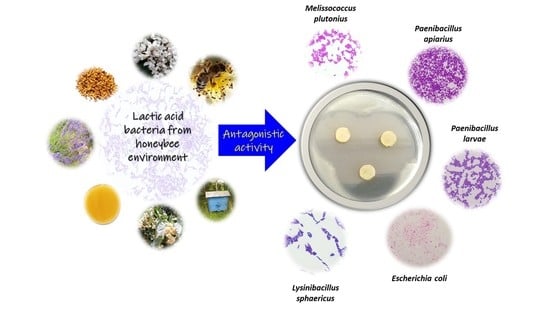
Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Vessels, and Other Materials
2.2. Biological Material
2.3. Culture, Propagation, Freezing, and Storage of Microorganisms
2.4. Antagonistic Activity Testing
2.4.1. Agar Slab Method
2.4.2. Microtitration Plate Method
2.5. Statistical Analysis
3. Results and Discussion
3.1. Determination of the Antagonistic Activity of LAB Using the Agar Slab Method
3.2. Antagonistic Activity of LAB Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, A.; Vaissière, B.; Cane, J.; Steffan-Dewenter, I.; Cunningham, S.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B Biol. Sci. 2006, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Van Engelsdorp, D.; Hayes, J.; Underwood, R.; Pettis, J. A Survey of Honey Bee Colony Losses in the U.S., Fall 2007 to Spring 2008. PLoS ONE 2008, 3, e4071. [Google Scholar] [CrossRef]
- Allsopp, M.H.; de Lange, W.J.; Veldtman, R. Valuing insect pollination services with cost of replacement. PLoS ONE 2008, 3, e3128. [Google Scholar] [CrossRef] [PubMed]
- Leska, A.; Nowak, A.; Nowak, I.; Górczyńska, A. Effects of Insecticides and Microbiological Contaminants on Apis mellifera Health. Molecules 2021, 26, 5080. [Google Scholar] [CrossRef] [PubMed]
- Piccini, C.; D’Alessandro, B.; Antúnez, K.; Zunino, P. Detection of Paenibacillus larvae subspecies larvae spores in naturally infected bee larvae and artificially contaminated honey by PCR. World J. Microbiol. Biotechnol. 2002, 18, 761–765. [Google Scholar] [CrossRef]
- Ansari, M.J.; Al-Ghamdi, A.; Nuru, A.; Ahmed, A.M.; Ayaad, T.H.; Al-Qarni, A.; Alattal, Y.; Al-Waili, N. Survey and molecular detection of Melissococcus plutonius, the causative agent of European Foulbrood in honeybees in Saudi Arabia. Saudi J. Biol. Sci. 2017, 4, 1327–1335. [Google Scholar] [CrossRef]
- Forsgren, E. European foulbrood in honey bees. J. Invertebr. Pathol. 2010, 103, S5–S9. [Google Scholar] [CrossRef]
- Keller, A.; Brandel, A.; Becker, M.C.; Balles, R.; Abdelmohsen, U.R.; Ankenbrand, M.J.; Sickel, W. Wild bees and their nests host Paenibacillus bacteria with functional potential of avail. Microbiome 2018, 6, 229. [Google Scholar] [CrossRef]
- Raymann, K.; Coon, K.; Shaffer, Z.; Salisbury, S.; Moran, N. Pathogenicity of Serratia marcescens Strains in Honey Bees. mBio 2018, 9, e01649-18. [Google Scholar] [CrossRef]
- Lakhman, A.; Galatiuk, O.; Romanishina, T.; Behas, V.; Zastulka, O. Bees Klebsiellosis: Key Aspects of Pathogenesis. Adv. Anim. Vet. Sci. 2021, 9, 1190–1193. [Google Scholar] [CrossRef]
- Poltev, V.I.; Neshatayeva, E.V. La melanose experimentale des abeilles cause par la champignon Aureobasidium pullulans (DeBary) Arnaud. Bull. Apic. 1969, 12, 189–198. [Google Scholar]
- Cheng, X.; Zhang, L.; Luo, J.; Yang, S.; Deng, Y.; Li, J.; Hou, C. Two Pathogenic Fungi Isolated From Chalkbrood Samples and Honey Bee Viruses They Carried. Front. Microbiol. 2022, 13, 806. [Google Scholar] [CrossRef]
- Lindström, A.; Korpela, S.; Fries, I. Horizontal transmission of Paenibacillus larvaes pores between honey bee (Apis mellifera) colonies through robbing. Apidologie 2008, 39, 515–522. [Google Scholar] [CrossRef]
