Transcriptome Profiling in Swine Macrophages Infected with African Swine Fever Virus (ASFV) Uncovers the Complex and Close Relationship with Host
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and ASFV Infection of PAMs
2.2. Real-Time Quantitative PCR (qPCR)
2.3. Transcriptome Sequencing and Data Analysis
2.4. Statistical Analysis of Alternative Splicing Events
3. Results
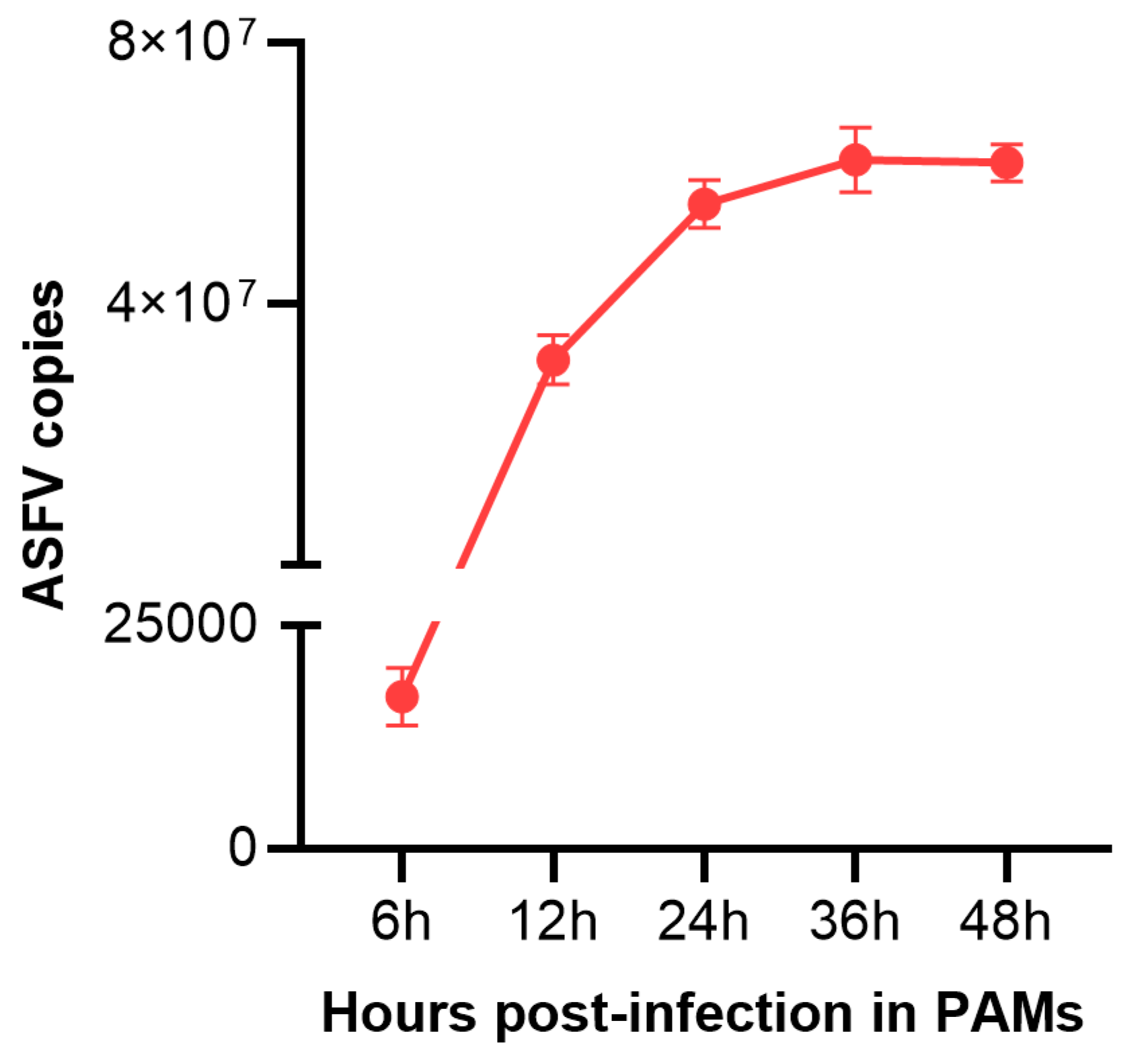
3.1. ASFV Infection Conditions in PAMs
