Parasitol. Today 1994, 10, 31–33. [Google Scholar] [CrossRef]
- USDA-APHIS. National Haemaphysalis longicornis Situation Report. Available online: https://www.aphis.usda.gov/animal_health/animal_diseases/tick/downloads/longhorned-tick-sitrep.pdf (accessed on 29 July 2022).
- Oakes, V.J.; Todd, S.M.; Carbonello, A.A.; Michalak, P.; Lahmers, K.K. Coinfection of cattle in Virginia with Theileria orientalis Ikeda genotype and Anaplasma marginale. J. Vet. Diagn. Investig. 2022, 34, 36–41. [Google Scholar] [CrossRef]
- Bogema, D.R.; Deutscher, A.T.; Fell, S.; Collins, D.; Eamens, G.J.; Jenkins, C. Development and validation of a quantitative PCR assay using multiplexed hydrolysis probes for detection and quantification of Theileria orientalis isolates and differentiation of clinically relevant subtypes. J. Clin. Microbiol. 2015, 53, 941–950. [Google Scholar] [CrossRef]
- Fay, M.P.; Hunsberger, S.A. Practical valid inferences for the two-sample binomial problem. Stat. Surv. 2021, 15, 72–110. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Fay, M.P. Two-sided exact tests and matching confidence intervals for discrete data. R J. 2010, 2, 53. [Google Scholar] [CrossRef] [Green Version]
- VDACS, Virginia Auction Summary Reports. Avaliable online: https://www.vdacs.virginia.gov/markets-and-finance-market-news-livestock-auctions.shtml (accessed on 5 June 2022).
- Friedman, J.H. Multivariate adaptive regression splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Harrell, F.E. General aspects of fitting regression models. In Regression Modeling Strategies; Springer: Berlin/Heidelberg, Germany, 2015; pp. 13–44. [Google Scholar]
- Milborrow, S. Derived from mda:mars by T. Hastie and R. Tibshirani., S. earth: Multivariate Adaptive Regression Splines 2011. Available online: https://cran.r-project.org/web/packages/earth/index.html (accessed on 11 July 2022).
- Hubert, L.J.; Golledge, R.G. Measuring association between spatially defined variables: Tjøstheim’s index and some extensions. Geogr. Anal. 1982, 14, 273–278. [Google Scholar] [CrossRef]
- Zuur, A.F.; Ieno, E.N.; Elphick, C.S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010, 1, 3–14. [Google Scholar] [CrossRef]
- Osorio, F.; Vallejos, R.; Cuevas, F. SpatialPack: Computing the association between two spatial processes. arXiv 2016, arXiv:1611.05289. [Google Scholar]
- Richardson, B. Virginia Market Cattle Data. 2022; unpublished raw data. [Google Scholar]
- Jacomy, M.; Venturini, T.; Heymann, S.; Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 2014, 9, e98679. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Girvan, M.; Newman, M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Warren, D.L.; Matzke, N.J.; Cardillo, M.; Baumgartner, J.B.; Beaumont, L.J.; Turelli, M.; Glor, R.E.; Huron, N.A.; Simões, M.; Iglesias, T.L.; et al. ENMTools 1.0: An R package for comparative ecological biogeography. Ecography 2021, 44, 504–511. [Google Scholar] [CrossRef]
- Vega, G.C.; Pertierra, L.R.; Olalla-Tárraga, M.Á. MERRAclim, a high-resolution global dataset of remotely sensed bioclimatic variables for ecological modelling. Sci. Data 2017, 4, 170078. [Google Scholar] [CrossRef] [PubMed]
- Dewitz, J. National Land Cover Database (NLCD) 2019 Products [Dataset]; US Geological Survey: Sioux Falls, SD, USA, 2021. Available online: https://www.mrlc.gov/data/nlcd-2019-land-cover-conus (accessed on 8 April 2022).
