The Swine IFN System in Viral Infections: Major Advances and Translational Prospects
Abstract
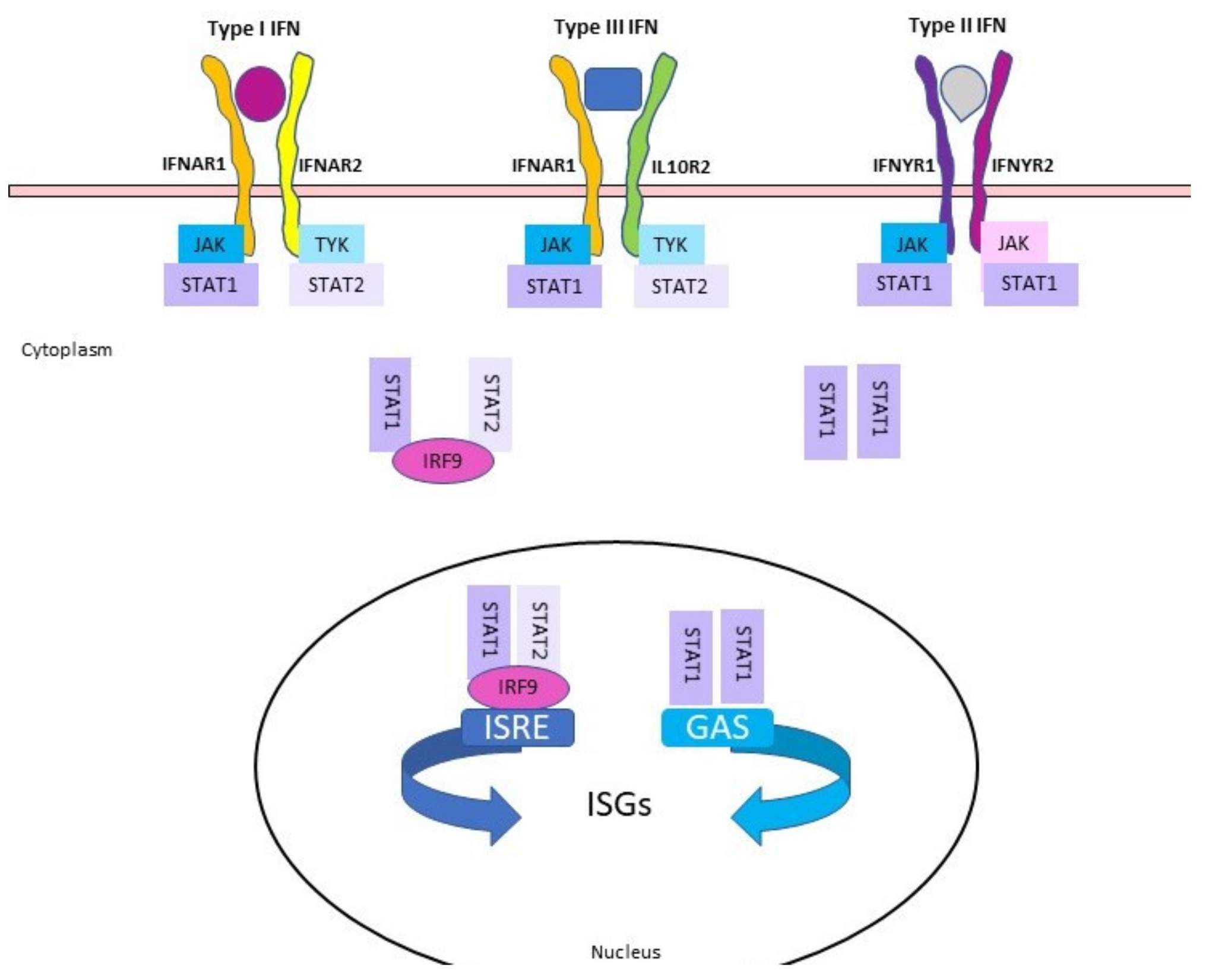
:1. Introduction
2. IFN Responses to RNA Viruses
2.1. Porcine Reproductive and Respiratory Syndrome
2.1.1. Innate Immune Responses to PRRSV
2.1.2. IFN Sensitivity of PRRSV
2.1.3. Type I IFN Responses
2.1.4. Type II IFN Responses
2.1.5. Type III IFN Responses
2.1.6. Translational Prospects
2.2. Foot-and-Mouth Disease
2.2.1. Innate Immune Response to FMDV
2.2.2. IFN Sensitivity of FMDV
2.2.3. Type I IFN Responses
2.2.4. Type II IFN Responses
2.2.5. Type III IFN Responses
2.2.6. Translational Prospects
2.3. Porcine Coronaviruses
2.3.1. Immune Response to PRCV
2.3.2. Translational Prospects
3. IFN Responses to DNA-Viruses
3.1. Porcine Circovirus 2
3.1.1. Innate Immune Response to PCV2
3.1.2. IFN Sensitivity of PCV2
3.1.3. Type I IFN Responses
3.1.4. Type II IFN Responses
3.1.5. Type III IFN Responses
3.1.6. Translational Prospects
3.2. Aujeszky’s Disease
3.2.1. Innate Immune Response to PRV
3.2.2. IFN Sensitivity of PRV
3.2.3. IFN Responses
3.2.4. Translational Prospects
3.3. African Swine Fever Virus
3.3.1. Innate Immune Responses to ASF
3.3.2. Sensitivity to IFN
3.3.3. IFN Response
3.3.4. Translational Prospects
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Isaacs, A.; Lindenmann, J. Virus Interference I The Interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957, 147, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takaoka, A.; Yanai, H. Interferon Signalling Network in Innate Defence. Cell. Microbiol. 2006, 8, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, C.; Razzuoli, E.; Crooke, H.; Soule, O.; Pezzoni, G.; Ferraris, M.; Ferrari, A.; Amadori, M. Differential Biological Activities of Swine Interferon-α Subtypes. J. Interferon Cytokine Res. 2015, 35, 990–1002. [Google Scholar] [CrossRef] [Green Version]
- Der, S.D.; Zhou, A.; Williams, B.R.G.; Silverman, R.H. Identification of Genes Differentially Regulated by Interferon α, β, or γ Using Oligonucleotide Arrays. Proc. Natl. Acad. Sci. USA 1998, 95, 15623–15628. [Google Scholar] [CrossRef] [Green Version]
- Flores, I.; Mariano, T.M.; Pestka, S. Human Interferon Omega (Omega) Binds to the Alpha/Beta Receptor. J. Biol. Chem. 1991, 266, 19875–19877. [Google Scholar] [CrossRef]
- Kotenko, S.V.; Gallagher, G.; Baurin, V.V.; Lewis-Antes, A.; Shen, M.; Shah, N.K.; Langer, J.A.; Sheikh, F.; Dickensheets, H.; Donnelly, R.P. IFN-Lambdas Mediate Antiviral Protection through a Distinct Class II Cytokine Receptor Complex. Nat. Immunol. 2003, 4, 69–77. [Google Scholar] [CrossRef]
- Donnelly, R.P.; Kotenko, S.V. Interferon-Lambda: A New Addition to an Old Family. J. Interferon Cytokine Res. 2010, 30, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Sang, Y.; Rowland, R.R.R.; Hesse, R.A.; Blecha, F. Differential Expression and Activity of the Porcine Type I Interferon Family. Physiol. Genom. 2010, 42, 248–258. [Google Scholar] [CrossRef] [Green Version]
- Yerle, M.; Gellin, J.; Echard, G.; Lefevre, F.; Gillois, M. Chromosomal Localization of Leukocyte Interferon Gene in the Pig (Sus Scrofa domestica L.) by in Situ Hybridization. Cytogenet. Cell Genet. 1986, 42, 129–132. [Google Scholar] [CrossRef]
- Cheng, G.; Chen, W.; Li, Z.; Yan, W.; Zhao, X.; Xie, J.; Liu, M.; Zhang, H.; Zhong, Y.; Zheng, Z. Characterization of the Porcine Alpha Interferon Multigene Family. Gene 2006, 382, 28–38. [Google Scholar] [CrossRef]
- Dafny, N. Is Interferon-α a Neuromodulator? Brain Res. Rev. 1998, 26, 1–15. [Google Scholar] [CrossRef]
- Li, T.; Chen, Z.J. The CGAS–CGAMP–STING Pathway Connects DNA Damage to Inflammation, Senescence, and Cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef]
- Pfeffer, L.M. The Role of Nuclear Factor ΚB in the Interferon Response. J. Interferon Cytokine Res. 2011, 31, 553–559. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [Green Version]
- Yum, S.; Li, M.; Fang, Y.; Chen, Z.J. TBK1 Recruitment to STING Activates Both IRF3 and NF-ΚB That Mediate Immune Defense against Tumors and Viral Infections. Proc. Natl. Acad. Sci. USA 2021, 118, e2100225118. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and Viruses: An Interplay between Induction, Signalling, Antiviral Responses and Virus Countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef]
- Chinsangaram, J.; Mason, P.W.; Grubman, M.J. Protection of Swine by Live and Inactivated Vaccines Prepared from a Leader Proteinase-Deficient Serotype A12 Foot-and-Mouth Disease Virus. Vaccine 1998, 16, 1516–1522. [Google Scholar] [CrossRef]
- Ronco, L.V.; Karpova, A.Y.; Vidal, M.; Howley, P.M. Human Papillomavirus 16 E6 Oncoprotein Binds to Interferon Regulatory Factor-3 and Inhibits Its Transcriptional Activity. Genes Dev. 1998, 12, 2061–2072. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Cho, Y.S.; Cho, M.C.; Shim, J.H.; Lee, K.A.; Ko, K.K.; Choe, Y.K.; Park, S.N.; Hoshino, T.; Kim, S.; et al. Both E6 and E7 Oncoproteins of Human Papillomavirus 16 Inhibit IL-18-Induced IFN-Gamma Production in Human Peripheral Blood Mononuclear and NK Cells. J. Immunol. 2001, 167, 497–504. [Google Scholar] [CrossRef]
- Lo Cigno, I.; Calati, F.; Albertini, S.; Gariglio, M. Subversion of Host Innate Immunity by Human Papillomavirus Oncoproteins. Pathogens 2020, 9, 292. [Google Scholar] [CrossRef] [Green Version]
- Yokota, S.; Yokosawa, N.; Okabayashi, T.; Suzutani, T.; Miura, S.; Jimbow, K.; Fujii, N. Induction of Suppressor of Cytokine Signaling-3 by Herpes Simplex Virus Type 1 Contributes to Inhibition of the Interferon Signaling Pathway. J. Virol. 2004, 78, 6282–6286. [Google Scholar] [CrossRef] [Green Version]
- Basler, C.F.; Mikulasova, A.; Martinez-Sobrido, L.; Paragas, J.; Mühlberger, E.; Bray, M.; Klenk, H.-D.; Palese, P.; García-Sastre, A. The Ebola Virus VP35 Protein Inhibits Activation of Interferon Regulatory Factor 3. J. Virol. 2003, 77, 7945–7956. [Google Scholar] [CrossRef] [Green Version]
- Brzózka, K.; Finke, S.; Conzelmann, K.-K. Identification of the Rabies Virus Alpha/Beta Interferon Antagonist: Phosphoprotein P Interferes with Phosphorylation of Interferon Regulatory Factor 3. J. Virol. 2005, 79, 7673–7681. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Metcalf, J.P. The Role of Type I IFNs in Influenza: Antiviral Superheroes or Immunopathogenic Villains? J. Innate Immun. 2020, 12, 437–447. [Google Scholar] [CrossRef]
- Holtkamp, D.J.; Kliebenstein, J.B.; Neumann, E.J.; Zimmerman, J.J.; Rotto, H.F.; Yoder, T.K.; Wang, C.; Yeske, P.E.; Mowrer, C.L.; Haley, C.A. Assessment of the Economic Impact of Porcine Reproductive and Respiratory Syndrome Virus on United States Pork Producers. J. Swine Health Prod. 2013, 21, 13. [Google Scholar]
- Zimmerman, J.J.; Dee, S.A.; Holtkamp, D.J.; Murtaugh, M.P.; Stadejek, T.; Stevenson, G.W.; Torremorell, M.; Yang, H.; Zhang, J. Porcine Reproductive and Respiratory Syndrome Viruses (Porcine Arteriviruses). In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 685–708. ISBN 9781119350859. [Google Scholar]
- Crisci, E.; Fraile, L.; Montoya, M. Cellular Innate Immunity against PRRSV and Swine Influenza Viruses. Vet. Sci. 2019, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org/ (accessed on 13 December 2021).
