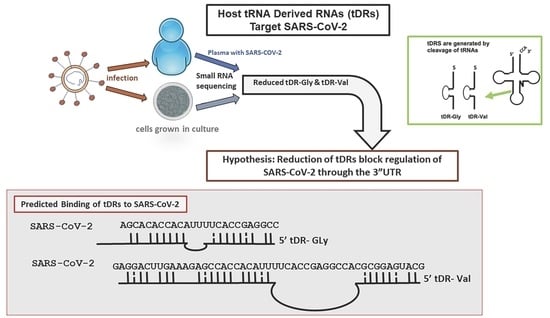
Host tRNA-Derived RNAs Target the 3′Untranslated Region of SARS-CoV-2
Abstract
:1. Introduction
2. Results
2.1. tDR Expression Is Altered in SARS-CoV-2 Infected Patients’ Plasma Compared to Uninfected Control Plasma
2.2. Identification of Altered tDR Expression in SARS-CoV-2-Infected Calu-3 Lung Cells
2.3. RNA Hybridization Analysis to Detect Potential Binding Sites in SARS-CoV-2 for Selected tDRs
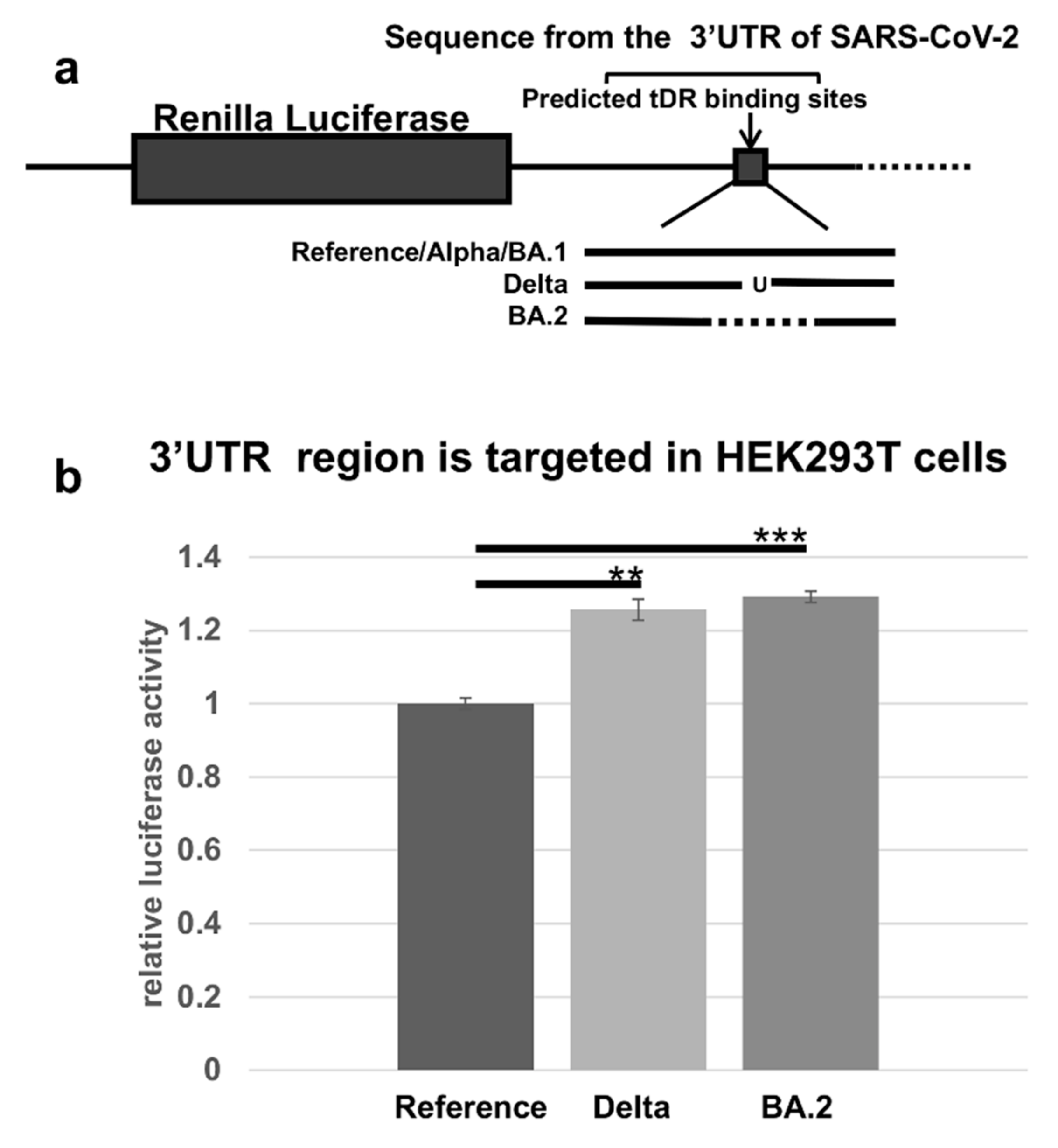
2.4. The Predicted tDR Binding Sequences from the SARS-CoV-2 Reference Genome and VOCs Were Placed Downstream of Luciferase to Determine if Host Factors Could Regulate Expression
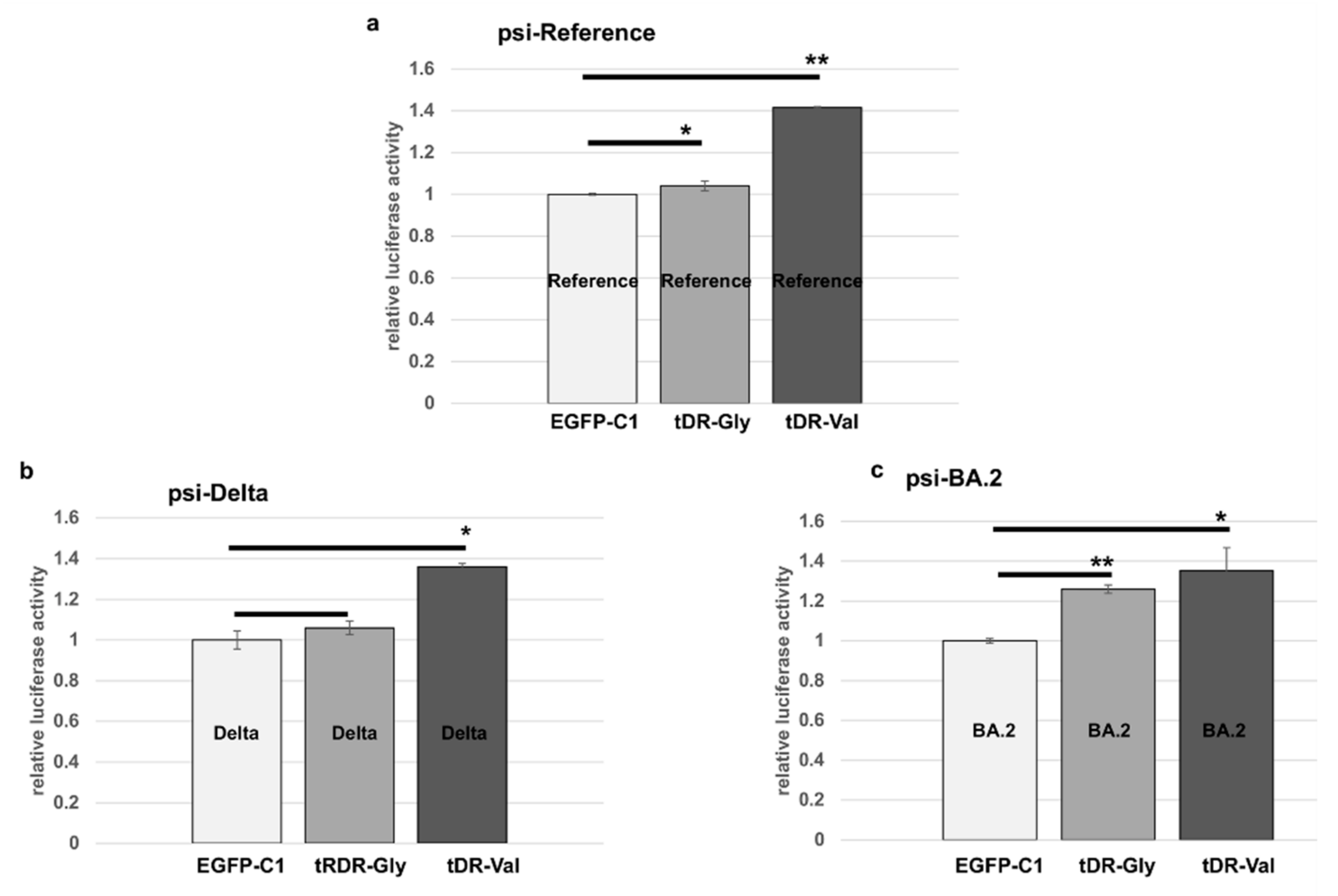
2.5. Sponge Constructs of tDR-Val and tDR-Gly Were Co-Transfected with the Psi-Reference and VOC Plasmids to Determine Direct Regulation by tDR-Gly and tDR-Val
3. Discussion
4. Materials and Methods
4.1. Data Analysis of Small RNAs in Patients’ Plasma
4.2. Data Analysis of Small RNA Libraries from Calu-3 Lung Cells Infected with SARS-CoV-2
4.3. RNAhybrid and RNA22 Analysis
4.4. Luciferase Plasmids Were Constructed Using a Region of the 3′UTR from the Index Virus Reference Sequence, Delta VOC and the Omicron BA.2 VOC
4.5. Sponge Constructs Were Designed to Bind Cellular tDR-Gly and tDR-Val to Inhibit Their Function
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Ghen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Coronavirus Disease (COVID-19) VOCs. Available online: https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern (accessed on 19 February 2022).
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Hale, C.; Saito, Y.; Blachere, N.E.; Bergh, M.; Conlon, E.G.; Schaefer-Babajew, D.J.; DaSilva, J.; Muecksch, F.; Gaebler, C.; et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N. Engl. J. Med. 2021, 384, 2212–2218. [Google Scholar] [CrossRef]
- Cao, L.; Xu, T.; Liu, X.; Ji, Y.; Huang, S.; Peng, H.; Li, C.; Guo, D. The Impact of Accumulated Mutations in SARS-CoV-2 Variants on the qPCR Detection Efficiency. Front. Cell Infect. Microbiol. 2022, 12, 823306. [Google Scholar] [CrossRef]