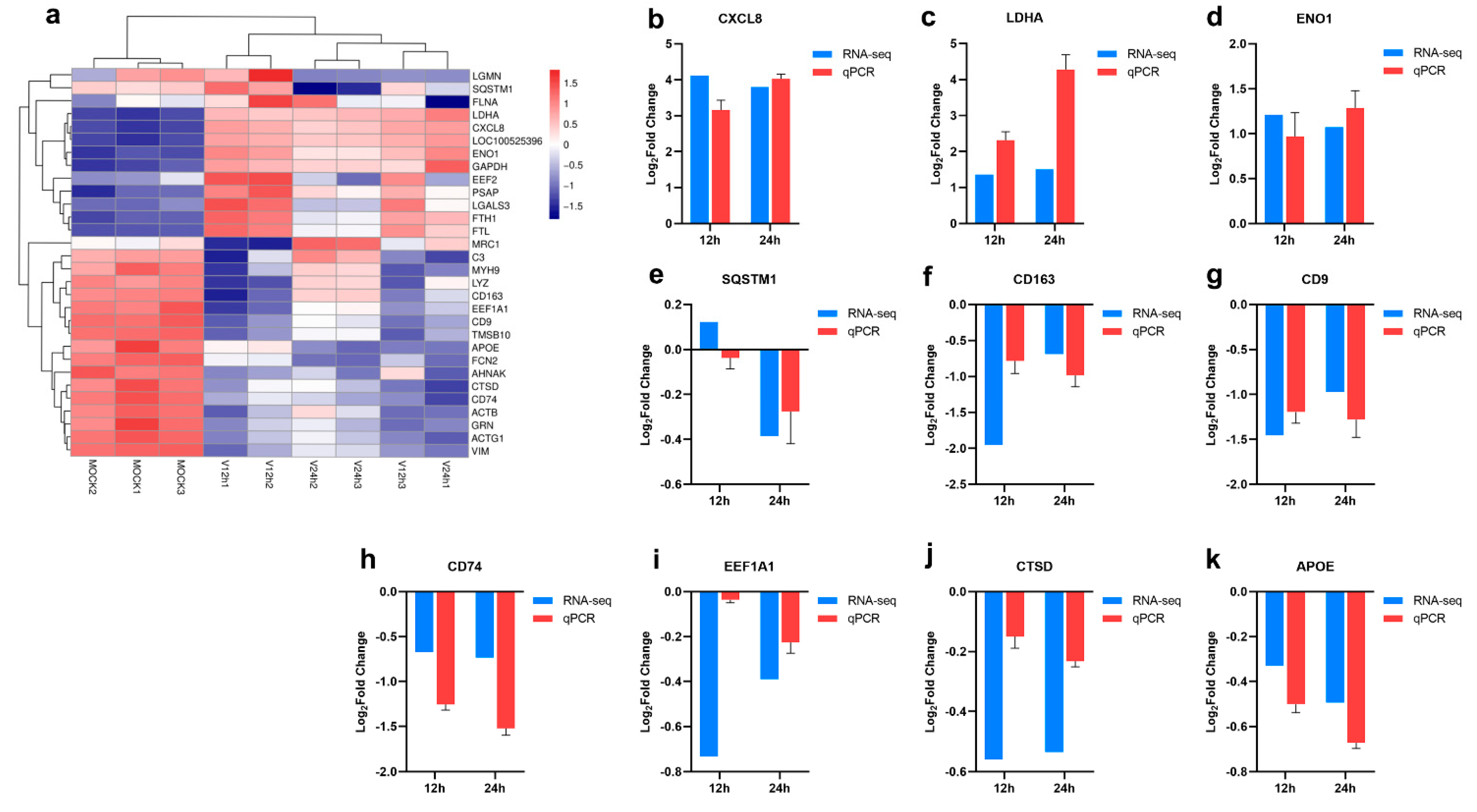
3.2. Gene Expression Statistics and Differential Analysis among Different Samples
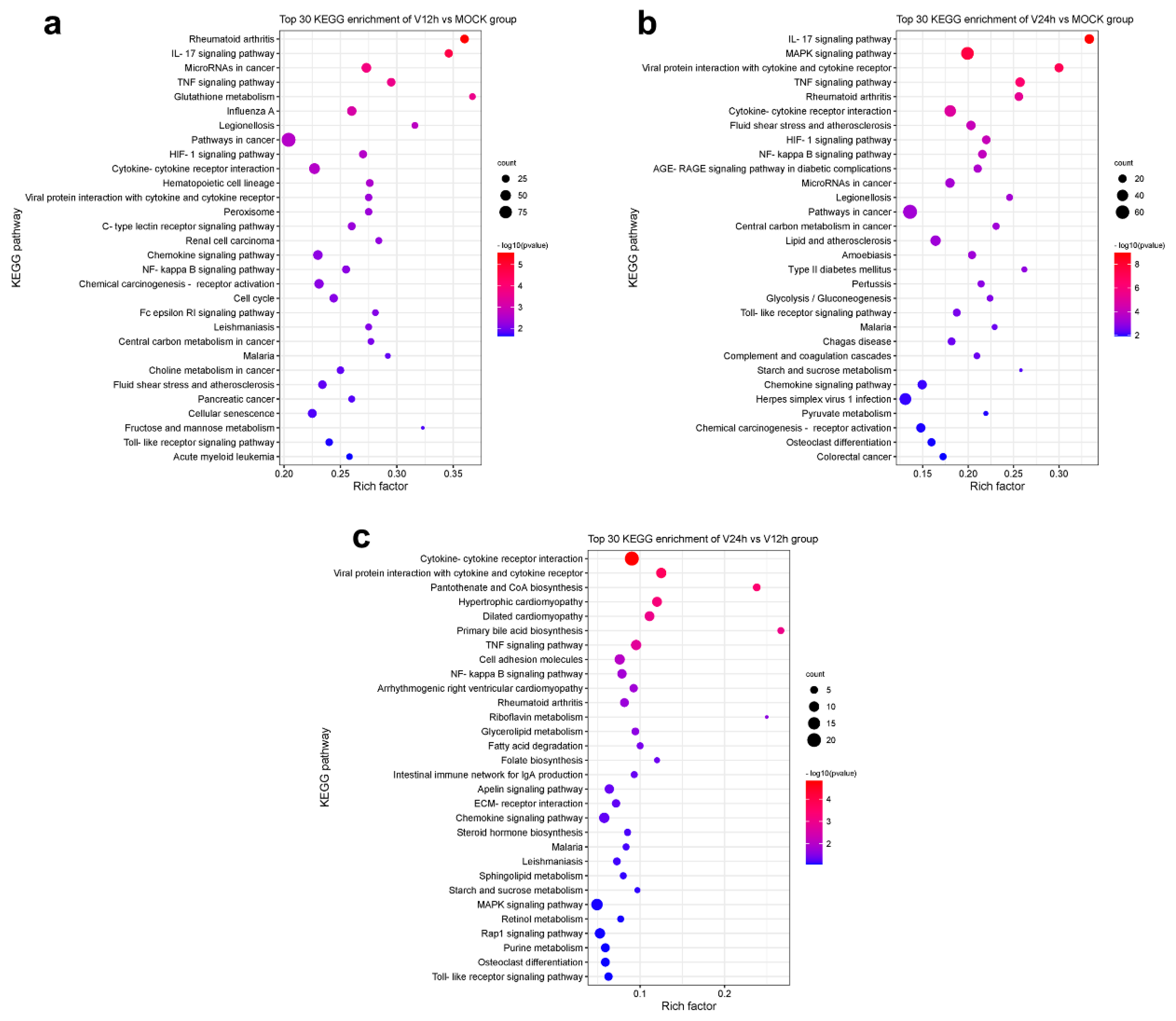
3.3. KEGG Pathway Analysis of DEGs
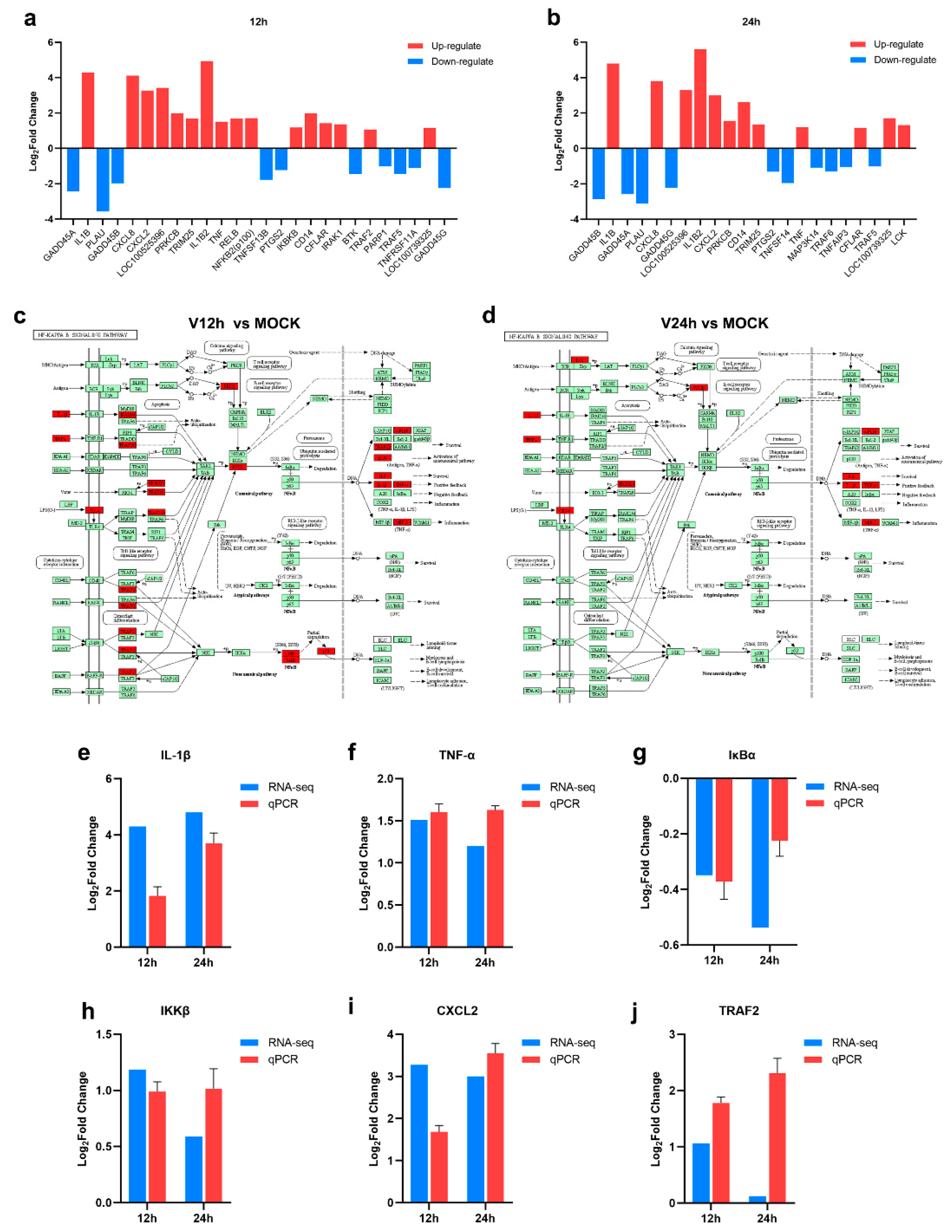
3.4. The Activation of the NF-κB Signaling Pathway in ASFV-Infected PAMs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African swine fever virus evasion of host defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef]
- Boklund, A.; Cay, B.; Depner, K.; Földi, Z.; Guberti, V.; Masiulis, M.; Miteva, A.; More, S.; Olsevskis, E.; Šatrán, P.; et al. Epidemiological analyses of African swine fever in the European Union (November 2017 until November 2018). EFSA J. Eur. Food Saf. Auth. 2018, 16, e05494. [Google Scholar]
- Wu, K.; Liu, J.; Wang, L.; Fan, S.; Li, Z.; Li, Y.; Yi, L.; Ding, H.; Zhao, M.; Chen, J. Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China. Vaccines 2020, 8, 531. [Google Scholar] [CrossRef]
- Ma, J.; Chen, H.; Gao, X.; Xiao, J.; Wang, H. African swine fever emerging in China: Distribution characteristics and high-risk areas. Prev. Vet. Med. 2020, 175, 104861. [Google Scholar] [CrossRef] [PubMed]
- The Global Economic Impact of ASF—WOAH Bulletin. Available online: https://bulletin.woah.org/?panorama=02-2-2-2020-1-economic (accessed on 29 August 2022).