- Amadori, M.; Razzuoli, E. Immune Control of PRRS: Lessons to Be Learned and Possible Ways Forward. Front. Vet. Sci. 2014, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Mateu, E.; Diaz, I. The Challenge of PRRS Immunology. Vet. J. 2008, 177, 345–351. [Google Scholar] [CrossRef]
- Yoo, D.; Song, C.; Sun, Y.; Du, Y.; Kim, O.; Liu, H.-C. Modulation of Host Cell Responses and Evasion Strategies for Porcine Reproductive and Respiratory Syndrome Virus. Virus Res. 2010, 154, 48–60. [Google Scholar] [CrossRef]
- Costers, S.; Delputte, P.L.; Nauwynck, H.J. Porcine Reproductive and Respiratory Syndrome Virus-Infected Alveolar Macrophages Contain No Detectable Levels of Viral Proteins in Their Plasma Membrane and Are Protected against Antibody-Dependent, Complement-Mediated Cell Lysis. J. Gen. Virol. 2006, 87, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Costers, S.; Lefebvre, D.J.; Goddeeris, B.; Delputte, P.L.; Nauwynck, H.J. Functional Impairment of PRRSV-Specific Peripheral CD3+CD8high Cells. Vet. Res. 2009, 40, 46. [Google Scholar] [CrossRef] [Green Version]
- Lunney, J.K.; Fritz, E.R.; Reecy, J.M.; Kuhar, D.; Prucnal, E.; Molina, R.; Christopher-Hennings, J.; Zimmerman, J.; Rowland, R.R.R. Interleukin-8, Interleukin-1beta, and Interferon-Gamma Levels Are Linked to PRRS Virus Clearance. Viral Immunol. 2010, 23, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, A.; Mateu, E.; Murtaugh, M.P.; Summerfield, A. Impact of Genotype 1 and 2 of Porcine Reproductive and Respiratory Syndrome Viruses on Interferon-α Responses by Plasmacytoid Dendritic Cells. Vet. Res. 2013, 44, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albina, E.; Carrat, C.; Charley, B. Interferon-Alpha Response to Swine Arterivirus (PoAV), the Porcine Reproductive and Respiratory Syndrome Virus. J. Interferon Cytokine Res. 1998, 18, 485–490. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Overend, C.; Ambrogio, J.; Maganti, R.J.; Grubman, M.J.; Garmendia, A.E. Marked Differences between MARC-145 Cells and Swine Alveolar Macrophages in IFNβ-Induced Activation of Antiviral State against PRRSV. Vet. Immunol. Immunopathol. 2011, 139, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Bautista, E.M.; Molitor, T.W. IFN Gamma Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication in Macrophages. Arch. Virol. 1999, 144, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- García-Nicolás, O.; Baumann, A.; Vielle, N.J.; Gómez-Laguna, J.; Quereda, J.J.; Pallarés, F.J.; Ramis, G.; Carrasco, L.; Summerfield, A. Virulence and Genotype-Associated Infectivity of Interferon-Treated Macrophages by Porcine Reproductive and Respiratory Syndrome Viruses. Virus Res. 2014, 179, 204–211. [Google Scholar] [CrossRef]
- Ogno, G.; Sautter, C.A.; Canelli, E.; García-Nicolás, O.; Stadejek, T.; Martelli, P.; Borghetti, P.; Summerfield, A. In vitro Characterization of PRRSV Isolates with Different in Vivo Virulence Using Monocyte-Derived Macrophages. Vet. Microbiol. 2019, 231, 139–146. [Google Scholar] [CrossRef]
- Zhao, J.; Zhu, L.; Xu, L.; Huang, J.; Sun, X.; Xu, Z. Porcine Interferon Lambda 3 (IFN-Λ3) Shows Potent Anti-PRRSV Activity in Primary Porcine Alveolar Macrophages (PAMs). BMC Vet. Res. 2020, 16, 408. [Google Scholar] [CrossRef]
- Van Reeth, K.; Labarque, G.; Nauwynck, H.; Pensaert, M. Differential Production of Proinflammatory Cytokines in the Pig Lung during Different Respiratory Virus Infections: Correlations with Pathogenicity. Res. Vet. Sci. 1999, 67, 47–52. [Google Scholar] [CrossRef]
- Li, L.; Wei, Z.; Zhou, Y.; Gao, F.; Jiang, Y.; Yu, L.; Zheng, H.; Tong, W.; Yang, S.; Zheng, H.; et al. Host MiR-26a Suppresses Replication of Porcine Reproductive and Respiratory Syndrome Virus by Upregulating Type I Interferons. Virus Res. 2015, 195, 86–94. [Google Scholar] [CrossRef]
- Chen, J.; Shi, X.; Zhang, X.; Wang, A.; Wang, L.; Yang, Y.; Deng, R.; Zhang, G.-P. MicroRNA 373 Facilitates the Replication of Porcine Reproductive and Respiratory Syndrome Virus by Its Negative Regulation of Type I Interferon Induction. J. Virol. 2017, 91, e01311-16. [Google Scholar] [CrossRef] [Green Version]
- Chang, X.; Shi, X.; Zhang, X.; Chen, J.; Fan, X.; Yang, Y.; Wang, L.; Wang, A.; Deng, R.; Zhou, E.; et al. MiR-382-5p Promotes Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Replication by Negatively Regulating the Induction of Type I Interferon. FASEB J. 2020, 34, 4497–4511. [Google Scholar] [CrossRef] [Green Version]
- Ladinig, A.; Ashley, C.; Detmer, S.E.; Wilkinson, J.M.; Lunney, J.K.; Plastow, G.; Harding, J.C. Maternal and Fetal Predictors of Fetal Viral Load and Death in Third Trimester, Type 2 Porcine Reproductive and Respiratory Syndrome Virus Infected Pregnant Gilts. Vet. Res. 2015, 46, 107. [Google Scholar] [CrossRef] [Green Version]
- Amadori, M.; Listorti, V.; Razzuoli, E. Reappraisal of PRRS Immune Control Strategies: The Way Forward. Pathogens 2021, 10, 1073. [Google Scholar] [CrossRef]
- Davidson, S.; Maini, M.K.; Wack, A. Disease-Promoting Effects of Type I Interferons in Viral, Bacterial, and Coinfections. J. Interferon Cytokine Res. 2015, 35, 252–264. [Google Scholar] [CrossRef]
- Summerfield, A.; Alves, M.; Ruggli, N.; de Bruin, M.G.M.; McCullough, K.C. High IFN-Alpha Responses Associated with Depletion of Lymphocytes and Natural IFN-Producing Cells during Classical Swine Fever. J. Interferon Cytokine Res. 2006, 26, 248–255. [Google Scholar] [CrossRef]
- Franzoni, G.; Edwards, J.C.; Kurkure, N.V.; Edgar, D.S.; Sanchez-Cordon, P.J.; Haines, F.J.; Salguero, F.J.; Everett, H.E.; Bodman-Smith, K.B.; Crooke, H.R.; et al. Partial Activation of Natural Killer and Γδ T Cells by Classical Swine Fever Viruses Is Associated with Type I Interferon Elicited from Plasmacytoid Dendritic Cells. Clin. Vaccine Immunol. 2014, 21, 1410–1420. [Google Scholar] [CrossRef] [Green Version]
- Candotti, P.; Dotti, S.; Guana, S.; Rota Nodari, S.; Amadori, M.; Villa, R.; Petrini, S.; Lombardi, G.; Ferrari, M. Susceptibility of Pure Bred Large White and Landrace Pigs to Experimental Infection with Porcine Reproductive and Respiratory Syndrome Virus. In Proceedings of the 20th IPVS Congress (PRRSV), Durban, South Africa, 22–26 June 2008; pp. 22–26. [Google Scholar]
- Razzuoli, E.; Dotti, S.; Villa, R.; Martinelli, N.; Ferrari, M.; Amadori, M. Early Immune Responses to Infection by Attenuated and Non-Attenuated, Type I PRRS Virus Strains. In Proceedings of the 4th European Veterinary Immunology Workshop (EVIW), Edinburgh, Scotland, 2–4 September 2012; p. 23. [Google Scholar]
- Aman, M.J.; Tretter, T.; Eisenbeis, I.; Bug, G.; Decker, T.; Aulitzky, W.E.; Tilg, H.; Huber, C.; Peschel, C. Interferon-Alpha Stimulates Production of Interleukin-10 in Activated CD4+ T Cells and Monocytes. Blood 1996, 87, 4731–4736. [Google Scholar] [CrossRef] [Green Version]
- Van Gucht, S.; Van Reeth, K.; Pensaert, M. Interaction between Porcine Reproductive-Respiratory Syndrome Virus and Bacterial Endotoxin in the Lungs of Pigs: Potentiation of Cytokine Production and Respiratory Disease. J. Clin. Microbiol. 2003, 41, 960–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimeno, M.; Darwich, L.; Diaz, I.; de la Torre, E.; Pujols, J.; Martín, M.; Inumaru, S.; Cano, E.; Domingo, M.; Montoya, M.; et al. Cytokine Profiles and Phenotype Regulation of Antigen Presenting Cells by Genotype-I Porcine Reproductive and Respiratory Syndrome Virus Isolates. Vet. Res. 2011, 42, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlazzo, G.; Ruggeri, J.; Boniotti, M.B.; Guarneri, F.; Barbieri, I.; Tonni, M.; Bertasio, C.; Alborali, G.L.; Amadori, M. In vitro Cytokine Responses to Virulent PRRS Virus Strains. Front. Vet. Sci. 2020, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Levings, M.K.; Sangregorio, R.; Galbiati, F.; Squadrone, S.; de Waal Malefyt, R.; Roncarolo, M.G. IFN-Alpha and IL-10 Induce the Differentiation of Human Type 1 T Regulatory Cells. J. Immunol. 2001, 166, 5530–5539. [Google Scholar] [CrossRef] [Green Version]
- Silva-Campa, E.; Flores-Mendoza, L.; Reséndiz, M.; Pinelli-Saavedra, A.; Mata-Haro, V.; Mwangi, W.; Hernández, J. Induction of T Helper 3 Regulatory Cells by Dendritic Cells Infected with Porcine Reproductive and Respiratory Syndrome Virus. Virology 2009, 387, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Bao, D.; Wang, R.; Qiao, S.; Wan, B.; Wang, Y.; Liu, M.; Shi, X.; Guo, J.; Zhang, G. Antibody-Dependent Enhancement of PRRSV Infection down-Modulates TNF-α and IFN-β Transcription in Macrophages. Vet. Immunol. Immunopathol. 2013, 156, 128–134. [Google Scholar] [CrossRef]