- Pierce, J.B.; Simion, V.; Icli, B.; Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs. Genes 2020, 11, 1354. [Google Scholar] [CrossRef]
- Plawgo, K.; Raczynska, K.D. Context-Dependent Regulation of Gene Expression by Non-Canonical Small RNAs. Noncoding RNA 2022, 8, 29. [Google Scholar] [CrossRef]
- Yu, X.; Xie, Y.; Zhang, S.; Song, X.; Xiao, B.; Yan, Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021, 11, 461–469. [Google Scholar] [CrossRef]
- Diebel, K.W.; Zhou, K.; Clarke, A.B.; Bemis, L.T. Beyond the Ribosome: Extra-translational Functions of tRNA Fragments. Biomark Insights 2016, 11 (Suppl. S1), 1–8. [Google Scholar] [CrossRef]
- Maillard, P.V.; van der Veen, A.G.; Poirier, E.Z.; Reis e Sousa, C. Slicing and dicing viruses: Antiviral RNA interference in mammals. EMBO J. 2019, 38, e100941. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Henzinger, H.; Barth, D.A.; Klec, C.; Pichler, M. Non-Coding RNAs and SARS-Related Coronaviruses. Viruses 2020, 12, 1374. [Google Scholar] [CrossRef]
- Wu, W.; Choi, E.J.; Lee, I.; Lee, Y.S.; Bao, X. Non-Coding RNAs and Their Role in Respiratory Syncytial Virus (RSV) and Human Metapneumovirus (hMPV) Infections. Viruses 2020, 12, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, J.T.; Varble, A.; Sachidanandam, R.; Zlatev, I.; Manoharan, M.; García-Sastre, A.; TenOever, B.R. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. USA 2010, 107, 11525–11530. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Choi, E.J.; Wang, B.; Zhang, K.; Adam, A.; Huang, G.; Tunkle, L.; Huang, P.; Goru, R.; Imirowicz, I.; et al. Changes of Small Non-coding RNAs by Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Front. Mol. Biosci. 2022, 9, 821137. [Google Scholar] [CrossRef]
- Grehl, C.; Schultheiß, C.; Hoffmann, K.; Binder, M.; Altmann, T.; Grosse, I.; Kuhlmann, M. Detection of SARS-CoV-2 Derived Small RNAs and Changes in Circulating Small RNAs Associated with COVID-19. Viruses 2021, 13, 1593. [Google Scholar] [CrossRef]