- Al-Waili, N.; Salom, K.; Al-Ghamdi, A.; Ansari, M.J. Antibiotic, pesticide, and microbial contaminants of honey: Human health hazards. Sci. World J. 2012, 2012, 930849. [Google Scholar] [CrossRef]
- Forfert, N.; Natsopoulou, M.; Frey, E.; Rosenkranz, P.; Paxton, R.; Moritz, R. Parasites and Pathogens of the Honeybee (Apis mellifera) and Their Influence on Inter-Colonial Transmission. PLoS ONE 2015, 10, e0140337. [Google Scholar] [CrossRef]
- Borum, A. Microbiota and Its Importance in Honey Bees. Bee Stud. J. Apic. Sci. 2021, 13, 23–30. [Google Scholar] [CrossRef]
- Kwong, W.; Moran, N. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Daisley, B.A.; Chmiel, J.A.; Pitek, A.P.; Thompson, G.J.; Reid, G. Missing Microbes in Bees: How Systematic Depletion of Key Symbionts Erodes Immunity. Trends Microbiol. 2020, 28, 1010–1021. [Google Scholar] [CrossRef]
- Lombogia, C.; Tulung, M.; Posangi, J.; Tallei, T. Bacterial Composition, Community Structure, and Diversity in Apis nigrocincta Gut. Int. J. Microbiol. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Pachla, A.; Ptaszyńska, A.; Wicha, M.; Kunat, M.; Wydrych, J.; Oleńska, E.; Małek, W. Insight into probiotic properties of lactic acid bacterial endosymbionts of Apis mellifera L. derived from the Polish apiary. Saudi J. Biol. Sci. 2021, 28, 1890–1899. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.; Merenstein, D.; Pot, B.; Morelli, L.; Canani, R.; Flint, H.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of Action of Probiotics. Adv. Nutr. 2020, 1, 1054. [Google Scholar] [CrossRef]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.; Sreenivasa, M. Probiotic Properties of Lactic Acid Bacteria Isolated from Neera: A Naturally Fermenting Coconut Palm Nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Ugras, S. Isolation, identification and characterization of probiotic properties of bacterium from the honey stomachs of Yigilca honeybees in Turkey. Türk. Entomol. Derg. 2017, 41, 253–261. [Google Scholar] [CrossRef]
- Moradi, M.; Kousheh, S.A.; Almasi, H.; Alizadeh, A.; Guimarães, J.T.; Yılmaz, N.; Lotfi, A. Postbiotics produced by lactic acid bacteria: The next frontier in food safety. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3390–3415. [Google Scholar] [CrossRef]
- Nowak, A.; Zakłos-Szyda, M.; Rosicka-Kaczmarek, J.; Motyl, I. Anticancer Potential of Post-Fermentation Media and Cell Extracts of Probiotic Strains: An In Vitro Study. Cancers 2022, 14, 1853. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.; Sanders, M.; Shamir, R.; Swann, J.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Wegh, C.A.M.; Geerlings, S.Y.; Knol, J.; Roeselers, G.; Belzer, C. Postbiotics and Their Potential Applications in Early Life Nutrition and Beyond. Int. J. Mol. Sci. 2019, 20, 4673. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.; Vesterlund, S. Antimicrobial Components from Lactic Acid Bacteria. Lact. Acid Bact. 2004, 139, 375–396. [Google Scholar] [CrossRef]