- Darwich, L.; Díaz, I.; Mateu, E. Certainties, Doubts and Hypotheses in Porcine Reproductive and Respiratory Syndrome Virus Immunobiology. Virus Res. 2010, 154, 123–132. [Google Scholar] [CrossRef]
- Meier, W.A.; Galeota, J.; Osorio, F.A.; Husmann, R.J.; Schnitzlein, W.M.; Zuckermann, F.A. Gradual Development of the Interferon-Gamma Response of Swine to Porcine Reproductive and Respiratory Syndrome Virus Infection or Vaccination. Virology 2003, 309, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Diaz, I.; Gimeno, M.; Darwich, L.; Navarro, N.; Kuzemtseva, L.; Galindo, I.; Segalés, J.; Martín, M.; Pujols, J.; Mateu, E. Characterization of Homologous and Heterologous Adaptive Immune Responses in Porcine Reproductive and Respiratory Syndrome Virus Infection. Vet. Res. 2012, 43, 30. [Google Scholar] [CrossRef] [Green Version]
- Lowe, J.E.; Husmann, R.; Firkins, L.D.; Zuckermann, F.A.; Goldberg, T.L. Correlation of Cell-Mediated Immunity against Porcine Reproductive and Respiratory Syndrome Virus with Protection against Reproductive Failure in Sows during Outbreaks of Porcine Reproductive and Respiratory Syndrome in Commercial Herds. J. Am. Vet. Med. Assoc. 2005, 226, 1707–1711. [Google Scholar] [CrossRef] [Green Version]
- Amadori, M.; Zanotti, C. Immunoprophylaxis in Intensive Farming Systems: The Way Forward. Vet. Immunol. Immunopathol. 2016, 181, 2–9. [Google Scholar] [CrossRef]
- Drigo, M.; Giacomini, E.; Lazzaro, M.; Pasotto, D.; Bilato, D.; Ruggeri, J.; Boniotti, M.B.; Alborali, G.L.; Amadori, M. Comparative Evaluation of Immune Responses of Swine in PRRS-Stable and Unstable Herds. Vet. Immunol. Immunopathol. 2018, 200, 32–39. [Google Scholar] [CrossRef]
- Salmon, H.; Berri, M.; Gerdts, V.; Meurens, F. Humoral and Cellular Factors of Maternal Immunity in Swine. Dev. Comp. Immunol. 2009, 33, 384–393. [Google Scholar] [CrossRef]
- Wesley, R.D.; Lager, K.M.; Kehrli, M.E. Infection with Porcine Reproductive and Respiratory Syndrome Virus Stimulates an Early Gamma Interferon Response in the Serum of Pigs. Can. J. Vet. Res. 2006, 70, 176–182. [Google Scholar]
- Welch, S.-K.W.; Calvert, J.G. A Brief Review of CD163 and Its Role in PRRSV Infection. Virus Res. 2010, 154, 98–103. [Google Scholar] [CrossRef]
- Karniychuk, U.U.; Nauwynck, H.J. Pathogenesis and Prevention of Placental and Transplacental Porcine Reproductive and Respiratory Syndrome Virus Infection. Vet. Res. 2013, 44, 95. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, V.; Manickam, C.; Binjawadagi, B.; Linhares, D.; Murtaugh, M.P.; Renukaradhya, G.J. Evaluation of Immune Responses to Porcine Reproductive and Respiratory Syndrome Virus in Pigs during Early Stage of Infection under Farm Conditions. Virol. J. 2012, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.-W.; Nam, E.; Lee, Y.J.; Noh, Y.-H.; Lee, S.-C.; Yoon, I.-J.; Kim, H.-S.; Kang, S.-Y.; Choi, Y.-K.; Lee, C. Genomic Analysis and Pathogenic Characteristics of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Nsp2 Deletion Strains Isolated in Korea. Vet. Microbiol. 2014, 170, 232–245. [Google Scholar] [CrossRef]
- Razzuoli, E.; Villa, R.; Ferrari, A.; Amadori, M. A Pig Tonsil Cell Culture Model for Evaluating Oral, Low-Dose IFN-α Treatments. Vet. Immunol. Immunopathol. 2014, 160, 244–254. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Lager, K.M.; Grubman, M.J.; Brough, D.E.; Ettyreddy, D.; Sacco, R.E.; Gauger, P.C.; Loving, C.L.; Vorwald, A.C.; Kehrli, M.E., Jr.; et al. Adenovirus-Mediated Expression of Interferon-α Delays Viral Replication and Reduces Disease Signs in Swine Challenged with Porcine Reproductive and Respiratory Syndrome Virus. Viral Immunol. 2009, 22, 173–180. [Google Scholar] [CrossRef]
- Charerntantanakul, W. Adjuvants for Porcine Reproductive and Respiratory Syndrome Virus Vaccines. Vet. Immunol. Immunopathol. 2009, 129, 1–13. [Google Scholar] [CrossRef]
- Ruggeri, J.; Ferlazzo, G.; Boniotti, M.B.; Capucci, L.; Guarneri, F.; Barbieri, I.; Alborali, G.L.; Amadori, M. Characterization of the IgA Response to PRRS Virus in Pig Oral Fluids. PLoS ONE 2020, 15, e0229065. [Google Scholar] [CrossRef] [Green Version]
- Grubman, M.J.; Baxt, B. Foot-and-Mouth Disease. Clin. Microbiol. Rev. 2004, 17, 465–493. [Google Scholar] [CrossRef] [Green Version]
- Kitching, R.P.; Alexandersen, S. Clinical Variation in Foot and Mouth Disease: Pigs. Rev. Sci. Tech. 2002, 21, 513–518. [Google Scholar] [CrossRef]
- Domingo, E.; Baranowski, E.; Escarmís, C.; Sobrino, F. Foot-and-Mouth Disease Virus. Comp. Immunol. Microbiol. Infect. Dis. 2002, 25, 297–308. [Google Scholar] [CrossRef]
- Singh, R.K.; Sharma, G.K.; Mahajan, S.; Dhama, K.; Basagoudanavar, S.H.; Hosamani, M.; Sreenivasa, B.P.; Chaicumpa, W.; Gupta, V.K.; Sanyal, A. Foot-and-Mouth Disease Virus: Immunobiology, Advances in Vaccines and Vaccination Strategies Addressing Vaccine Failures-An Indian Perspective. Vaccines 2019, 7, 90. [Google Scholar] [CrossRef] [Green Version]
- Poonsuk, K.; Giménez-Lirola, L.; Zimmerman, J.J. A Review of Foot-and-Mouth Disease Virus (FMDV) Testing in Livestock with an Emphasis on the Use of Alternative Diagnostic Specimens. Anim. Health Res. Rev. 2018, 19, 100–112. [Google Scholar] [CrossRef]
- Toka, F.N.; Golde, W.T. Cell Mediated Innate Responses of Cattle and Swine Are Diverse during Foot-and-Mouth Disease Virus (FMDV) Infection: A Unique Landscape of Innate Immunity. Immunol. Lett. 2013, 152, 135–143. [Google Scholar] [CrossRef]
- Golde, W.T.; Pacheco, J.M.; Duque, H.; Doel, T.; Penfold, B.; Ferman, G.S.; Gregg, D.R.; Rodriguez, L.L. Vaccination against Foot-and-Mouth Disease Virus Confers Complete Clinical Protection in 7 Days and Partial Protection in 4 Days: Use in Emergency Outbreak Response. Vaccine 2005, 23, 5775–5782. [Google Scholar] [CrossRef]
- Barnard, A.L.; Arriens, A.; Cox, S.; Barnett, P.; Kristensen, B.; Summerfield, A.; McCullough, K.C. Immune Response Characteristics Following Emergency Vaccination of Pigs against Foot-and-Mouth Disease. Vaccine 2005, 23, 1037–1047. [Google Scholar] [CrossRef]
- Barnett, P.V.; Cox, S.J.; Aggarwal, N.; Gerber, H.; McCullough, K.C. Further Studies on the Early Protective Responses of Pigs Following Immunisation with High Potency Foot and Mouth Disease Vaccine. Vaccine 2002, 20, 3197–3208. [Google Scholar] [CrossRef]
- Dudnikov, A.I.; Fao, R.; Vladimir, E.C.; Mikhalishin, V.V.; Gusev, A.A.; Ulupov, N.A.; Mamkov, N.S. Inactivated FMD (Foot-and-Mouth Disease) Vaccine with New Immunobiological Characteristics Produced at the All-Russian Research Institute for Animal Health; FAO: Rome, Italy, 1995. [Google Scholar]
- Takamatsu, H.-H.; Denyer, M.S.; Stirling, C.; Cox, S.; Aggarwal, N.; Dash, P.; Wileman, T.E.; Barnett, P.V. Porcine Gammadelta T Cells: Possible Roles on the Innate and Adaptive Immune Responses Following Virus Infection. Vet. Immunol. Immunopathol. 2006, 112, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yi, J.; Yang, W.; Ren, J.; Wen, Y.; Zheng, H.; Li, D. Advances in Foot-and-Mouth Disease Virus Proteins Regulating Host Innate Immunity. Front. Microbiol. 2020, 11, 2046. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, X.; Zhang, D.; Hou, J.; Xu, G.; Sheng, C.; Choudhury, S.M.; Zhu, Z.; Li, D.; Zhang, K.; et al. Molecular Mechanisms of Immune Escape for Foot-and-Mouth Disease Virus. Pathogens 2020, 9, 729. [Google Scholar] [CrossRef]
- Ma, X.; Ma, L.; Chang, Q.; Ma, P.; Li, L.-J.; Wang, Y.; Ma, Z.; Cao, X. Type I Interferon Induced and Antagonized by Foot-and-Mouth Disease Virus. Front. Microbiol. 2018, 9, 1862. [Google Scholar] [CrossRef] [Green Version]
- Stenfeldt, C.; Heegaard, P.M.; Stockmarr, A.; Tjørnehøj, K.; Belsham, G.J. Analysis of the Acute Phase Responses of Serum Amyloid A, Haptoglobin and Type 1 Interferon in Cattle Experimentally Infected with Foot-and-Mouth Disease Virus Serotype O. Vet. Res. 2011, 42, 66. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, A.I.; Alexandersen, S. Predicting the Spread of Foot and Mouth Disease by Airborne Virus. Rev. Sci. Tech. 2002, 21, 569–575. [Google Scholar] [CrossRef]