- Battaglia, R.; Alonzo, R.; Pennisi, C.; Caponnetto, A.; Ferrara, C.; Stella, M.; Barbagallo, C.; Barbagallo, D.; Ragusa, M.; Purrello, M.; et al. MicroRNA-Mediated Regulation of the Virus Cycle and Pathogenesis in the SARS-CoV-2 Disease. Int. J. Mol. Sci. 2021, 22, 13192. [Google Scholar] [CrossRef]
- Srivastava, M.; Hall, D.; Omoru, O.B.; Gill, H.M.; Smith, S.; Janga, S.C. Mutational Landscape and Interaction of SARS-CoV-2 with Host Cellular Components. Microorganisms 2021, 9, 1794. [Google Scholar] [CrossRef]
- Wang, Q.; Lee, I.; Ren, J.; Ajay, S.S.; Lee, Y.S.; Bao, X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013, 21, 368–379. [Google Scholar] [CrossRef]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wen, Y.Z.; Huang, Z.L.; Shen, X.; Wang, J.H.; Luo, Y.H.; Chen, W.X.; Lun, Z.R.; Li, H.B.; Qu, L.H.; et al. SARS-CoV-2 causes a significant stress response mediated by small RNAs in the blood of COVID-19 patients. Mol. Ther. Nucleic Acids 2022, 27, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Peña, N.; Zhang, W.; Watkins, C.; Halucha, M.; Alshammary, H.; Hernandez, M.M.; Liu, W.C.; Albrecht, R.A.; Garcia-Sastre, A.; Simon, V.; et al. Profiling Selective Packaging of Host RNA and Viral RNA Modification in SARS-CoV-2 Viral Preparations. Front. Cell Dev. Biol. 2022, 10, 768356. [Google Scholar] [CrossRef] [PubMed]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef] [PubMed]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Manning, A.C.; Bagi, A.; Yang, X.; Gokulnath, P.; Spanos, M.; Howard, J.; Chan, P.P.; Sweeney, T.; Kitchen, R.; et al. Distinct Stress-Dependent Signatures of Cellular and Extracellular tRNA-Derived Small RNAs. Adv. Sci. 2022, 9, e2200829. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadi-Dehcheshmeh, M.; Moghbeli, S.M.; Rahimirad, S.; Alanazi, I.O.; Shehri, Z.S.A.; Ebrahimie, E. A Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs. Cells 2021, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Wacker, A.; Weigand, J.E.; Akabayov, S.R.; Altincekic, N.; Bains, J.K.; Banijamali, E.; Binas, O.; Castillo-Martinez, J.; Cetiner, E.; Ceylan, B.; et al. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids. Res. 2020, 48, 12415–12435. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Tengs, T. No species-level losses of s2m suggests critical role in replication of SARS-related coronaviruses. Sci. Rep. 2021, 11, 16145. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bemis, L.; Su, L.J.; Gao, D.; Flaig, T.W. miR-125b Regulation of Androgen Receptor Signaling Via Modulation of the Receptor Complex Co-Repressor NCOR2. Biores Open Access 2012, 1, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Arena, E.T.; Rueden, C.T.; Hiner, M.C.; Wang, S.; Yuan, M.; Eliceiri, K.W. Quantitating the cell: Turning images into numbers with ImageJ. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e260. [Google Scholar] [CrossRef]
- Nunes, A.; Ribeiro, D.R.; Marques, M.; Santos, M.A.S.; Ribeiro, D.; Soares, A.R. Emerging Roles of tRNAs in RNA Virus Infections. Trends Biochem. Sci. 2020, 45, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Torbati, E.; Krause, K.L.; Ussher, J.E. The Immune Response to SARS-CoV-2 and Variants of Concern. Viruses 2021, 3, 1911. [Google Scholar] [CrossRef]
- Mousavi, S.R.; Sajjadi, M.S.; Khosravian, F.; Feizbakhshan, S.; Salmanizadeh, S.; Esfahani, Z.T.; Beni, F.A.; Arab, A.; Kazemi, M.; Shahzamani, K.; et al. Dysregulation of RNA interference components in COVID-19 patients. BMC Res. Notes 2021, 14, 401. [Google Scholar] [CrossRef]
- Zlotorynski, E. Dicing viral RNA in stem cells. Nature reviews. Mol. Cell Biol. 2021, 22, 586. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Buck, M.D.; Chakravarty, P.; Carvalho, J.; Frederico, B.; Cardoso, A.; Healy, L.; Ulferts, R.; Beale, R.; Reis e Sousa, C. An isoform of Dicer protects mammalian stem cells against multiple RNA viruses. Science 2021, 373, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Plowman, T.; Lagos, D. Non-Coding RNAs in COVID-19: Emerging Insights and Current Questions. Noncoding RNA 2021, 7, 54. [Google Scholar] [CrossRef]