- ASF Distribution and the Situation in 2020–2022 (Based on INs, FURs and SMRs). Available online: https://www.woah.org/app/uploads/2022/02/asf-report6.pdf (accessed on 20 July 2022).
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Song, J.; Kang, L.; Huang, L.; Zhou, S.; Hu, L.; Zheng, J.; Li, C.; Zhang, X.; He, X.; et al. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PLoS Pathog. 2021, 17, e1009733. [Google Scholar] [CrossRef]
- Correia, S.; Ventura, S.; Parkhouse, R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013, 173, 87–100. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Z.; Feng, T.; Ma, Z.; Xue, Q.; Wu, P.; Li, P.; Li, S.; Yang, F.; Cao, W.; et al. African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation. J. Virol. 2021, 95, e82421. [Google Scholar] [CrossRef]
- Whittall, J.T.; Parkhouse, R.M. Changes in swine macrophage phenotype after infection with African swine fever virus: Cytokine production and responsiveness to interferon-gamma and lipopolysaccharide. Immunology 1997, 91, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, J.; Wu, Y.; Chen, H.; Zhang, S.; Li, J.; Xin, T.; Jia, H.; Hou, S.; Jiang, Y.; et al. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 2018, 506, 437–443. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, X.; Nie, Y.; Li, H.; Chen, W.; Lin, W.; Chen, F.; Xie, Q. African Swine Fever Virus Protein E199L Promotes Cell Autophagy through the Interaction of PYCR2. Virol. Sin. 2021, 36, 196–206. [Google Scholar] [CrossRef]
- Sun, M.; Yu, S.; Ge, H.; Wang, T.; Li, Y.; Zhou, P.; Pan, L.; Han, Y.; Yang, Y.; Sun, Y.; et al. The A137R Protein of African Swine Fever Virus Inhibits Type I Interferon Production via the Autophagy-Mediated Lysosomal Degradation of TBK1. J. Virol. 2022, 96, e195721. [Google Scholar] [CrossRef] [PubMed]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [Green Version]
- Ju, X.; Li, F.; Li, J.; Wu, C.; Xiang, G.; Zhao, X.; Nan, Y.; Zhao, D.; Ding, Q. Genome-wide transcriptomic analysis of highly virulent African swine fever virus infection reveals complex and unique virus host interaction. Vet. Microbiol. 2021, 261, 109211. [Google Scholar] [CrossRef] [PubMed]
- African Swine Fever—WOAH—World Organisation for Animal Health. Available online: https://www.woah.org/en/disease/african-swine-fever/ (accessed on 30 August 2022).
- Yang, B.; Shen, C.; Zhang, D.; Zhang, T.; Shi, X.; Yang, J.; Hao, Y.; Zhao, D.; Cui, H.; Yuan, X.; et al. Mechanism of interaction between virus and host is inferred from the changes of gene expression in macrophages infected with African swine fever virus CN/GS/2018 strain. Virol. J. 2021, 18, 170. [Google Scholar] [CrossRef]
- Bao, Z.Q.; Liao, T.T.; Yang, W.R.; Wang, Y.; Luo, H.Y.; Wang, X.Z. Heat stress-induced autophagy promotes lactate secretion in cultured immature boar Sertoli cells by inhibiting apoptosis and driving SLC2A3, LDHA, and SLC16A1 expression. Theriogenology 2017, 87, 339–348. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, X.; Dong, W.; Wang, X.; He, S.; Zhang, H.; Wang, X.; Wei, R.; Chen, Y.; Liu, X.; et al. TREM2 suppresses the proinflammatory response to facilitate PRRSV infection via PI3K/NF-kappaB signaling. PLoS Pathog. 2020, 16, e1008543. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, C.; Cheng, S.; Wang, X.; Li, W.; Charreyre, C.; Audonnet, J.C.; He, Q. Porcine CD74 is involved in the inflammatory response activated by nuclear factor kappa B during porcine circovirus type 2 (PCV-2) infection. Arch. Virol. 2013, 158, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, Y.; Feng, Y.; Quan, W.; Luo, Y.; Wang, H.; Zheng, J.; Chen, X.; Huang, Z.; Chen, X.; et al. Effects of the NF-kappaB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV. Viruses 2022, 14, 297. [Google Scholar] [CrossRef]