- Pacheco, J.M.; Smoliga, G.R.; O’Donnell, V.; Brito, B.P.; Stenfeldt, C.; Rodriguez, L.L.; Arzt, J. Persistent Foot-and-Mouth Disease Virus Infection in the Nasopharynx of Cattle; Tissue-Specific Distribution and Local Cytokine Expression. PLoS ONE 2015, 10, e0125698. [Google Scholar] [CrossRef]
- Amadori, M.; Bugnetti, M.; Tironi, D. Induction and Characterization of Swine Beta Interferon. Comp. Immunol. Microbiol. Infect. Dis. 1987, 10, 71–78. [Google Scholar] [CrossRef]
- Dash, P.; Barnett, P.V.; Denyer, M.S.; Jackson, T.; Stirling, C.M.A.; Hawes, P.C.; Simpson, J.L.; Monaghan, P.; Takamatsu, H.-H. Foot-and-Mouth Disease Virus Replicates Only Transiently in Well-Differentiated Porcine Nasal Epithelial Cells. J. Virol. 2010, 84, 9149–9160. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhao, F.; Gong, M.; Shao, J.; Xie, Y.; Chang, H.; Zhang, Y. Antiviral Activity of Porcine Interferon Omega 7 against Foot-and-mouth Disease Virus in vitro. J. Med. Virol. 2019, 91, 208–214. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Shao, J.; Zhao, F.; Gong, M.; Xie, Y.; Chang, H.; Zhang, Y. Antiviral Activity of Porcine Interferon Delta 8 against Foot-and-Mouth Disease Virus in vitro. Int. Immunopharmacol. 2018, 59, 47–52. [Google Scholar] [CrossRef]
- Li, S.-F.; Gong, M.-J.; Xie, Y.-L.; Shao, J.-J.; Zhao, F.-R.; Zhang, Y.-G.; Chang, H.-Y. A Novel Type I Interferon, Interferon Alphaomega, Shows Antiviral Activity against Foot-and-Mouth Disease Virus in vitro. Microb. Pathog. 2019, 127, 79–84. [Google Scholar] [CrossRef]
- Zhang, Z.D.; Hutching, G.; Kitching, P.; Alexandersen, S. The Effects of Gamma Interferon on Replication of Foot-and-Mouth Disease Virus in Persistently Infected Bovine Cells. Arch. Virol. 2002, 147, 2157–2167. [Google Scholar] [CrossRef]
- Kim, S.-M.; Park, J.-H.; Lee, K.-N.; Kim, S.-K.; You, S.-H.; Kim, T.; Tark, D.; Lee, H.-S.; Seo, M.-G.; Kim, B. Robust Protection against Highly Virulent Foot-and-Mouth Disease Virus in Swine by Combination Treatment with Recombinant Adenoviruses Expressing Porcine Alpha and Gamma Interferons and Multiple Small Interfering RNAs. J. Virol. 2015, 89, 8267–8279. [Google Scholar] [CrossRef] [Green Version]
- Díaz-San Segundo, F.; Weiss, M.; Perez-Martín, E.; Koster, M.J.; Zhu, J.; Grubman, M.J.; de los Santos, T. Antiviral Activity of Bovine Type III Interferon against Foot-and-Mouth Disease Virus. Virology 2011, 413, 283–292. [Google Scholar] [CrossRef] [Green Version]
- Ahl, R. Interferon and Foot-and-Mouth Disease Virus. In Proceedings of the Session of the Research Group of the European Commission for the Control of Foot-and-Mouth Disease, Borovets, Bulgaria, 5–8 September 2000; FAO: Rome, Italy, 2000; pp. 139–147. [Google Scholar]
- Ahl, R.; Gottschalk, M. Production and Purification of Bovine Interferon Beta. Methods Enzymol. 1986, 119, 211–220. [Google Scholar] [CrossRef]
- Grubman, M.J.; Moraes, M.P.; Diaz-San Segundo, F.; Pena, L.; de los Santos, T. Evading the Host Immune Response: How Foot-and-Mouth Disease Virus Has Become an Effective Pathogen. FEMS Immunol. Med. Microbiol. 2008, 53, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Amadori, M.; Cristiano, A.; Ferrari, M. Constitutive Expression of Interferons in Swine Leukocytes. Res. Vet. Sci. 2010, 88, 64–71. [Google Scholar] [CrossRef]
- Razzuoli, E.; Villa, R.; Sossi, E.; Amadori, M. Reverse Transcription Real-Time PCR for Detection of Porcine Interferon α and β Genes. Scand. J. Immunol. 2011, 74, 412–418. [Google Scholar] [CrossRef]
- Bautista, E.M.; Ferman, G.S.; Gregg, D.; Brum, M.C.S.; Grubman, M.J.; Golde, W.T. Constitutive Expression of Alpha Interferon by Skin Dendritic Cells Confers Resistance to Infection by Foot-and-Mouth Disease Virus. J. Virol. 2005, 79, 4838–4847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, E.; Juleff, N.; Gubbins, S.; Prentice, H.; Seago, J.; Charleston, B. Bovine Plasmacytoid Dendritic Cells Are the Major Source of Type I Interferon in Response to Foot-and-Mouth Disease Virus in vitro and in Vivo. J. Virol. 2011, 85, 4297–4308. [Google Scholar] [CrossRef] [Green Version]
- Yadin, H.; Chai, D.; Bril, A.; Gelman, B.; Amadori, M. Experimental Vaccination against FMDV with Immunocomplexes. In Proceedings of the Session of the Research Group of the Standing Technical Committee of FAO EUFMD, Borovets, Bulgaria, 5–8 September 2000; FAO: Rome, Italy, 2000; pp. 263–269. [Google Scholar]
- Amadori, M. Influence of FMD Vaccine Potency and Formulation on the Immune Response to Heterologous FMDV Strains. In Proceedings of the Session of the Research Group of the European Commission for the Control of Foot-and-Mouth Disease, Aldershot, UK, 5–8 September 1998; FAO: Rome, Italy, 1998; pp. 261–265. [Google Scholar]
- Bucafusco, D.; Di Giacomo, S.; Pega, J.; Schammas, J.M.; Cardoso, N.; Capozzo, A.V.; Perez-Filgueira, M. Foot-and-Mouth Disease Vaccination Induces Cross-Reactive IFN-γ Responses in Cattle That Are Dependent on the Integrity of the 140S Particles. Virology 2015, 476, 11–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, Y.; Fleming, L.; Statham, B.; Hamblin, P.; Barnett, P.; Paton, D.J.; Park, J.-H.; Joo, Y.S.; Parida, S. Interferon-γ Induced by in vitro Re-Stimulation of CD4+ T-Cells Correlates with in Vivo FMD Vaccine Induced Protection of Cattle against Disease and Persistent Infection. PLoS ONE 2012, 7, e44365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.K.; Bhatt, M.; Sankar, M.; Mohapatra, J.K.; Dash, B.B.; Gowane, G.R.; Subramaniam, S.; Ranjan, R.; Pattnaik, B. Kinetics of Interferon Gamma and Interleukin-21 Response Following Foot and Mouth Disease Virus Infection. Microb. Pathog. 2018, 125, 20–25. [Google Scholar] [CrossRef]
- Stenfeldt, C.; Eschbaumer, M.; Rekant, S.I.; Pacheco, J.M.; Smoliga, G.R.; Hartwig, E.J.; Rodriguez, L.L.; Arzt, J. The Foot-and-Mouth Disease Carrier State Divergence in Cattle. J. Virol. 2016, 90, 6344–6364. [Google Scholar] [CrossRef] [Green Version]
- Dias, C.C.A.; Moraes, M.P.; Segundo, F.D.-S.; de los Santos, T.; Grubman, M.J. Porcine Type I Interferon Rapidly Protects Swine against Challenge with Multiple Serotypes of Foot-and-Mouth Disease Virus. J. Interferon Cytokine Res. 2011, 31, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Carvajal, L.; Diaz-San Segundo, F.; Ramirez-Medina, E.; Rodríguez, L.L.; de Los Santos, T. Constitutively Active IRF7/IRF3 Fusion Protein Completely Protects Swine against Foot-and-Mouth Disease. J. Virol. 2016, 90, 8809–8821. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Huang, Q.; Cao, Y.; Qian, P.; Chen, H. Porcine Interferon-Gamma Protects Swine from Foot-and-Mouth Disease Virus (FMDV). Vet. Immunol. Immunopathol. 2008, 122, 309–311. [Google Scholar] [CrossRef]
- Cheng, G.; Zhao, X.; Yan, W.; Wang, W.; Zuo, X.; Huang, K.; Liu, Y.; Chen, J.; Wang, J.; Cong, W.; et al. Alpha Interferon Is a Powerful Adjuvant for a Recombinant Protein Vaccine against Foot-and-Mouth Disease Virus in Swine, and an Effective Stimulus of in Vivo Immune Response. Vaccine 2007, 25, 5199–5208. [Google Scholar] [CrossRef]
- Moraes, M.P.; Chinsangaram, J.; Brum, M.C.S.; Grubman, M.J. Immediate Protection of Swine from Foot-and-Mouth Disease: A Combination of Adenoviruses Expressing Interferon Alpha and a Foot-and-Mouth Disease Virus Subunit Vaccine. Vaccine 2003, 22, 268–279. [Google Scholar] [CrossRef]
- Cao, Y.; Lu, Z.; Li, Y.; Sun, P.; Li, D.; Li, P.; Bai, X.; Fu, Y.; Bao, H.; Zhou, C.; et al. Poly(I:C) Combined with Multi-Epitope Protein Vaccine Completely Protects against Virulent Foot-and-Mouth Disease Virus Challenge in Pigs. Antiviral Res. 2013, 97, 145–153. [Google Scholar] [CrossRef]
- Perez-Martin, E.; Weiss, M.; Diaz-San Segundo, F.; Pacheco, J.M.; Arzt, J.; Grubman, M.J.; de los Santos, T. Bovine Type III Interferon Significantly Delays and Reduces the Severity of Foot-and-Mouth Disease in Cattle. J. Virol. 2012, 86, 4477–4487. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-M.; Park, J.-H.; Lee, K.-N.; Kim, S.-K.; Ko, Y.-J.; Lee, H.-S.; Cho, I.-S. Enhanced Inhibition of Foot-and-Mouth Disease Virus by Combinations of Porcine Interferon-α and Antiviral Agents. Antiviral Res. 2012, 96, 213–220. [Google Scholar] [CrossRef]
- Saif, L.; Wang, Q.; Vlasova, A.; Jung, K.; Xiao, S. Coronaviruses. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar]