- Tengs, T.; Kristoffersen, A.B.; Bachvaroff, T.R.; Jonassen, C.M. A mobile genetic element with unknown function found in distantly related viruses. Virol. J. 2013, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 21 March 2022).
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Li, Z.; Ji, M.; Tan, A.C.; Bemis, J.; Tse, J.V.; Huang, G.; Park, J.; Ji, C.; Chen, J.; et al. MIR29B regulates expression of MLLT11 (AF1Q), an MLL fusion partner, and low MIR29B expression associates with adverse cytogenetics and poor overall survival in AML. Br. J. Haematol. 2011, 153, 753–757. [Google Scholar] [CrossRef]



| Treatment Condition | Control Condition | tRF ID | Log2 Fold Change | Adjusted p-Value |
|---|---|---|---|---|
| SARS-CoV-2 24 h | mock 4 h | 5028c | 2.97 | 0.001 |
| SARS-CoV-2 24 h | mock 4 h | 5027c | −4.89 | <0.001 |
| SARS-CoV-2 24 h | mock 4 h | 5020a | 2.34 | 0.005 |
| SARS-CoV-2 24 h | mock 4 h | 5008c | −1.60 | 0.025 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hendrickson, E.N.; Ericson, M.E.; Bemis, L.T. Host tRNA-Derived RNAs Target the 3′Untranslated Region of SARS-CoV-2. Pathogens 2022, 11, 1479. https://doi.org/10.3390/pathogens11121479
Hendrickson EN, Ericson ME, Bemis LT. Host tRNA-Derived RNAs Target the 3′Untranslated Region of SARS-CoV-2. Pathogens. 2022; 11(12):1479. https://doi.org/10.3390/pathogens11121479
Chicago/Turabian StyleHendrickson, Emily N., Marna E. Ericson, and Lynne T. Bemis. 2022. "Host tRNA-Derived RNAs Target the 3′Untranslated Region of SARS-CoV-2" Pathogens 11, no. 12: 1479. https://doi.org/10.3390/pathogens11121479
APA StyleHendrickson, E. N., Ericson, M. E., & Bemis, L. T. (2022). Host tRNA-Derived RNAs Target the 3′Untranslated Region of SARS-CoV-2. Pathogens, 11(12), 1479. https://doi.org/10.3390/pathogens11121479