- Iorizzo, M.; Letizia, F.; Ganassi, S.; Testa, B.; Petrarca, S.; Albanese, G.; Di Criscio, D.; De Cristofaro, A. Functional Properties and Antimicrobial Activity from Lactic Acid Bacteria as Resources to Improve the Health and Welfare of Honey Bees. Insects 2022, 13, 308. [Google Scholar] [CrossRef]
- Wang, J.; Wei, X.; Fan, M. Assessment of Antibiotic Susceptibility within Lactic Acid Bacteria and Coagulase-Negative Staphylococci Isolated from Hunan Smoked Pork, a Naturally Fermented Meat Product in China. J. Food Sci. 2018, 83, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Zendo, T.; Ohashi, C.; Maeno, S.; Piao, X.; Salminen, S.; Sonomoto, K.; Endo, A. Kunkecin A, a New Nisin Variant Bacteriocin Produced by the Fructophilic Lactic Acid Bacterium, Apilactobacillus kunkeei FF30-6 Isolated From Honey Bees. Front. Microbiol. 2020, 11, 1903. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Kjos, M.; Nes, I.; Diep, D.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef]
- Nowak, A.; Szczuka, D.; Górczyńska, A.; Motyl, I.; Kręgiel, D. Characterization of Apis mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review. Cells 2021, 10, 701. [Google Scholar] [CrossRef]
- Ortiz-Alvarado, Y.; Clark, D.R.; Vega-Melendez, C.J.; Flores-Cruz, Z.; Domingez-Bello, M.G.; Giray, T. Antibiotics in hives and their effects on honey bee physiology and behavioral development. Biol. Open 2020, 9, bio053884. [Google Scholar] [CrossRef]
- Leska, A.; Nowak, A.; Motyl, I. Isolation and Some Basic Characteristics of Lactic Acid Bacteria from Honeybee (Apis mellifera L.) Environment—A Preliminary Study. Agriculture 2022, 12, 1562. [Google Scholar] [CrossRef]
- Sakandar, H.; Kubow, S.; Sadiq, F. Isolation and in-vitro probiotic characterization of fructophilic lactic acid bacteria from Chinese fruits and flowers. LWT 2019, 104, 70–75. [Google Scholar] [CrossRef]
- Pajor, M.; Worobo, R.W.; Milewski, S.; Szweda, P. The Antimicrobial Potential of Bacteria Isolated from Honey Samples Produced in the Apiaries Located in Pomeranian Voivodeship in Northern Poland. Int. J. Environ. Res. Public Health 2018, 15, 2002. [Google Scholar] [CrossRef]
- Kowalska, J.D.; Nowak, A.; Śliżewska, K.; Stańczyk, M.; Łukasiak, M.; Dastych, J. Anti-Salmonella Potential of New Lactobacillus Strains with the Application in the Poultry Industry. Pol. J. Microbiol. 2020, 69, 5–18. [Google Scholar] [CrossRef]
- Strus, M. A new method for testing antagonistic activity of lactic acid bacteria (LAB) on selected pathogenic indicator bacteria (in Polish). Med. Dosw. Mikrobiol. 1998, 50, 123–130. [Google Scholar]
- Choi, A.; Patra, J.; Kim, W.; Kang, S. Antagonistic Activities and Probiotic Potential of Lactic Acid Bacteria Derived from a Plant-Based Fermented Food. Front. Microbiol. 2018, 9, 1963. [Google Scholar] [CrossRef] [PubMed]
- Dejene, F.; Regasa Dadi, B.; Tadesse, D. In Vitro Antagonistic Effect of Lactic Acid Bacteria Isolated from Fermented Beverage and Finfish on Pathogenic and Foodborne Pathogenic Microorganism in Ethiopia. Int. J. Microbiol. 2021, 2021, 5370556. [Google Scholar] [CrossRef]