- Domingo, E.; Baranowski, E.; Ruiz-Jarabo, C.M.; Martín-Hernández, A.M.; Sáiz, J.C.; Escarmís, C. Quasispecies Structure and Persistence of RNA Viruses. Emerg. Infect. Dis. 1998, 4, 521–527. [Google Scholar] [CrossRef] [Green Version]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and Re-Emerging Coronaviruses in Pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Zhang, X.; Hasoksuz, M.; Nagesha, H.S.; Haynes, L.M.; Fang, Y.; Lu, S.; Saif, L.J. Two-Way Antigenic Cross-Reactivity between Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Group 1 Animal CoVs Is Mediated through an Antigenic Site in the N-Terminal Region of the SARS-CoV Nucleoprotein. J. Virol. 2007, 81, 13365–13377. [Google Scholar] [CrossRef] [Green Version]
- Saif, L.J.; Jung, K. Comparative Pathogenesis of Bovine and Porcine Respiratory Coronaviruses in the Animal Host Species and SARS-CoV-2 in Humans. J. Clin. Microbiol. 2020, 58, e01355-20. [Google Scholar] [CrossRef]
- Jung, K.; Renukaradhya, G.J.; Alekseev, K.P.; Fang, Y.; Tang, Y.; Saif, L.J. Porcine Reproductive and Respiratory Syndrome Virus Modifies Innate Immunity and Alters Disease Outcome in Pigs Subsequently Infected with Porcine Respiratory Coronavirus: Implications for Respiratory Viral Co-Infections. J. Gen. Virol. 2009, 90, 2713–2723. [Google Scholar] [CrossRef]
- Charley, B.; Riffault, S.; Van Reeth, K. Porcine Innate and Adaptative Immune Responses to Influenza and Coronavirus Infections. Ann. N. Y. Acad. Sci. 2006, 1081, 130–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Alekseev, K.; Jung, K.; Vlasova, A.; Hadya, N.; Saif, L.J. Cytokine Responses in Porcine Respiratory Coronavirus-Infected Pigs Treated with Corticosteroids as a Model for Severe Acute Respiratory Syndrome. J. Virol. 2008, 82, 4420–4428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Reeth, K.; Pensaert, M.B. Production of Interferon-Alpha, Tumor Necrosis Factor-Alpha and Interleukin-1 in the Lungs of Pigs Infected with the Porcine Respiratory Coronavirus. In Proceedings of the 3rd Congress of the European Society for Veterinary Virology, Interlaken, Switzerland, 4–7 September 1995; pp. 197–201. [Google Scholar]
- Baudoux, P.; Carrat, C.; Besnardeau, L.; Charley, B.; Laude, H. Coronavirus Pseudoparticles Formed with Recombinant M and E Proteins Induce Alpha Interferon Synthesis by Leukocytes. J. Virol. 1998, 72, 8636–8643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baudoux, P.; Besnardeau, L.; Carrat, C.; Rottier, P.; Charley, B.; Laude, H. Interferon Alpha Inducing Property of Coronavirus Particles and Pseudoparticles. Adv. Exp. Med. Biol. 1998, 440, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Van Reeth, K.; Nauwynck, H. Proinflammatory Cytokines and Viral Respiratory Disease in Pigs. Vet. Res. 2000, 31, 187–213. [Google Scholar] [CrossRef] [Green Version]
- Atanasova, K.; Gucht, S.V.; Barbé, F.; Lefebvre, D.J.; Chiers, K.; Reeth, K.V. Lung Cell Tropism and Inflammatory Cytokine-Profile of Porcine Respiratory Coronavirus Infection. Open Vet. Sci. J. 2008, 2, 117–126. [Google Scholar] [CrossRef]
- Van Reeth, K.; Van Gucht, S.; Pensaert, M. In Vivo Studies on Cytokine Involvement during Acute Viral Respiratory Disease of Swine: Troublesome but Rewarding. Vet. Immunol. Immunopathol. 2002, 87, 161–168. [Google Scholar] [CrossRef]
- Van Reeth, K.; Van Gucht, S.; Pensaert, M. Correlations between Lung Proinflammatory Cytokine Levels, Virus Replication, and Disease after Swine Influenza Virus Challenge of Vaccination-Immune Pigs. Viral Immunol. 2002, 15, 583–594. [Google Scholar] [CrossRef]
- Jung, K.; Alekseev, K.P.; Zhang, X.; Cheon, D.-S.; Vlasova, A.N.; Saif, L.J. Altered Pathogenesis of Porcine Respiratory Coronavirus in Pigs Due to Immunosuppressive Effects of Dexamethasone: Implications for Corticosteroid Use in Treatment of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2007, 81, 13681–13693. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.K.; Lam, C.W.K.; Wu, A.K.L.; Ip, W.K.; Lee, N.L.S.; Chan, I.H.S.; Lit, L.C.W.; Hui, D.S.C.; Chan, M.H.M.; Chung, S.S.C.; et al. Plasma Inflammatory Cytokines and Chemokines in Severe Acute Respiratory Syndrome. Clin. Exp. Immunol. 2004, 136, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Xu, J.; Zhou, C.; Wu, Z.; Zhong, S.; Liu, J.; Luo, W.; Chen, T.; Qin, Q.; Deng, P. Characterization of Cytokine/Chemokine Profiles of Severe Acute Respiratory Syndrome. Am. J. Respir. Crit. Care Med. 2005, 171, 850–857. [Google Scholar] [CrossRef]
- Nicholls, J.M.; Poon, L.L.M.; Lee, K.C.; Ng, W.F.; Lai, S.T.; Leung, C.Y.; Chu, C.M.; Hui, P.K.; Mak, K.L.; Lim, W.; et al. Lung Pathology of Fatal Severe Acute Respiratory Syndrome. Lancet 2003, 361, 1773–1778. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.S.; Lin, C.F.; Fang, Y.T.; Kuo, Y.M.; Liao, P.C.; Yeh, T.M.; Hwa, K.Y.; Shieh, C.C.K.; Yen, J.H.; Wang, H.J.; et al. Antibody to Severe Acute Respiratory Syndrome (SARS)-Associated Coronavirus Spike Protein Domain 2 Cross-Reacts with Lung Epithelial Cells and Causes Cytotoxicity. Clin. Exp. Immunol. 2005, 141, 500–508. [Google Scholar] [CrossRef] [Green Version]
- Cameron, M.J.; Ran, L.; Xu, L.; Danesh, A.; Bermejo-Martin, J.F.; Cameron, C.M.; Muller, M.P.; Gold, W.L.; Richardson, S.E.; Poutanen, S.M.; et al. Interferon-Mediated Immunopathological Events Are Associated with Atypical Innate and Adaptive Immune Responses in Patients with Severe Acute Respiratory Syndrome. J. Virol. 2007, 81, 8692–8706. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, J.; Zhan, Y.; Wu, L.; Yu, X.; Zhang, W.; Ye, L.; Xu, S.; Sun, R.; Wang, Y.; et al. Analysis of Serum Cytokines in Patients with Severe Acute Respiratory Syndrome. Infect. Immun. 2004, 72, 4410–4415. [Google Scholar] [CrossRef] [Green Version]
- Cameron, M.J.; Bermejo-Martin, J.F.; Danesh, A.; Muller, M.P.; Kelvin, D.J. Human Immunopathogenesis of Severe Acute Respiratory Syndrome (SARS). Virus Res. 2008, 133, 13–19. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Heegaard, P.M.H.; Sturek, M.; Alloosh, M.; Belsham, G.J. Animal Models for COVID-19: More to the Picture Than ACE2, Rodents, Ferrets, and Non-Human Primates. A Case for Porcine Respiratory Coronavirus and the Obese Ossabaw Pig. Front. Microbiol. 2020, 11, 573756. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting Potential Drivers of COVID-19: Neutrophil Extracellular Traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef]
- Runft, S.; Färber, I.; Krüger, J.; Armando, F.; Rocha, C.; Pöhlmann, S.; Burigk, L.; Leitzen, E.; Ciurkiewicz, M.; Braun, A.; et al. Alternatives to Animal Models and Their Application in the Discovery of Species Susceptibility to SARS-CoV-2 and Other Respiratory Infectious Pathogens—A Review. Vet. Pathol. 2022; online ahead of print. [Google Scholar]
- Meekins, D.A.; Morozov, I.; Trujillo, J.D.; Gaudreault, N.N.; Bold, D.; Carossino, M.; Artiaga, B.L.; Indran, S.V.; Kwon, T.; Balaraman, V.; et al. Susceptibility of Swine Cells and Domestic Pigs to SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 2278–2288. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in Fruit Bats, Ferrets, Pigs, and Chickens: An Experimental Transmission Study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of Ferrets, Cats, Dogs, and Other Domesticated Animals to SARS-Coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Pandey, K.; Acharya, A.; Mohan, M.; Ng, C.L.; Reid, S.P.; Byrareddy, S.N. Animal Models for SARS-CoV-2 Research: A Comprehensive Literature Review. Transbound. Emerg. Dis. 2021, 68, 1868–1885. [Google Scholar] [CrossRef]
- Ciurkiewicz, M.; Armando, F.; Schreiner, T.; de Burh, N.; Pilchova, V.; Krupp-Buzimikic, V.; Gabriel, G.; Von Köckritz-Blickwede, M.; Baumgärtner, W.; Schulz, C.; et al. Ferrets Are Valuable Models for SARS-CoV-2 Research. Vet. Pathol. 2022, online ahead of print. [CrossRef]
- Krüger, N.; Rocha, C.; Runft, S.; Krüger, J.; Färber, I.; Armando, F.; Leitzen, E.; Brogden, G.; Gerold, G.; Pöhlmann, S.; et al. The Upper Respiratory Tract of Felids Is Highly Susceptible to SARS-CoV-2 Infection. Int. J. Mol. Sci. 2021, 22, 10636. [Google Scholar] [CrossRef]
- Vergara-Alert, J.; Rodon, J.; Carrillo, J.; Te, N.; Izquierdo-Useros, N.; Rodríguez de la Concepción, M.L.; Ávila-Nieto, C.; Guallar, V.; Valencia, A.; Cantero, G.; et al. Pigs Are Not Susceptible to SARS-CoV-2 Infection but Are a Model for Viral Immunogenicity Studies. Transbound. Emerg. Dis. 2021, 68, 1721–1725. [Google Scholar] [CrossRef]