- Zhu, E.; Wu, H.; Chen, W.; Qin, Y.; Liu, J.; Fan, S.; Ma, S.; Wu, K.; Mao, Q.; Luo, C.; et al. Classical swine fever virus employs the PERK- and IRE1-dependent autophagy for viral replication in cultured cells. Virulence 2021, 12, 130–149. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Villamandos, J.C.; Bautista, M.J.; Sanchez-Cordon, P.J.; Carrasco, L. Pathology of African swine fever: The role of monocyte-macrophage. Virus Res. 2013, 173, 140–149. [Google Scholar] [CrossRef]
- Chen, C.; Gao, H.; Su, X. Autophagy-related signaling pathways are involved in cancer (Review). Exp. Ther. Med. 2021, 22, 710. [Google Scholar] [CrossRef]
- Li, L.; Fu, J.; Li, J.; Guo, S.; Chen, Q.; Zhang, Y.; Liu, Z.; Tan, C.; Chen, H.; Wang, X. African Swine Fever Virus pI215L Inhibits Type I Interferon Signaling by Targeting Interferon Regulatory Factor 9 for Autophagic Degradation. J. Virol. 2022, 96, e94422. [Google Scholar] [CrossRef]
- Hernaez, B.; Cabezas, M.; Munoz-Moreno, R.; Galindo, I.; Cuesta-Geijo, M.A.; Alonso, C. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr. Mol. Med. 2013, 13, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Banjara, S.; Shimmon, G.L.; Dixon, L.K.; Netherton, C.L.; Hinds, M.G.; Kvansakul, M. Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin. Viruses 2019, 11, 789. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.F.; Liu, Y.; Fan, Y.; Hua, S.N.; Qu, H.Y.; Dong, S.W.; Li, R.L.; Zhao, M.Y.; Zhen, Y.; Yu, X.L.; et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J. Hematol. Oncol. 2015, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Yin, N.; Peng, M.; Stamatiades, E.G.; Shyu, A.; Li, P.; Zhang, X.; Do, M.H.; Wang, Z.; Capistrano, K.J.; et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 2021, 371, 405–410. [Google Scholar] [CrossRef]
- Zhao, Y.; Chahar, H.S.; Komaravelli, N.; Dossumbekova, A.; Casola, A. Human metapneumovirus infection of airway epithelial cells is associated with changes in core metabolic pathways. Virology 2019, 531, 183–191. [Google Scholar] [CrossRef]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passalacqua, K.D.; Lu, J.; Goodfellow, I.; Kolawole, A.O.; Arche, J.R.; Maddox, R.J.; Carnahan, K.E.; O’Riordan, M.; Wobus, C.E. Glycolysis Is an Intrinsic Factor for Optimal Replication of a Norovirus. mbio 2019, 10, e02175-18. [Google Scholar] [CrossRef] [Green Version]
- Fan, S.; Wu, K.; Zhao, M.; Yuan, J.; Ma, S.; Zhu, E.; Chen, Y.; Ding, H.; Yi, L.; Chen, J. LDHB inhibition induces mitophagy and facilitates the progression of CSFV infection. Autophagy 2021, 17, 2305–2324. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, W.; Zhang, J.; Zhang, J.; Zhang, H.; Zhu, Y.; Meng, X.; Yi, Z.; Wang, R. Influenza A Virus (H1N1) Infection Induces Glycolysis to Facilitate Viral Replication. Virol. Sin. 2021, 36, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Song, Y.; Li, Z.; Xue, Z.; Cao, W.; Liu, X.; Zheng, H. African Swine Fever Virus Regulates Host Energy and Amino Acid Metabolism To Promote Viral Replication. J. Virol. 2022, 96, e191921. [Google Scholar] [CrossRef]