- Thompson, J.; Weaver, M.; Lupatsch, I.; Shields, R.; Plummer, S.; Coates, C.; Rowley, A. Antagonistic Activity of Lactic Acid Bacteria Against Pathogenic Vibrios and Their Potential Use as Probiotics in Shrimp (Penaeus vannamei) Culture. Front. Mar. Sci. 2022, 9. [Google Scholar] [CrossRef]
- Ebeling, J.; Knispel, H.; Hertlein, G.; Fünfhaus, A.; Genersch, E. Biology of Paenibacillus larvae, a deadly pathogen of honey bee larvae. Appl. Microbiol. Biotechnol. 2016, 100, 7387–7395. [Google Scholar] [CrossRef]
- Katznelson, H. Bacillus apiarius, n. sp., an aerobac spore-forming organism isolated from honeybee larvae. J. Bacteriol. 1955, 70, 635–636. [Google Scholar] [CrossRef]
- Adeniyi, B.A.; Adetoye, A.; Ayeni, F.A. Antibacterial activities of lactic acid bacteria isolated from cow faeces against potential enteric pathogens. Afr. Health Sci. 2015, 15, 888–895. [Google Scholar] [CrossRef]
- Ren, D.; Zhu, J.; Gong, S.; Liu, H.; Yu, H. Antimicrobial Characteristics of Lactic Acid Bacteria Isolated from Homemade Fermented Foods. Biomed. Res. Int. 2018, 2018, 1–9. [Google Scholar] [CrossRef]
- Forsgren, E.; Olofsson, T.; Vásquez, A.; Fries, I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 2009, 41, 99–108. [Google Scholar] [CrossRef]
- Kieliszek, M.; Piwowarek, K.; Kot, A.; Błażejak, S.; Chlebowska-Śmigiel, A.; Wolska, I. Pollen and bee bread as new health-oriented products: A review. Trends Food Sci. Technol. 2018, 71, 170–180. [Google Scholar] [CrossRef]
- Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics 2020, 9, 262. [Google Scholar] [CrossRef]
- Lindenfelser, L. In vivo activity of propolis against Bacillus larvae. J. Invertebr. Pathol. 1968, 12, 129–131. [Google Scholar] [CrossRef]
- Yoshiyama, M.; Wu, M.; Sugimura, Y.; Takaya, N.; Kimoto-Nira, H.; Suzuki, C. Inhibition of Paenibacillus larvae by lactic acid bacteria isolated from fermented materials. J. Invertebr. Pathol. 2013, 112, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, R.; Cao, Y.; Chen, C.; Bai, F.; Xu, T.; Zhao, R.; Zhang, X.; Zhao, J.; Cheng, C. Identification and antagonistic activity of endophytic bacterial strain Paenibacillus sp. 5 L8 isolated from the seeds of maize (Zea mays L., Jingke 968). J. Microbiol. 2015, 66, 653–660. [Google Scholar] [CrossRef]
- Fünfhaus, A.; Ebeling, J.; Genersch, E. Bacterial pathogens of bees. Curr. Opin. Insect Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Pietropaoli, M.; Carpana, E.; Milito, M.; Palazzetti, M.; Guarducci, M.; Croppi, S.; Formato, G. Use of Lactobacillus plantarum in Preventing Clinical Cases of American and European Foulbrood in Central Italy. Appl. Sci. 2022, 12, 1388. [Google Scholar] [CrossRef]
- Azzami, K.; Ritter, W.; Tautz, J.; Beier, H. Infection of honey bees with acute bee paralysis virus does not trigger humoral or cellular immune responses. Arch. Virol. 2012, 157, 689–702. [Google Scholar] [CrossRef]
- Chang, R.; Chen, J.; Zhong, Z.; Li, Y.; Wu, K.; Zheng, H.; Yang, Y. Inflammatory bowel disease-associated Escherichia coli strain LF82 in the damage of gut and cognition of honeybees. Front. Cell Infect. Microbiol. 2022, 12. [Google Scholar] [CrossRef]