- Graham, S.P.; McLean, R.K.; Spencer, A.J.; Belij-Rammerstorfer, S.; Wright, D.; Ulaszewska, M.; Edwards, J.C.; Hayes, J.W.P.; Martini, V.; Thakur, N.; et al. Evaluation of the Immunogenicity of Prime-Boost Vaccination with the Replication-Deficient Viral Vectored COVID-19 Vaccine Candidate ChAdOx1 NCoV-19. NPJ Vaccines 2020, 5, 69. [Google Scholar] [CrossRef]
- Segalés, J. Porcine Circovirus Type 2 (PCV2) Infections: Clinical Signs, Pathology and Laboratory Diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef]
- Segalés, J.; Olvera, A.; Grau-Roma, L.; Charreyre, C.; Nauwynck, H.; Larsen, L.; Dupont, K.; McCullough, K.; Ellis, J.; Krakowka, S.; et al. PCV-2 Genotype Definition and Nomenclature. Vet. Rec. 2008, 162, 867–868. [Google Scholar] [CrossRef]
- Segalés, J.; Kekarainen, T.; Cortey, M. The Natural History of Porcine Circovirus Type 2: From an Inoffensive Virus to a Devastating Swine Disease? Vet. Microbiol. 2013, 165, 13–20. [Google Scholar] [CrossRef]
- Larochelle, R.; Magar, R.; D’Allaire, S. Comparative Serologic and Virologic Study of Commercial Swine Herds with and without Postweaning Multisystemic Wasting Syndrome. Can. J. Vet. Res. 2003, 67, 114–120. [Google Scholar]
- Vincent, I.E.; Carrasco, C.P.; Guzylack-Piriou, L.; Herrmann, B.; McNeilly, F.; Allan, G.M.; Summerfield, A.; McCullough, K.C. Subset-Dependent Modulation of Dendritic Cell Activity by Circovirus Type 2. Immunology 2005, 115, 388–398. [Google Scholar] [CrossRef]
- Darwich, L.; Segalés, J.; Mateu, E. Pathogenesis of Postweaning Multisystemic Wasting Syndrome Caused by Porcine Circovirus 2: An Immune Riddle. Arch. Virol. 2004, 149, 857–874. [Google Scholar] [CrossRef]
- Alarcon, P.; Velasova, M.; Mastin, A.; Nevel, A.; Stärk, K.D.C.; Wieland, B. Farm Level Risk Factors Associated with Severity of Post-Weaning Multi-Systemic Wasting Syndrome. Prev. Vet. Med. 2011, 101, 182–191. [Google Scholar] [CrossRef]
- Segalés, J.; Calsamiglia, M.; Olvera, A.; Sibila, M.; Badiella, L.; Domingo, M. Quantification of Porcine Circovirus Type 2 (PCV2) DNA in Serum and Tonsillar, Nasal, Tracheo-Bronchial, Urinary and Faecal Swabs of Pigs with and without Postweaning Multisystemic Wasting Syndrome (PMWS). Vet. Microbiol. 2005, 111, 223–229. [Google Scholar] [CrossRef]
- Segalés, J. Best Practice and Future Challenges for Vaccination against Porcine Circovirus Type 2. Expert Rev. Vaccines 2015, 14, 473–487. [Google Scholar] [CrossRef]
- Harmon, K.M.; Gauger, P.C.; Zhang, J.; Piñeyro, P.E.; Dunn, D.D.; Chriswell, A.J. Whole-Genome Sequences of Novel Porcine Circovirus Type 2 Viruses Detected in Swine from Mexico and the United States. Genome Announc. 2015, 3, e01315-15. [Google Scholar] [CrossRef] [Green Version]
- Park, K.H.; Oh, T.; Yang, S.; Cho, H.; Kang, I.; Chae, C. Evaluation of a Porcine Circovirus Type 2a (PCV2a) Vaccine Efficacy against Experimental PCV2a, PCV2b, and PCV2d Challenge. Vet. Microbiol. 2019, 231, 87–92. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine Circovirus 2 Genotypes, Immunity and Vaccines: Multiple Genotypes but One Single Serotype. Pathogens 2020, 9, 1049. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, C.; Martinelli, N.; Lelli, D.; Amadori, M. Correlates of Protection Following Vaccination with Inactivated Porcine Circovirus 2 Vaccines. Viral Immunol. 2015, 28, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Kekarainen, T.; Segalés, J. Porcine Circovirus 2 Immunology and Viral Evolution. Porcine Health Manag. 2015, 1, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghetti, P.; Morganti, M.; Saleri, R.; Ferrari, L.; De Angelis, E.; Cavalli, V.; Cacchioli, A.; Corradi, A.; Martelli, P. Innate Pro-Inflammatory and Adaptive Immune Cytokines in PBMC of Vaccinated and Unvaccinated Pigs Naturally Exposed to Porcine Circovirus Type 2 (PCV2) Infection Vary with the Occurrence of the Disease and the Viral Burden. Vet. Microbiol. 2013, 163, 42–53. [Google Scholar] [CrossRef]
- Baumann, A.; McCullough, K.C.; Summerfield, A. Porcine Circovirus Type 2 Stimulates Plasmacytoid Dendritic Cells in the Presence of IFN-Gamma. Vet. Immunol. Immunopathol. 2013, 156, 223–228. [Google Scholar] [CrossRef]
- Kekarainen, T.; Montoya, M.; Dominguez, J.; Mateu, E.; Segalés, J. Porcine Circovirus Type 2 (PCV2) Viral Components Immunomodulate Recall Antigen Responses. Vet. Immunol. Immunopathol. 2008, 124, 41–49. [Google Scholar] [CrossRef]
- Kixmöller, M.; Ritzmann, M.; Eddicks, M.; Saalmüller, A.; Elbers, K.; Fachinger, V. Reduction of PMWS-Associated Clinical Signs and Co-Infections by Vaccination against PCV2. Vaccine 2008, 26, 3443–3451. [Google Scholar] [CrossRef]
- Bordon, Y. Macrophages: Innate Memory Training. Nat. Rev. Immunol. 2014, 14, 713. [Google Scholar] [CrossRef]
- Crisci, E.; Fraile, L.; Moreno, N.; Blanco, E.; Cabezón, R.; Costa, C.; Mussá, T.; Baratelli, M.; Martinez-Orellana, P.; Ganges, L.; et al. Chimeric Calicivirus-like Particles Elicit Specific Immune Responses in Pigs. Vaccine 2012, 30, 2427–2439. [Google Scholar] [CrossRef]
- Madec, F.; Rose, N.; Grasland, B.; Cariolet, R.; Jestin, A. Post-Weaning Multisystemic Wasting Syndrome and Other PCV2-Related Problems in Pigs: A 12-Year Experience. Transbound. Emerg. Dis. 2008, 55, 273–283. [Google Scholar] [CrossRef]
- Meerts, P.; Misinzo, G.; Nauwynck, H.J. Enhancement of Porcine Circovirus 2 Replication in Porcine Cell Lines by IFN-Gamma before and after Treatment and by IFN-Alpha after Treatment. J. Interferon Cytokine Res. 2005, 25, 684–693. [Google Scholar] [CrossRef]
- Dvorak, C.M.T.; Puvanendiran, S.; Murtaugh, M.P. Porcine Circovirus 2 Infection Induces IFNβ Expression through Increased Expression of Genes Involved in RIG-I and IRF7 Signaling Pathways. Virus Res. 2018, 253, 38–47. [Google Scholar] [CrossRef]
- Ramamoorthy, S.; Huang, F.F.; Huang, Y.W.; Meng, X.J. Interferon-Mediated Enhancement of in vitro Replication of Porcine Circovirus Type 2 Is Influenced by an Interferon-Stimulated Response Element in the PCV2 Genome. Virus Res. 2009, 145, 236–243. [Google Scholar] [CrossRef]
- Wikström, F.H.; Meehan, B.M.; Berg, M.; Timmusk, S.; Elving, J.; Fuxler, L.; Magnusson, M.; Allan, G.M.; McNeilly, F.; Fossum, C. Structure-Dependent Modulation of Alpha Interferon Production by Porcine Circovirus 2 Oligodeoxyribonucleotide and CpG DNAs in Porcine Peripheral Blood Mononuclear Cells. J. Virol. 2007, 81, 4919–4927. [Google Scholar] [CrossRef] [Green Version]
- Franzoni, G.; Graham, S.P.; Dei Giudici, S.; Oggiano, A. Porcine Dendritic Cells and Viruses: An Update. Viruses 2019, 11, 445. [Google Scholar] [CrossRef] [Green Version]
- Vincent, I.E.; Balmelli, C.; Meehan, B.; Allan, G.; Summerfield, A.; McCullough, K.C. Silencing of Natural Interferon Producing Cell Activation by Porcine Circovirus Type 2 DNA. Immunology 2007, 120, 47–56. [Google Scholar] [CrossRef]
- Stevenson, L.S.; McCullough, K.; Vincent, I.; Gilpin, D.F.; Summerfield, A.; Nielsen, J.; McNeilly, F.; Adair, B.M.; Allan, G.M. Cytokine and C-Reactive Protein Profiles Induced by Porcine Circovirus Type 2 Experimental Infection in 3-Week-Old Piglets. Viral Immunol. 2006, 19, 189–195. [Google Scholar] [CrossRef]
- Darwich, L.; Segalés, J.; Resendes, A.; Balasch, M.; Plana-Durán, J.; Mateu, E. Transient Correlation between Viremia Levels and IL-10 Expression in Pigs Subclinically Infected with Porcine Circovirus Type 2 (PCV2). Res. Vet. Sci. 2008, 84, 194–198. [Google Scholar] [CrossRef]
- Hasslung, F.; Wallgren, P.; Ladekjaer-Hansen, A.-S.; Bøtner, A.; Nielsen, J.; Wattrang, E.; Allan, G.M.; McNeilly, F.; Ellis, J.; Timmusk, S.; et al. Experimental Reproduction of Postweaning Multisystemic Wasting Syndrome (PMWS) in Pigs in Sweden and Denmark with a Swedish Isolate of Porcine Circovirus Type 2. Vet. Microbiol. 2005, 106, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-W.; Jeng, C.-R.; Liu, J.J.; Lin, T.-L.; Chang, C.-C.; Chia, M.-Y.; Tsai, Y.-C.; Pang, V.F. Reduction of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Infection in Swine Alveolar Macrophages by Porcine Circovirus 2 (PCV2)-Induced Interferon-Alpha. Vet. Microbiol. 2005, 108, 167–177. [Google Scholar] [CrossRef]
- Huang, B.; Li, J.; Zhang, X.; Zhao, Q.; Lu, M.; Lv, Y. RIG-1 and MDA-5 Signaling Pathways Contribute to IFN-β Production and Viral Replication in Porcine Circovirus Virus Type 2-Infected PK-15 Cells in vitro. Vet. Microbiol. 2017, 211, 36–42. [Google Scholar] [CrossRef]