- Fishbourne, E.; Abrams, C.C.; Takamatsu, H.H.; Dixon, L.K. Modulation of chemokine and chemokine receptor expression following infection of porcine macrophages with African swine fever virus. Vet. Microbiol. 2013, 162, 937–943. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Niu, Q.; Yang, J.; Zhao, Y.; Tian, Z.; Fan, J.; Zhang, Z.; Wang, Y.; Geng, S.; Zhang, Y.; et al. Transcriptome Profiling Reveals Features of Immune Response and Metabolism of Acutely Infected, Dead and Asymptomatic Infection of African Swine Fever Virus in Pigs. Front. Immunol. 2021, 12, 808545. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; McLaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Popescu, L.; Gaudreault, N.N.; Whitworth, K.M.; Murgia, M.V.; Nietfeld, J.C.; Mileham, A.; Samuel, M.; Wells, K.D.; Prather, R.S.; Rowland, R. Genetically edited pigs lacking CD163 show no resistance following infection with the African swine fever virus isolate, Georgia 2007/1. Virology 2017, 501, 102–106. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Na, N.; Zhang, X.; Zhao, Y. The biological function and significance of CD74 in immune diseases. Inflamm. Res. 2017, 66, 209–216. [Google Scholar] [CrossRef]
- Abbas, W.; Kumar, A.; Herbein, G. The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 5, 75. [Google Scholar] [CrossRef] [Green Version]
- Payton, A.; van den Boogerd, E.; Davidson, Y.; Gibbons, L.; Ollier, W.; Rabbitt, P.; Worthington, J.; Horan, M.; Pendleton, N. Influence and interactions of cathepsin D, HLA-DRB1 and APOE on cognitive abilities in an older non-demented population. Genes Brain Behav. 2006, 5 (Suppl. 1), 23–31. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72 Pt A, 3–12. [Google Scholar] [CrossRef]
- Salguero, F.J.; Ruiz-Villamor, E.; Bautista, M.J.; Sanchez-Cordon, P.J.; Carrasco, L.; Gomez-Villamandos, J.C. Changes in macrophages in spleen and lymph nodes during acute African swine fever: Expression of cytokines. Vet. Immunol. Immunopathol. 2002, 90, 11–22. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2020, 7, 601641. [Google Scholar] [CrossRef] [PubMed]
- Dorrington, M.G.; Fraser, I. NF-kappaB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cackett, G.; Portugal, R.; Matelska, D.; Dixon, L.; Werner, F. African Swine Fever Virus and Host Response: Transcriptome Profiling of the Georgia 2007/1 Strain and Porcine Macrophages. J. Virol. 2022, 96, e193921. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, L.; Wang, J.; Su, D.; Li, D.; Du, Y.; Yang, G.; Zhang, G.; Chu, B. African Swine Fever Virus K205R Induces ER Stress and Consequently Activates Autophagy and the NF-kappaB Signaling Pathway. Viruses 2022, 14, 394. [Google Scholar] [CrossRef]
- Powell, P.P.; Dixon, L.K.; Parkhouse, R.M. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 1996, 70, 8527–8533. [Google Scholar] [CrossRef] [Green Version]
- Ayanwale, A.; Trapp, S.; Guabiraba, R.; Caballero, I.; Roesch, F. New Insights in the Interplay Between African Swine Fever Virus and Innate Immunity and Its Impact on Viral Pathogenicity. Front. Microbiol. 2022, 13, 958307. [Google Scholar] [CrossRef] [PubMed]
- Thai, M.; Graham, N.A.; Braas, D.; Nehil, M.; Komisopoulou, E.; Kurdistani, S.K.; McCormick, F.; Graeber, T.G.; Christofk, H.R. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 2014, 19, 694–701. [Google Scholar] [CrossRef] [Green Version]