- Telionis, P.A. Lyme Disease and Forest Fragmentation in the Peridomestic Environment. Master’s Thesis, Virginia Tech, Blacksburg, VA, USA, 2020. [Google Scholar]
- Warren, D.L.; Glor, R.E.; Turelli, M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution 2008, 62, 2868–2883. [Google Scholar] [CrossRef] [PubMed]
- Cliff, A.D.; Ord, J.K. Spatial Processes: Models & Applications; Pion Ltd.: London, UK, 1981. [Google Scholar]
- Bivand, R. R packages for analyzing spatial data: A comparative case study with areal data. Geogr. Anal. 2022, 54, 488–518. [Google Scholar] [CrossRef]
- Hungerford, L.L. Use of spatial statistics to identify and test significance in geographic disease patterns. Prev. Vet. Med. 1991, 11, 237–242. [Google Scholar] [CrossRef]
- Ejigu, B.A.; Wencheko, E. Introducing covariate dependent weighting matrices in fitting autoregressive models and measuring spatio-environmental autocorrelation. Spat. Stat. 2020, 38, 100454. [Google Scholar] [CrossRef]
- Whittier, W.D.; Currin, N.; Currin, J.F. Anaplasmosis in Beef Cattle; Virginia Cooperative Extension: Blacksburg, VA, USA, 2005; Avaliable online: https://vtechworks.lib.vt.edu/bitstream/handle/10919/50705/400-465.pdf (accessed on 3 March 2022).
- Lawrence, K.; Gedye, K.; McFadden, A.; Pulford, D.; Heath, A.; Pomroy, W. Review of the New Zealand Theileria orientalis Ikeda type epidemic and epidemiological research since 2012. Pathogens 2021, 10, 1346. [Google Scholar] [CrossRef]
- Lakew, B.T.; Kheravii, S.K.; Wu, S.B.; Eastwood, S.; Andrew, N.R.; Jenkins, C.; Walkden-Brown, S.W. Endemic infection of cattle with multiple genotypes of Theileria orientalis on the Northern Tablelands of New South Wales despite limited presence of ticks. Ticks Tick. Borne. Dis. 2021, 12, 101645. [Google Scholar] [CrossRef]
- USDA-NASS, Census of Agriculture. 2017. Available online: https://www.nass.usda.gov/AgCensus (accessed on 15 May 2022).
- Laing, W.N. Cattle in Early Virginia. Ph.D. Thesis, University of Virginia, Charlottesville, VA, USA, 1952. [Google Scholar]
- Burmeister, C.A.; Conway, H.M.; Brodell, A.P. Economic Factors Affecting the Beef-Cattle Industry of Virginia; US Department of Agriculture: Washington, DC, USA, 1931.