- Raftari, M.; Jalilian, F.A.; Abdulamir, A.S.; Son, R.; Sekawi, Z.; Fatimah, A.B. Effect of organic acids on Escherichia coli O157:H7 and Staphylococcus aureus contaminated meat. Open Microbiol. J. 2009, 3, 121–127. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, K.M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef]
- Loncaric, I.; Heigl, H.; Licek, E.; Moosbeckhofer, R.; Busse, H.; Rosengarten, R. Typing of Pantoea agglomerans isolated from colonies of honey bees (Apis mellifera) and culturability of selected strains from honey. Apidologie 2009, 40, 40–54. [Google Scholar] [CrossRef]
- Hoffmann, H.; Roggenkamp, A. Population genetics of the nomen species Enterobacter cloacae. Appl. Environ. Microbiol. 2003, 69, 5306–5318. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Pagès, J.M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 2015, 6, 392. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.; Ali Khan, K.; Javed Ansari, M.; Almasaudi, S.; Al-Kahtani, S. Effect of gut bacterial isolates from Apis mellifera jemenitica on Paenibacillus larvae infected bee larvae. Saudi J. Biol. Sci. 2018, 25, 383–387. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, H.S.; Yong, D.; Jeong, S.H.; Lee, K.; Chong, Y. Bacteroides faecis and Bacteroides intestinalis recovered from clinical specimens of human intestinal origin. Yonsei Med. J. 2015, 56, 292–294. [Google Scholar] [CrossRef]
- Chen, S.; Tang, Q.; Geng, J.; Liu, Y.; Jiang, J.; Cai, X.; Cao, H.; Wu, Y.; Ren, Y.; Liu, K.; et al. Detection of Viable Zygosaccharomyces rouxii in Honey and Honey Products via PMAXX-qPCR. J. Food Qual. 2022, 2022, 1–8. [Google Scholar] [CrossRef]
- Detry, R.; Simon-Delso, N.; Bruneau, E.; Daniel, H.M. Specialisation of Yeast Genera in Different Phases of Bee Bread Maturation. Microorganisms 2020, 8, 1789. [Google Scholar] [CrossRef]
- Rosa, C.; Lachance, M.; Silva, J.; Teixeira, A.; Marini, M.; Antonini, Y.; Martins, R.P. Yeast communities associated with stingless bees. FEMS Yeast Res. 2003, 4, 271–275. [Google Scholar] [CrossRef]
- Lo Cascio, G.; Dalle Carbonare, L.; Maccacaro, L.; Caliari, F.; Ligozzi, M.; Lo Cascio, V.; Fontana, R. First case of bloodstream infection due to Candida magnoliae in a Chinese oncological patient. J. Clin. Microbiol. 2007, 45, 3470–3473. [Google Scholar] [CrossRef][Green Version]
- Bereczki, L.; Bartha, N.; Kocsubé, S.; Sóki, J.; Lengyel, G.; Tálosi, G.; Máder, K.; Deák, J.; Dóczi, I. Fungaemia caused by Candida pulcherrima. Med. Mycol. 2012, 50, 522–524. [Google Scholar] [CrossRef][Green Version]
- Pakshir, K.; Zomorodian, K.; Zakaei, A.; Motamedi, M.; Rahimi Ghiasi, M.; Karamitalab, M. Molecular identification and in-vitro antifungal susceptibility testing of Candida species isolated from patients with onychomycosis. Curr. Med. Mycol. 2015, 1, 26–32. [Google Scholar] [CrossRef][Green Version]