- Seo, H.W.; Oh, Y.; Han, K.; Park, C.; Chae, C. Reduction of Porcine Circovirus Type 2 (PCV2) Viremia by a Reformulated Inactivated Chimeric PCV1-2 Vaccine-Induced Humoral and Cellular Immunity after Experimental PCV2 Challenge. BMC Vet. Res. 2012, 8, 194. [Google Scholar] [CrossRef] [Green Version]
- Koinig, H.C.; Talker, S.C.; Stadler, M.; Ladinig, A.; Graage, R.; Ritzmann, M.; Hennig-Pauka, I.; Gerner, W.; Saalmüller, A. PCV2 Vaccination Induces IFN-γ/TNF-α Co-Producing T Cells with a Potential Role in Protection. Vet. Res. 2015, 46, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guarneri, F.; Tonni, M.; Sarli, G.; Boniotti, M.B.; Lelli, D.; Barbieri, I.; D’Annunzio, G.; Alborali, G.L.; Bacci, B.; Amadori, M. Non-Assembled ORF2 Capsid Protein of Porcine Circovirus 2b Does Not Confer Protective Immunity. Pathogens 2021, 10, 1161. [Google Scholar] [CrossRef] [PubMed]
- Hayday, A.C. Gammadelta T Cells and the Lymphoid Stress-Surveillance Response. Immunity 2009, 31, 184–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, P.D.; Christiansen, D.; Winterhalter, A.; Brooks, A.; Gorrell, M.; Lilienfeld, B.G.; Seebach, J.D.; Sandrin, M.; Sharland, A. Porcine Cells Express More than One Functional Ligand for the Human Lymphocyte Activating Receptor NKG2D. Xenotransplantation 2008, 15, 321–332. [Google Scholar] [CrossRef]
- Masopust, D.; Picker, L.J. Hidden Memories: Frontline Memory T Cells and Early Pathogen Interception. J. Immunol. 2012, 188, 5811–5817. [Google Scholar] [CrossRef]
- Sang, Y.; Rowland, R.R.R.; Blecha, F. Molecular Characterization and Antiviral Analyses of Porcine Type III Interferons. J. Interferon Cytokine Res. 2010, 30, 801–807. [Google Scholar] [CrossRef] [Green Version]
- Amadori, M. Issues and Possible Intervention Strategies Relating to Early Weaning of Piglets. Anim. Sci. Rev. 2012, 125–137. [Google Scholar] [CrossRef]
- Fan, H.; Xiao, S.; Tong, T.; Wang, S.; Xie, L.; Jiang, Y.; Chen, H.; Fang, L. Immunogenicity of Porcine Circovirus Type 2 Capsid Protein Targeting to Different Subcellular Compartments. Mol. Immunol. 2008, 45, 653–660. [Google Scholar] [CrossRef]
- Lauw, F.N.; Pajkrt, D.; Hack, C.E.; Kurimoto, M.; van Deventer, S.J.; van der Poll, T. Proinflammatory Effects of IL-10 during Human Endotoxemia. J. Immunol. 2000, 165, 2783–2789. [Google Scholar] [CrossRef]
- Tilg, H.; van Montfrans, C.; van den Ende, A.; Kaser, A.; van Deventer, S.J.H.; Schreiber, S.; Gregor, M.; Ludwiczek, O.; Rutgeerts, P.; Gasche, C.; et al. Treatment of Crohn’s Disease with Recombinant Human Interleukin 10 Induces the Proinflammatory Cytokine Interferon Gamma. Gut 2002, 50, 191–195. [Google Scholar] [CrossRef] [Green Version]
- Mettenleiter, T.C. Aujeszky’s Disease and the Development of the Marker/DIVA Vaccination Concept. Pathogens 2020, 9, 563. [Google Scholar] [CrossRef]
- Wei, J.; Ma, Y.; Wang, L.; Chi, X.; Yan, R.; Wang, S.; Li, X.; Chen, X.; Shao, W.; Chen, J.-L. Alpha/Beta Interferon Receptor Deficiency in Mice Significantly Enhances Susceptibility of the Animals to Pseudorabies Virus Infection. Vet. Microbiol. 2017, 203, 234–244. [Google Scholar] [CrossRef]
- Grob, P.; Schijns, V.E.; van den Broek, M.F.; Cox, S.P.; Ackermann, M.; Suter, M. Role of the Individual Interferon Systems and Specific Immunity in Mice in Controlling Systemic Dissemination of Attenuated Pseudorabies Virus Infection. J. Virol. 1999, 73, 4748–4754. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.-K.; Ming, S.-L.; Zeng, L.; Chang, W.-R.; Pan, J.-J.; Zhang, C.; Wan, B.; Wang, J.; Su, Y.; Yang, G.-Y.; et al. Inhibition of Histone Deacetylase 1 Suppresses Pseudorabies Virus Infection through CGAS-STING Antiviral Innate Immunity. Mol. Immunol. 2021, 136, 55–64. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.-L.; Ming, S.-L.; Wang, C.-F.; Shi, L.-J.; Su, B.-Q.; Wu, H.-T.; Zeng, L.; Han, Y.-Q.; Liu, Z.-H.; et al. BRD4 Inhibition Exerts Anti-Viral Activity through DNA Damage-Dependent Innate Immune Responses. PLoS Pathog. 2020, 16, e1008429. [Google Scholar] [CrossRef] [Green Version]
- Brukman, A.; Enquist, L.W. Suppression of the Interferon-Mediated Innate Immune Response by Pseudorabies Virus. J. Virol. 2006, 80, 6345–6356. [Google Scholar] [CrossRef] [Green Version]
- Bo, Z.; Miao, Y.; Xi, R.; Zhong, Q.; Bao, C.; Chen, H.; Sun, L.; Qian, Y.; Jung, Y.-S.; Dai, J. PRV UL13 Inhibits CGAS–STING-Mediated IFN-β Production by Phosphorylating IRF3. Vet. Res. 2020, 51, 118. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, M.; Ye, C.; Ruan, K.; Xu, A.; Gao, F.; Tong, G.; Zheng, H. Inhibition of the DNA-Sensing Pathway by Pseudorabies Virus UL24 Protein via Degradation of Interferon Regulatory Factor 7. Vet. Microbiol. 2021, 255, 109023. [Google Scholar] [CrossRef]
- Lv, L.; Cao, M.; Bai, J.; Jin, L.; Wang, X.; Gao, Y.; Liu, X.; Jiang, P. PRV-Encoded UL13 Protein Kinase Acts as an Antagonist of Innate Immunity by Targeting IRF3-Signaling Pathways. Vet. Microbiol. 2020, 250, 108860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R.; et al. Pseudorabies Virus DUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Response. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Chen, S.; Zhang, Y.; Wang, M.; Qin, C.; Yu, C.; Zhang, Y.; Li, Y.; Chen, L.; Zhang, X.; et al. Pseudorabies Virus DNA Polymerase Processivity Factor UL42 Inhibits Type I IFN Response by Preventing ISGF3-ISRE Interaction. J. Immunol. 2021, 207, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Lomniczi, B. Biological Properties of Aujeszky’s Disease (Pseudorabies) Virus Strains with Special Regard to Interferon Production and Interferon Sensitivity. Archiv für die Gesamte Virusforschung 1974, 44, 205–214. [Google Scholar] [CrossRef]
- Pol, J.M.; Broekhuysen-Davies, J.M.; Wagenaar, F.; La Bonnardière, C. The Influence of Porcine Recombinant Interferon-Alpha 1 on Pseudorabies Virus Infection of Porcine Nasal Mucosa in vitro. J. Gen. Virol. 1991, 72 Pt 4, 933–938. [Google Scholar] [CrossRef]
- Wang, P.; Xia, L.; Liang, X.; Han, F.; Ren, H.; Zhang, Y.; Wei, Z. Expression of Porcine Interferon-α and Its Bioactivity Analysis in vitro and in Vivo. Bioprocess Biosyst. Eng. 2021, 44, 473–482. [Google Scholar] [CrossRef]
- Liu, H.; Li, S.; Yang, X.; Wang, X.; Li, Y.; Wang, C.; Chen, L.; Chang, H. Porcine ISG15 Modulates the Antiviral Response during Pseudorabies Virus Replication. Gene 2018, 679, 212–218. [Google Scholar] [CrossRef]
- Chen, X.; Sun, D.; Dong, S.; Zhai, H.; Kong, N.; Zheng, H.; Tong, W.; Li, G.; Shan, T.; Tong, G. Host Interferon-Stimulated Gene 20 Inhibits Pseudorabies Virus Proliferation. Virol. Sin. 2021, 36, 1027–1035. [Google Scholar] [CrossRef]
- Yao, Q.; Qian, P.; Cao, Y.; He, Y.; Si, Y.; Xu, Z.; Chen, H. Synergistic Inhibition of Pseudorabies Virus Replication by Porcine Alpha/Beta Interferon and Gamma Interferon in vitro. Eur. Cytokine Netw. 2007, 18, 71–77. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, J.; Feng, H.; Wei, Q.; Zhao, S.; Yang, S.; Ma, H.; Liu, D.; et al. Antiviral Activity of Porcine Interferon Delta 8 against Pesudorabies Virus in vitro. Int. J. Biol. Macromol. 2021, 177, 10–18. [Google Scholar] [CrossRef]
- De Regge, N.; Van Opdenbosch, N.; Nauwynck, H.J.; Efstathiou, S.; Favoreel, H.W. Interferon Alpha Induces Establishment of Alphaherpesvirus Latency in Sensory Neurons In vitro. PLoS ONE 2010, 5, e13076. [Google Scholar] [CrossRef]
- Calzada-Nova, G.; Schnitzlein, W.; Husmann, R.; Zuckermann, F.A. Characterization of the Cytokine and Maturation Responses of Pure Populations of Porcine Plasmacytoid Dendritic Cells to Porcine Viruses and Toll-like Receptor Agonists. Vet. Immunol. Immunopathol. 2010, 135, 20–33. [Google Scholar] [CrossRef]
- Lamote, J.A.S.; Kestens, M.; Van Waesberghe, C.; Delva, J.; De Pelsmaeker, S.; Devriendt, B.; Favoreel, H.W. The Pseudorabies Virus Glycoprotein GE/GI Complex Suppresses Type I Interferon Production by Plasmacytoid Dendritic Cells. J. Virol. 2017, 91, e02276-16. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Qiu, S.; Zhang, L.; Sun, Y.; Bao, E.; Lv, Y. Pseudorabies Virus Glycoprotein GE Suppresses Interferon-β Production via CREB-Binding Protein Degradation. Virus Res. 2021, 291, 198220. [Google Scholar] [CrossRef]