- Hungerford, L.L.; Smith, R.D. Variations in seroprevalence and host factors for bovine Anaplasmosis in Illinois. Vet. Res. Commun. 1997, 21, 9–18. [Google Scholar] [CrossRef]
- Cumbie, A.N.; Trimble, R.N.; Eastwood, G. Pathogen spillover to an invasive tick species: First detection of Bourbon virus in Haemaphysalis longicornis in the United States. Pathogens 2022, 11, 454. [Google Scholar] [CrossRef]
- Selim, A.; Attia, K.; AlKahtani, M.D.F.; Albohairy, F.M.; Shoulah, S. Molecular epidemiology and genetic characterization of Theileria orientalis in cattle. Trop. Anim. Health Prod. 2022, 54, 178. [Google Scholar] [CrossRef]
- Jenkins, C.; Micallef, M.; Alex, S.M.; Collins, D.; Djordjevic, S.P.; Bogema, D.R. Temporal dynamics and subpopulation analysis of Theileria orientalis genotypes in cattle. Infect. Genet. Evol. 2015, 32, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Almazán, C.; Scimeca, R.C.; Reichard, M.V.; Mosqueda, J. Babesiosis and Theileriosis in North America. Pathogens 2022, 11, 168. [Google Scholar] [CrossRef] [PubMed]