- Foley, K.; Fazio, G.; Jensen, A.; Hughes, W. The distribution of Aspergillus spp. opportunistic parasites in hives and their pathogenicity to honey bees. Vet. Microbiol. 2014, 169, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D. Antifungal Activity of Lactic Acid Bacteria Isolated from Kimchi Against Aspergillus fumigatus. Mycobiology 2005, 33, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Mani-López, E.; Arrioja-Bretón, D.; López-Malo, A. The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Compr. Rev. Food Sci. Food Saf. 2021, 21, 604–641. [Google Scholar] [CrossRef] [PubMed]
- George-Okafor, U.; Ozoani, U.; Tasie, F.; Mba-Omeje, K. The efficacy of cell-free supernatants from Lactobacillus plantarum Cs and Lactobacillus acidophilus ATCC 314 for the preservation of home-processed tomato-paste. Sci. Afr. 2020, 8, e00395. [Google Scholar] [CrossRef]
- Lim, H.S.; Yeu, J.E.; Hong, S.P.; Kang, M.S. Characterization of Antibacterial Cell-Free Supernatant from Oral Care Probiotic Weissella cibaria, CMU. Molecules 2018, 23, 1984. [Google Scholar] [CrossRef]
- Yazgan, H. Lactobacillus reuteri ATCC55730 ve Lactobacillus plantarum FI8595 Supernatantlarının Bazı Balık Bozucu ve Gıda Kaynaklı Patojen Bakterilerine Karşı Antimikrobiyal Etkisi. Eur. J. Lipid Sci. Technol. 2020, 485–489. [Google Scholar] [CrossRef]
- Siedler, S.; Balti, R.; Neves, A. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef]
- Rouse, S.; Harnett, D.; Vaughan, A.; Sinderen, D. Lactic acid bacteria with potential to eliminate fungal spoilage in foods. J. Appl. Microbiol. 2008, 104, 915–923. [Google Scholar] [CrossRef]
- Betesho Babrud, R.; Kasra Kermanshahi, R.; Motamedi Sede, F.; Moosavinejad, S.Z. The effect of Lactobacillus reuteri cell free supernatant on growth and biofilm formation of Paenibacillus larvae. Iran J. Vet. Res. 2019, 20, 192–198. [Google Scholar]
- Carina Audisio, M.; Torres, M.; Sabaté, D.; Ibarguren, C.; Apella, M. Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol. Res. 2011, 166, 1–13. [Google Scholar] [CrossRef]
- Morita, H.; Toh, H.; Fukuda, S.; Horikawa, H.; Oshima, K.; Suzuki, T.; Murakami, M.; Hisamatsu, S.; Kato, Y.; Takizawa, T.; et al. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 2008, 15, 151–161. [Google Scholar] [CrossRef]
- Chen, L.; Bromberger, P.D.; Nieuwenhuiys, G.; Hatti-Kaul, R. Redox Balance in Lactobacillus reuteri DSM20016: Roles of Iron-Dependent Alcohol Dehydrogenases in Glucose/Glycerol Metabolism. PLoS ONE 2016, 11, e0168107. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; Ganassi, S.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antimicrobial Activity against Paenibacillus larvae and Functional Properties of Lactiplantibacillus plantarum Strains: Potential Benefits for Honeybee Health. Antibiotics 2020, 9, 442. [Google Scholar] [CrossRef]
- Portella, A.; Karp, S.; Scheidt, G.; Woiciechwski, A.; Parada, J.; Soccol, C. Modelling antagonic effect of lactic acid bacteria supernatants on some pathogenic bacteria. Braz. Arch. Biol. Technol. 2009, 52, 29–36. [Google Scholar] [CrossRef][Green Version]
- Bian, L.; Molan, A.; Maddox, I.; Shu, Q. Antimicrobial activity of Lactobacillus reuteri DPC16 supernatants against selected food borne pathogens. World J. Microbiol. Biotechnol. 2010, 27, 991–998. [Google Scholar] [CrossRef]