- Thai, M.; Thaker, S.K.; Feng, J.; Du, Y.; Hu, H.; Ting, W.T.; Graeber, T.G.; Braas, D.; Christofk, H.R. MYC-induced reprogramming of glutamine catabolism supports optimal virus replication. Nat. Commun. 2015, 6, 8873. [Google Scholar] [CrossRef] [Green Version]
- Heaton, N.S.; Randall, G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 2010, 8, 422–432. [Google Scholar] [CrossRef]

| Primers | Sequence (5′-3′) | References or Genbank |
|---|---|---|
| ASFV-B646L-F | CTGCTCATGGTATCAATCTTATCGA | [17] |
| ASFV-B646L-R | GATACCACAAGATCRGCCGT | |
| CXCL8-F | AGC CCG TGT CAA CAT GAC TT | [18] |
| CXCL8-R | TGG AAA GGT GTG GAA TGC GT | |
| LDHA-F | CGTCAGCAAGAGGGAGA | [19] |
| LDHA-R | AAGCACTGGATTGGAAGCAACAA | |
| ENO1-F | AAGCCCTGGAGCTGCTGA | XM_021095279.1 |
| ENO1-R | CGTACTTGCCCGACCTGTAGAA | |
| SQSTM1-F | GCTGCTCTTCCGACCCT | XM_003123639.4 |
| SQSTM1-R | GCGATCTTATTCATTTGCTCC | |
| CD163-F | ATTCATCATCCTCGGACCCAT | [20] |
| CD163-R | CCCAGCACAACGACCACCT | |
| CD9-F | CCAGGATTTCTACAGGGACA | NM_214006.1 |
| CD9-R | GCATAGTGGATGGCTTTCAG | |
| CD74-F | AGAGCAAGTGCAGCCGTGGAG | [21] |
| CD74-R | GGTACAGGAAGTAGGCGGTGGTG | |
| EEF1A1-F | AGTGCTAATATGCCTTGGTT | NM_001097418.2 |
| EEF1A1-R | TTGTCAGTTGGACGAGTTG | |
| CTSD-F | ACGTGAAGAACGGCACCACC | NM_001037721.1 |
| CTSD-R | GCCCAACAACGCAGAATTACA | |
| APOE-F | TGGGAGGAGTCCAAGTGGCA | NM_214308.1 |
| APOE-R | GCTCCGTCAGTTCCTGGGTGA | |
| IL-1β-F | ACCTGGACCTTGGTTCTCTG | [22] |
| IL-1β-R | CATCTGCCTGATGCTCTTG | |
| TNF-α-F | TGGCCCAAGGACTCAGATCAT | [23] |
| TNF-α-R | TCGGCTTTGACATTGGCTACA | |
| IκBα-F | ACCAACCAGCCAGAAATCG | NM_001005150.1 |
| IκBα-R | CACAGGCAAGGTGTAGAGGG | |
| IKKβ-F | AGAGGATCTTCTGCGAGTA | NM_001244129.1 |
| IKKβ-R | CTTTGGGTGCGTAACTG | |
| CXCL2-F | GTGGAAACAGCAACTGCTCA | [18] |
| CXCL2-R | AGGGCTTGGTAGTTGTCAGG | |
| TRAF2-F | CCACCGCTACTGCTCCTACTGC | XM_005652719.2 |
| TRAF2-R | CGCCTTCTTCATAAATGCCCTC | |
| GAPDH-F | TGGAGTCCACTGGTGTCTTCAC | [23] |
| GAPDH-R | TTCACGCCCATCACAAACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Chen, W.; Li, X.; Wu, K.; Wang, X.; Wang, W.; Li, Y.; Yi, L.; Zhao, M.; Ding, H.; et al. Transcriptome Profiling in Swine Macrophages Infected with African Swine Fever Virus (ASFV) Uncovers the Complex and Close Relationship with Host. Pathogens 2022, 11, 1411. https://doi.org/10.3390/pathogens11121411
Li Z, Chen W, Li X, Wu K, Wang X, Wang W, Li Y, Yi L, Zhao M, Ding H, et al. Transcriptome Profiling in Swine Macrophages Infected with African Swine Fever Virus (ASFV) Uncovers the Complex and Close Relationship with Host. Pathogens. 2022; 11(12):1411. https://doi.org/10.3390/pathogens11121411
Chicago/Turabian StyleLi, Zhaoyao, Wenxian Chen, Xiaowen Li, Keke Wu, Xinyan Wang, Weijun Wang, Yuwan Li, Lin Yi, Mingqiu Zhao, Hongxing Ding, and et al. 2022. "Transcriptome Profiling in Swine Macrophages Infected with African Swine Fever Virus (ASFV) Uncovers the Complex and Close Relationship with Host" Pathogens 11, no. 12: 1411. https://doi.org/10.3390/pathogens11121411
APA StyleLi, Z., Chen, W., Li, X., Wu, K., Wang, X., Wang, W., Li, Y., Yi, L., Zhao, M., Ding, H., Fan, S., & Chen, J. (2022). Transcriptome Profiling in Swine Macrophages Infected with African Swine Fever Virus (ASFV) Uncovers the Complex and Close Relationship with Host. Pathogens, 11(12), 1411. https://doi.org/10.3390/pathogens11121411