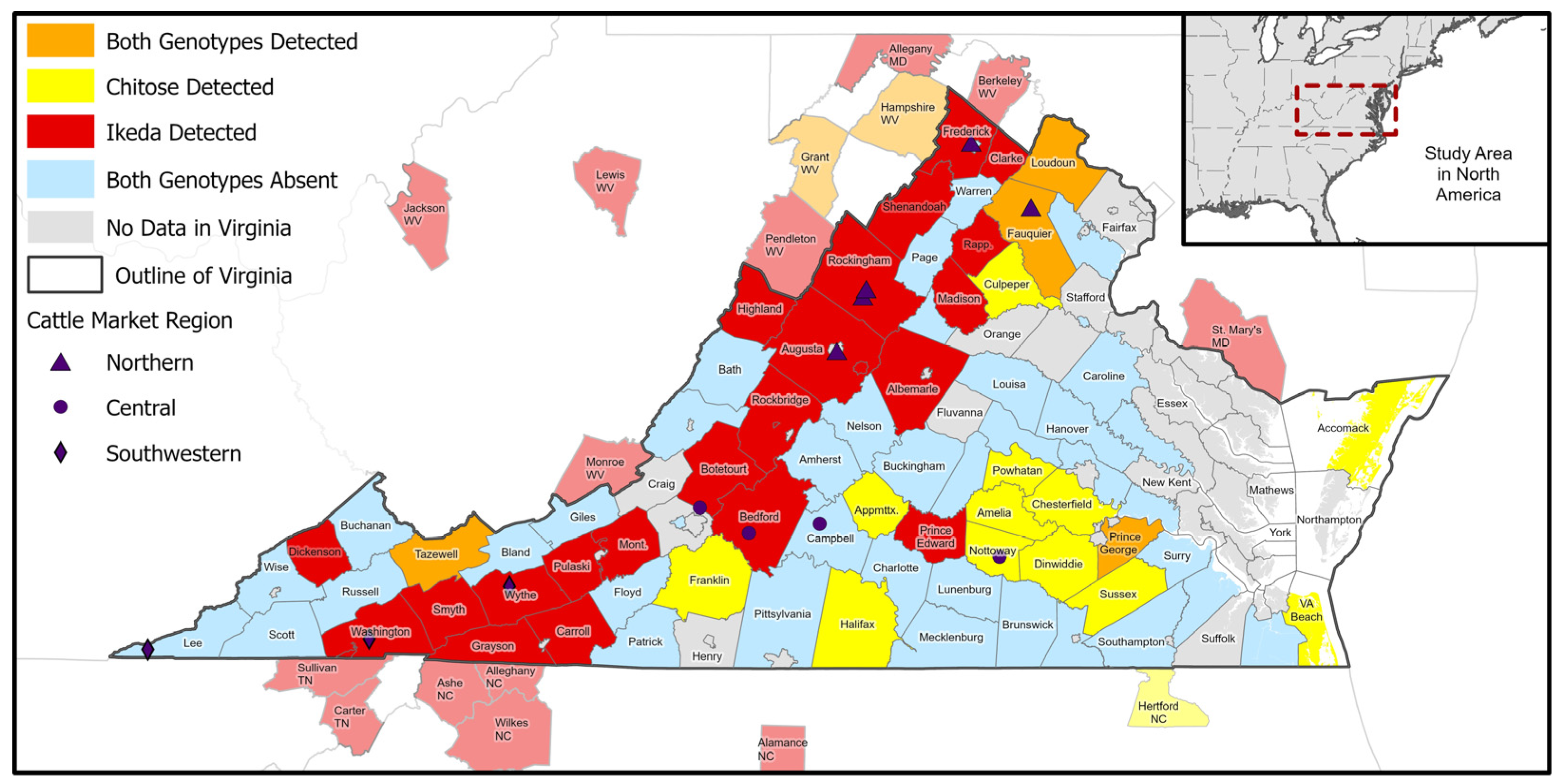
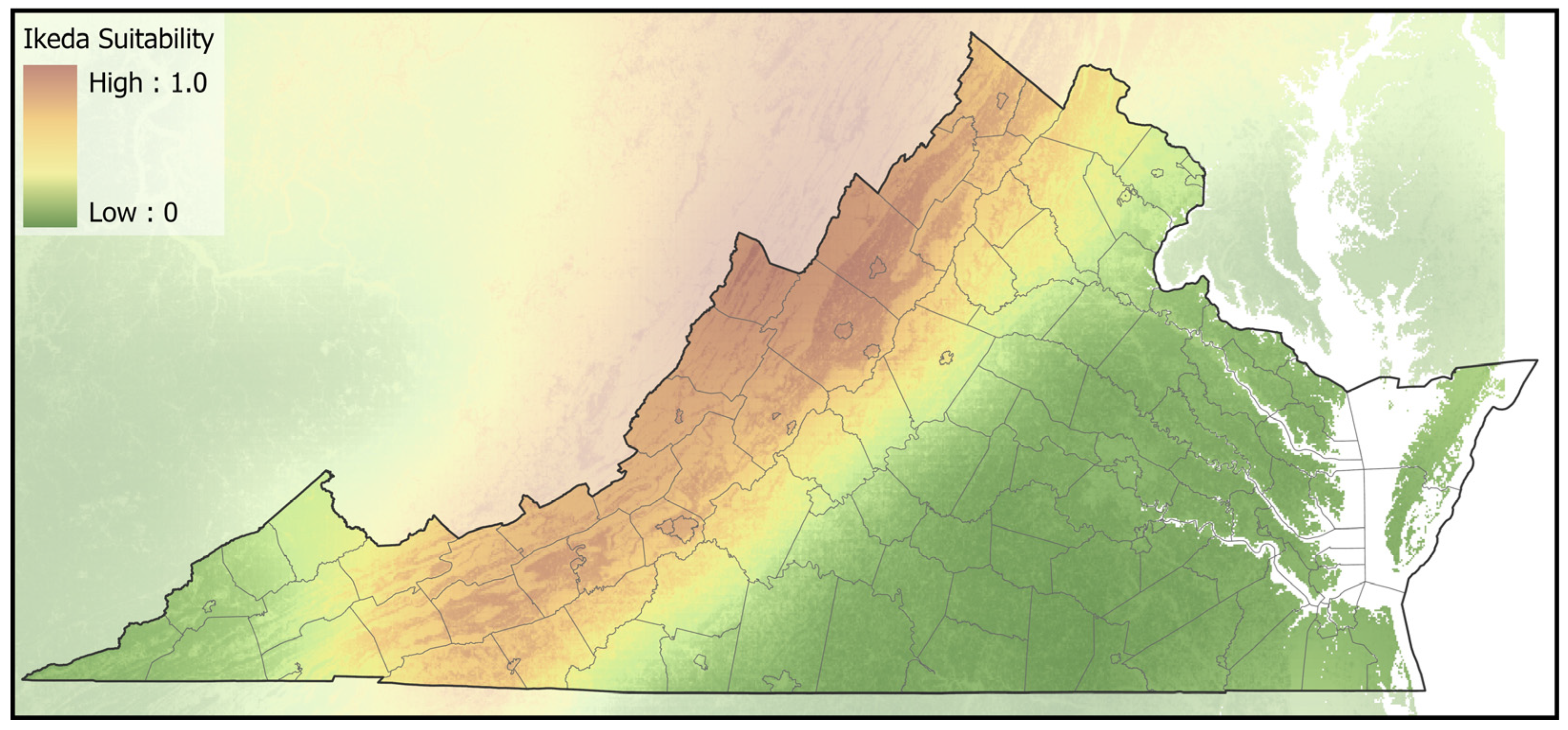
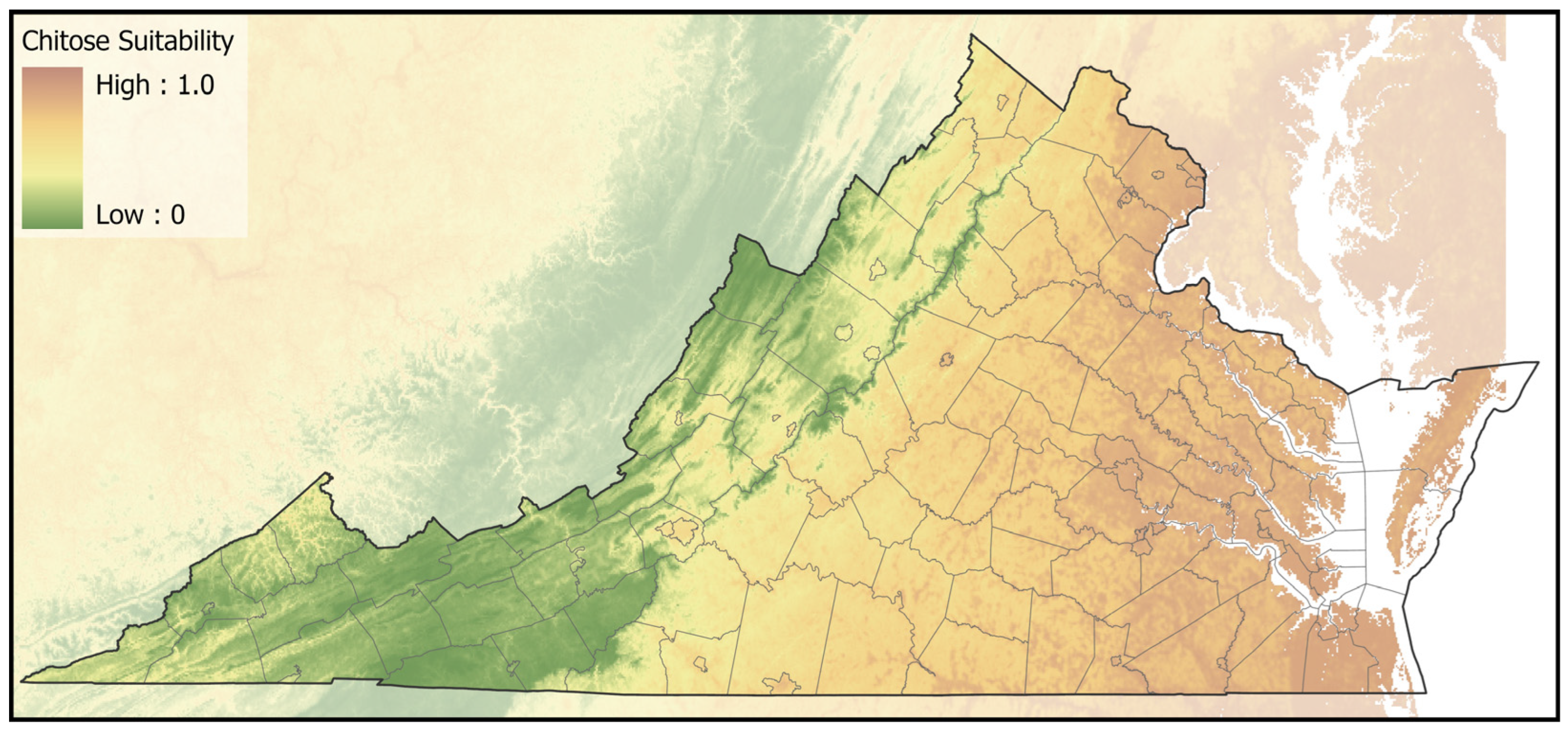
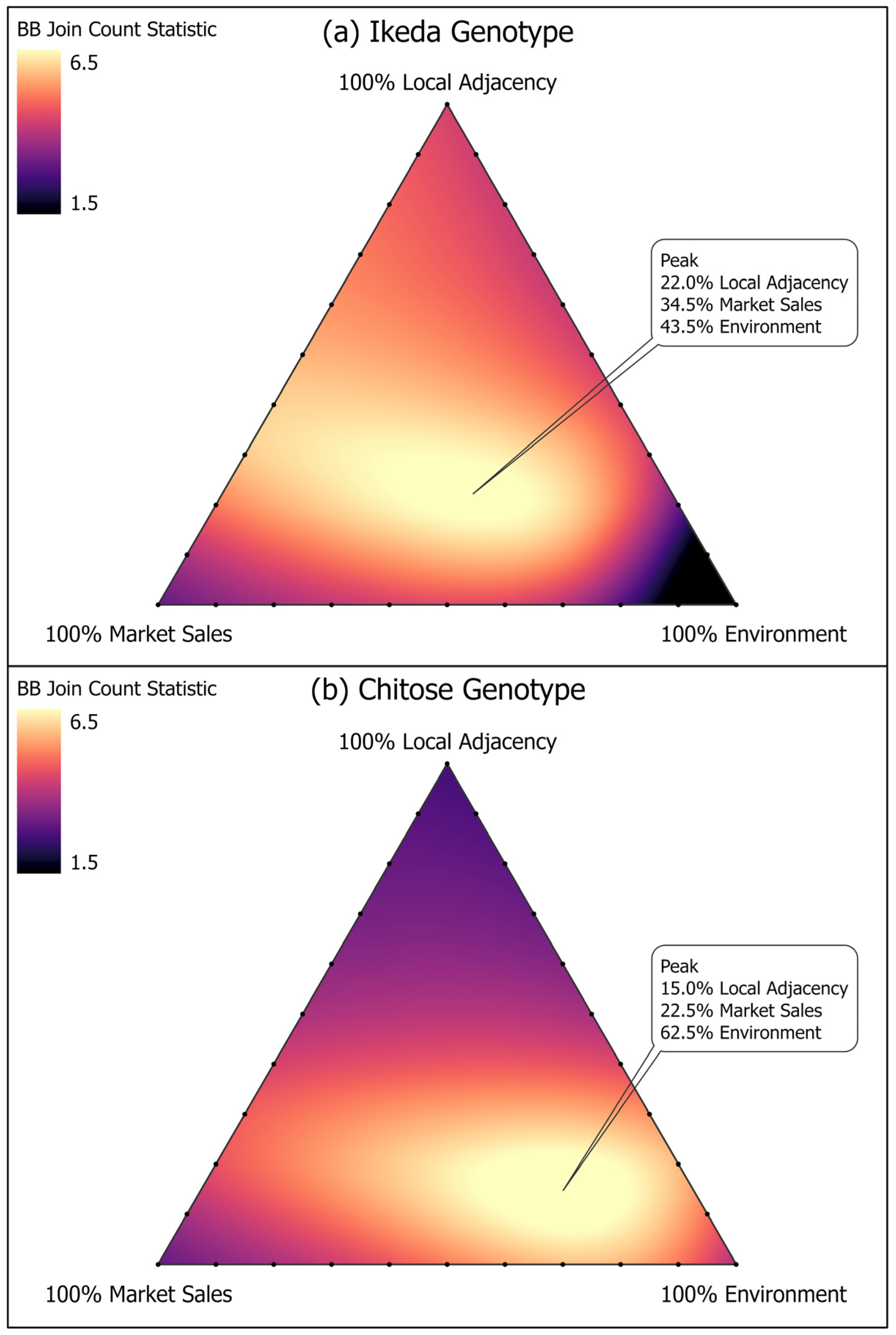
| Characteristic | Ikeda Positive (n = 172) | Chitose Positive (n = 36) | Theileria Negative (n = 1768) | Ikeda | Chitose | ||
|---|---|---|---|---|---|---|---|
| PR 1 | 95% CI 2 | PR 1 | 95% CI 2 | ||||
| Sex | |||||||
| Female | 172 | 36 | 1746 | Referent 3 | Referent 3 | ||
| Male | 0 | 0 | 7 | 0.00 | (0.00, 4.98) | 0.00 | (0.00, 20.67) |
| Unknown | 0 | 0 | 15 | 0.00 | (0.00, 2.82) | 0.00 | (0.00, 11.45) |
| Purpose | |||||||
| Beef | 154 | 36 | 1635 | Referent 3 | Referent 3 | ||
| Dairy | 18 | 0 | 107 | 1.67 | (0.97, 2.61) | 0.00 | (0.00, 1.72) |
| Unknown | 0 | 0 | 26 | 0.00 | (0.00, 1.83) | 0.00 | (0.00, 6.62) |
| Breed | |||||||
| Angus | 95 | 24 | 1093 | Referent 3 | Referent 3 | ||
| Cross-Bred | 49 | 8 | 337 | 1.59 * | (1.12, 2.19) | 1.08 | (0.44, 2.34) |
| Charolais | 4 | 3 | 77 | 0.62 | (0.16, 1.59) | 1.75 | (0.39, 5.32) |