- Caro Velez, C.; León Peláez, Á. Capacidad antifúngica de sobrenadantes libres de células obtenidos de la fermentación de un sustrato de “panela” con gránulos de kefir de agua. Rev. Colomb. Biotecnol. 2015, 17, 22–32. [Google Scholar] [CrossRef]








| Isolates from Honeybee Environment | Collection Strains | p-Value (U Mann–Whitney Test) | ||
|---|---|---|---|---|
| P. larvae ATCC 25367 | 16.92 ± 2.92 | 5.74 ± 1.85 | 0.0000 | ENV * |
| P. larvae ATCC 49843 | 7.82 ± 1.74 | 6.29 ± 1.39 | 0.0000 | ENV |
| P. apiaries DSM 5582 | 31.83 ± 4.84 | 6.42 ± 2.15 | 0.0000 | ENV |
| P. alvei DSM 29 | 29.45 ± 4.47 | 5.74 ± 1.29 | 0.0000 | ENV |
| L. sphaericus DSM 1866 | 4.72 ± 1.89 | 5.55 ± 1.88 | 0.0001 | |
| M. plutonius DSM 29964 | 6.57 ± 5.09 | 4.89 ± 5.91 | 0.0236 | ENV |
| E. coli ATCC 25922 | 6.46 ± 2.13 | 7.73 ± 1.94 | 0.0000 | |
| E. persicina 40 | 9.54 ± 2.28 | 15.55 ± 6.89 | 0.0000 | |
| P. agglomerans 43 | 10.44 ± 2.61 | 18.12 ± 7.06 | 0.0000 | |
| E. kobei 40 | 19.22 ± 6.29 | 7.69 ± 4.31 | 0.0000 | ENV |
| E. cloacae 41 | 8.28 ± 2.52 | 20.05 ± 10.23 | 0.0000 | |
| B. faecis DSM 24798 | 0.00 | 4.65 ± 4.51 | 0.0000 | |
| B. intestinalis DSM 17393 | 0.00 | 0.92 ± 2.35 | 0.0000 | |
| A. pullulans DSM 3042 | 0.00 | 0.00 | p > 0.05 | |
| Z. rouxii 26D | 0.00 | 2.75 ± 4.20 | 0.0000 | |
| C. magnoliae 27D | 0.00 | 0.33 ± 1.68 | 0.0127 | |
| Z. rouxii 28D | 0.00 | 0.67 ± 1.99 | 0.0000 | |
| M. pulcherrima 40D | 0.00 | 0.35 ± 1.57 | 0.0007 | |
| A. niger ATCC 16404 | 0.00 | 0.00 | p > 0.05 | |
| A. fumigatus ŁOCK CPC 1097 | 0.00 | 0.00 | p > 0.05 | |
| A. flavus ŁOCK CPC 0600 | 0.00 | 0.00 | p > 0.05 |
| LAB Strains (n = 103) | Bacteria | Yeasts | Molds | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-Positive | Gram-Negative | ||||||||||||||||||||
| P. larvae ATCC 25367 | P. larvae ATCC 49843 | P. apiaries DSM 5582 | P. alvei DSM 29 | L. sphaericus DSM 1866 | M. plutonius DSM 29964 | E. coli ATCC 25922 | E. persicina 40 | P. agglomerans 43 | E. kobei 40 | E. cloacae 41 | B. faecis DSM 24798 | B. intestinalis DSM 17393 | A. pullulans DSM 3042 | Z. rouxii 26D | C. magnoliae 27D | Z. rouxii 28D | M. pulcherrima 40D | A. niger ATCC 16404 | A. fumigatus ŁOCK CPC 1097 | A. flavus ŁOCK CPC 0600 | |
| P. acidilactici (n = 17) | 17 | 17 | 17 | 17 | 17 | 14 | 17 | 17 | 17 | 17 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. plantarum (n = 38) | 38 | 38 | 38 | 38 | 38 | 19 | 38 | 38 | 38 | 37 | 38 | 14 | 3 | 0 | 10 | 2 | 4 | 2 | 0 | 0 | 0 |
| P. pentosaceus (n = 20) | 20 | 20 | 20 | 20 | 20 | 11 | 20 | 20 | 20 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. brevis (n = 9) | 9 | 9 | 9 | 9 | 8 | 3 | 9 | 9 | 9 | 6 | 8 | 3 | 7 | 0 | 6 | 0 | 3 | 2 | 0 | 0 | 0 |
| L. paracasei (n = 3) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| L. rhamnosus (n = 2) | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. coryniformis (n = 2) | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| L. acidophilus (n = 2) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| L. casei (n = 2) | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| L. mesenteroides (n = 2) | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| L. delbrueckii (n = 1) | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. salivarius (n = 1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P. parvulus (n = 1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. fermentum (n = 1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. farraginis (n = 1) | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. kunkeei (n = 1) | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAB Strains | P. larvae ATCC 25367 | P. apiarius DSM 5582 | L. sphaericus DSM 1866 |
|---|---|---|---|
| A. kunkeei DSM 12361 | 9.22 ± 5.72 | 15.86 ± 6.75 | 3.20 ± 1.50 |
| P. acidilactici 4/1 | 22.31 ± 1.06 * | 5.39 ± 1.35 * | 22.28 ± 2.58 * |
| P. acidilactici 5/2 | 21.58 ± 4.21 * | 3.10 ± 1.31 * | 26.26 ± 1.55 * |
| P. acidilactici 6/1 | 23.84 ± 2.59 * | 12.97 ± 0.68 | 20.12 ± 1.95 * |
| P. acidilactici 7/1 | 24.73 ± 0.99 * | 9.72 ± 0.34 | 27.89 ± 3.37 * |
| P. acidilactici 8/1 | 26.96 ± 1.70 * | 5.18 ± 1.27 * | 33.91 ± 10.17 * |
| P. pentosaceus 9/3 | 21.14 ± 3.24 * | 8.03 ± 0.45 | 18.92 ± 1.37 * |
| L. plantarum 10/2 | 24.57 ± 1.66 * | 7.99 ± 1.39 | 24.57 ± 1.26 * |
| P. pentosaceus 11/3 | 26.18 ± 1.14 * | 9.24 ± 1.30 | 24.29 ± 1.09 * |
| P. pentosaceus 14/1 | 24.94 ± 0.65 * | 7.61 ± 0.49 | 27.71 ± 2.17 * |
| L. plantarum 14/3 | 16.23 ± 2.40 | 4.78 ± 1.24 * | 13.29 ± 3.23 * |
| L. plantarum 18/1 | 19.08 ± 3.46 * | 10.05 ± 1.58 | 19.48 ± 1.78 * |
| P. pentosaceus 19/1 | 19.76 ± 3.65 * | 10.28 ± 0.90 | 19.69 ± 1.87 * |
| L. plantarum 21/1 | 27.49 ± 4.53 * | 15.60 ± 7.47 | 25.65 ± 3.01 * |
| P. acidilactici 22/1 | 25.86 ± 3.16 * | 13.09 ± 10.25 | 34.99 ± 0.93 * |
| P. acidilactici 25/1 | 19.61 ± 4.51 * | 13.90 ± 2.91 | 26.90 ± 3.93 * |
| P. acidilactici 35/1 | 24.82 ± 2.19 * | 8.41 ± 0.80 | 36.25 ± 12.74 * |
| P. parvulus OK-S | 26.29 ± 2.53 * | 7.49 ± 0.52 | 17.42 ± 3.28 * |
| L. brevis KKA | 27.35 ± 4.33 * | 7.33 ± 2.18 * | 30.37 ± 1.13 * |
| L. plantarum 8AN | 26.60 ± 0.86 * | 10.28 ± 0.86 | 18.62 ± 3.36 * |
| L. salivarius 9AN | 23.82 ± 1.75 * | 9.88 ± 1.23 | 33.18 ± 2.60 * |
| L. casei 12AN | 27.27 ± 2.30 * | 7.37 ± 1.45 * | 20.15 ± 1.34 * |
| L. plantarum 145 | 24.08 ± 3.65 * | 8.74 ± 1.09 | 21.63 ± 5.72 * |
| L. acidophilus 573 | 18.94 ± 3.13 * | 5.49 ± 1.59 * | 24.06 ± 3.64 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leska, A.; Nowak, A.; Szulc, J.; Motyl, I.; Czarnecka-Chrebelska, K.H. Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens. Pathogens 2022, 11, 1367. https://doi.org/10.3390/pathogens11111367
Leska A, Nowak A, Szulc J, Motyl I, Czarnecka-Chrebelska KH. Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens. Pathogens. 2022; 11(11):1367. https://doi.org/10.3390/pathogens11111367
Chicago/Turabian StyleLeska, Aleksandra, Adriana Nowak, Justyna Szulc, Ilona Motyl, and Karolina Henryka Czarnecka-Chrebelska. 2022. "Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens" Pathogens 11, no. 11: 1367. https://doi.org/10.3390/pathogens11111367
APA StyleLeska, A., Nowak, A., Szulc, J., Motyl, I., & Czarnecka-Chrebelska, K. H. (2022). Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens. Pathogens, 11(11), 1367. https://doi.org/10.3390/pathogens11111367