| Holstein | 11 | 0 | 69 | 1.72 | (0.84, 3.02) | 0.00 | (0.00, 2.61) |
| Hereford | 5 | 1 | 72 | 0.81 | (0.26, 1.88) | 0.64 | (0.03, 3.67) |
| Other | 8 | 0 | 94 | 0.98 | (0.39, 1.93) | 0.00 | (0.00, 1.94) |
| Unknown | 0 | 0 | 26 | 0.00 | (0.00, 1.91) | 0.00 | (0.00, 6.56) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Telionis, A.; Lahmers, K.; Todd, M.; Carbonello, A.; Broaddus, C.C.; Bissett, C.J.; Hungerford, L.L. Distribution of Theileria orientalis in Virginia Market Cattle, 2018–2020. Pathogens 2022, 11, 1353. https://doi.org/10.3390/pathogens11111353
Telionis A, Lahmers K, Todd M, Carbonello A, Broaddus CC, Bissett CJ, Hungerford LL. Distribution of Theileria orientalis in Virginia Market Cattle, 2018–2020. Pathogens. 2022; 11(11):1353. https://doi.org/10.3390/pathogens11111353
Chicago/Turabian StyleTelionis, Alex, Kevin Lahmers, Michelle Todd, Amanda Carbonello, Charles C. Broaddus, Carolynn J. Bissett, and Laura L. Hungerford. 2022. "Distribution of Theileria orientalis in Virginia Market Cattle, 2018–2020" Pathogens 11, no. 11: 1353. https://doi.org/10.3390/pathogens11111353