- Artursson, K.; Lindersson, M.; Varela, N.; Scheynius, A.; Alm, G.V. Interferon-Alpha Production and Tissue Localization of Interferon-Alpha/Beta Producing Cells after Intradermal Administration of Aujeszky’s Disease Virus-Infected Cells in Pigs. Scand. J. Immunol. 1995, 41, 121–129. [Google Scholar] [CrossRef]
- Wattrang, E.; Edfors-Lilja, I.; Andersson, L.; Fossum, C. Genetic and Stress-Mediated Influence on Aujeszky’s Disease Virus Induced Interferon-Alpha Production in Porcine Leukocytes. Acta Vet. Hung. 1994, 42, 331–336. [Google Scholar]
- Hoegen, B.; Saalmüller, A.; Röttgen, M.; Rziha, H.-J.; Geldermann, H.; Reiner, G.; Pfaff, E.; Büttner, M. Interferon-Gamma Response of PBMC Indicates Productive Pseudorabies Virus (PRV) Infection in Swine. Vet. Immunol. Immunopathol. 2004, 102, 389–397. [Google Scholar] [CrossRef]
- Dotti, S.; Guadagnini, G.; Salvini, F.; Razzuoli, E.; Ferrari, M.; Alborali, G.L.; Amadori, M. Time-Course of Antibody and Cell-Mediated Immune Responses to Porcine Reproductive and Respiratory Syndrome Virus under Field Conditions. Res. Vet. Sci. 2013, 94, 510–517. [Google Scholar] [CrossRef]
- Schijns, V.; van Giersbergen, P.; Schellekens, H.; Horzinek, M.C. Recombinant Interferon-Gamma Applied to the Brain Ventricular System Protects Rats against Pseudorabies. J. Neuroimmunol. 1990, 28, 1–7. [Google Scholar] [CrossRef]
- Vandenbroeck, K.; Nauwynck, H.; Vanderpooten, A.; Van Reeth, K.; Goddeeris, B.; Billiau, A. Recombinant Porcine IFN-Gamma Potentiates the Secondary IgG and IgA Responses to an Inactivated Suid Herpesvirus-1 Vaccine and Reduces Postchallenge Weight Loss and Fever in Pigs. J. Interferon Cytokine Res. 1998, 18, 739–744. [Google Scholar] [CrossRef]
- Njau, E.P.; Domelevo Entfellner, J.-B.; Machuka, E.M.; Bochere, E.N.; Cleaveland, S.; Shirima, G.M.; Kusiluka, L.J.; Upton, C.; Bishop, R.P.; Pelle, R.; et al. The First Genotype II African Swine Fever Virus Isolated in Africa Provides Insight into the Current Eurasian Pandemic. Sci. Rep. 2021, 11, 13081. [Google Scholar] [CrossRef] [PubMed]
- Blome, S.; Franzke, K.; Beer, M. African Swine Fever—A Review of Current Knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef] [PubMed]
- OIE WAHIS Interface. Available online: https://www.oie.int/App/Uploads/2021/12/Report-65-Current-Situation-of-Asf.Pdf (accessed on 13 December 2021).
- Martins, C.; Boinas, F.S.; Iacolina, L.; Ruiz-Fons, F.; Gavier-Widén, D. 1. African Swine Fever (ASF), the Pig Health Challenge of the Century. In Understanding and Combatting African Swine Fever; Wageningen Academic Publishers: Wageningen, The Netherlannds, 2021; pp. 11–24. ISBN 9789086863570. [Google Scholar]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M. ICTV Virus Taxonomy Profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Chapman, D.A.G.; Netherton, C.L.; Upton, C. African Swine Fever Virus Replication and Genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Montoya, M.; Franzoni, G.; Pérez-Nuñez, D.; Revilla, Y.; Galindo, I.; Alonso, C.; Netherton, C.L.; Blohm, U. 3. Immune Responses against African Swine Fever Virus Infection. In Understanding and Combatting African Swine Fever; Wageningen Academic Publishers: Wageningen, The Netherlannds, 2021; pp. 63–85. ISBN 9789086863570. [Google Scholar]
- Bisimwa, P.N.; Ongus, J.R.; Steinaa, L.; Bisimwa, E.B.; Bochere, E.; Machuka, E.M.; Entfellner, J.-B.D.; Okoth, E.; Pelle, R. The First Complete Genome Sequence of the African Swine Fever Virus Genotype X and Serogroup 7 Isolated in Domestic Pigs from the Democratic Republic of Congo. Virol. J. 2021, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African Swine Fever Viruses Emerged in Domestic Pigs in China and Caused Chronic Infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Guinat, C.; Gogin, A.; Blome, S.; Keil, G.; Pollin, R.; Pfeiffer, D.U.; Dixon, L. Transmission Routes of African Swine Fever Virus to Domestic Pigs: Current Knowledge and Future Research Directions. Vet. Rec. 2016, 178, 262–267. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Gomez-Villamandos, J.C.; Carrasco, L. An Update on the Epidemiology and Pathology of African Swine Fever. J. Comp. Pathol. 2015, 152, 9–21. [Google Scholar] [CrossRef]
- Dixon, L.K.; Abrams, C.C.; Bowick, G.; Goatley, L.C.; Kay-Jackson, P.C.; Chapman, D.; Liverani, E.; Nix, R.; Silk, R.; Zhang, F. African Swine Fever Virus Proteins Involved in Evading Host Defence Systems. Vet. Immunol. Immunopathol. 2004, 100, 117–134. [Google Scholar] [CrossRef]
- Franzoni, G.; Graham, S.P.; Sanna, G.; Angioi, P.; Fiori, M.S.; Anfossi, A.; Amadori, M.; Dei Giudici, S.; Oggiano, A. Interaction of Porcine Monocyte-Derived Dendritic Cells with African Swine Fever Viruses of Diverse Virulence. Vet. Microbiol. 2018, 216, 190–197. [Google Scholar] [CrossRef]
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African Swine Fever Virus Evasion of Host Defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef]
- Franzoni, G.; Graham, S.P.; Giudici, S.D.; Bonelli, P.; Pilo, G.; Anfossi, A.G.; Pittau, M.; Nicolussi, P.S.; Laddomada, A.; Oggiano, A. Characterization of the Interaction of African Swine Fever Virus with Monocytes and Derived Macrophage Subsets. Vet. Microbiol. 2017, 198, 88–98. [Google Scholar] [CrossRef] [Green Version]
- García-Belmonte, R.; Pérez-Núñez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the CGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef] [Green Version]
- Gil, S.; Sepúlveda, N.; Albina, E.; Leitão, A.; Martins, C. The Low-Virulent African Swine Fever Virus (ASFV/NH/P68) Induces Enhanced Expression and Production of Relevant Regulatory Cytokines (IFNalpha, TNFalpha and IL12p40) on Porcine Macrophages in Comparison to the Highly Virulent ASFV/L60. Arch. Virol. 2008, 153, 1845–1854. [Google Scholar] [CrossRef] [Green Version]
- Reis, A.L.; Abrams, C.C.; Goatley, L.C.; Netherton, C.; Chapman, D.G.; Sanchez-Cordon, P.; Dixon, L.K. Deletion of African Swine Fever Virus Interferon Inhibitors from the Genome of a Virulent Isolate Reduces Virulence in Domestic Pigs and Induces a Protective Response. Vaccine 2016, 34, 4698–4705. [Google Scholar] [CrossRef] [Green Version]
- Mccullough, K.C.; Basta, S.; Knötig, S.; Gerber, H.; Schaffner, R.; Kim, Y.B.; Saalmüller, A.; Summerfield, A. Intermediate Stages in Monocyte–Macrophage Differentiation Modulate Phenotype and Susceptibility to Virus Infection. Immunology 1999, 98, 203–212. [Google Scholar] [CrossRef]
- Franzoni, G.; Razzuoli, E.; Dei Giudici, S.; Carta, T.; Galleri, G.; Zinellu, S.; Ledda, M.; Angioi, P.; Modesto, P.; Graham, S.P.; et al. Comparison of Macrophage Responses to African Swine Fever Viruses Reveals That the NH/P68 Strain Is Associated with Enhanced Sensitivity to Type I IFN and Cytokine Responses from Classically Activated Macrophages. Pathogens 2020, 9, 209. [Google Scholar] [CrossRef] [Green Version]
- Lanier, L.L. Nk Cell Recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef]
- Leitão, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigário, J.D.; Martins, C.L.V.Y. The Non-Haemadsorbing African Swine Fever Virus Isolate ASFV/NH/P68 Provides a Model for Defining the Protective Anti-Virus Immune Response. J. Gen. Virol. 2001, 82, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Lithgow, P.; Takamatsu, H.; Werling, D.; Dixon, L.; Chapman, D. Correlation of Cell Surface Marker Expression with African Swine Fever Virus Infection. Vet. Microbiol. 2014, 168, 413–419. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.J.; Ramanathan, P.; Bishop, E.A.; O’Donnell, V.; Gladue, D.P.; Borca, M.V. Mechanisms of African Swine Fever Virus Pathogenesis and Immune Evasion Inferred from Gene Expression Changes in Infected Swine Macrophages. PLoS ONE 2019, 14, e0223955. [Google Scholar] [CrossRef] [PubMed]
- Gregg, D.A.; Mebus, C.A.; Schlafer, D.H. African Swine Fever Interference with Foot-and-Mouth Disease Infection and Seroconversion in Pigs. J. Vet. Diagn. Investig. 1995, 7, 31–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregg, D.A.; Schlafer, D.H.; Mebus, C.A. African Swine Fever Virus Infection of Skin-Derived Dendritic Cells in vitro Causes Interference with Subsequent Foot-and-Mouth Disease Virus Infection. J. Vet. Diagn. Investig. 1995, 7, 44–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golding, J.P.; Goatley, L.; Goodbourn, S.; Dixon, L.K.; Taylor, G.; Netherton, C.L. Sensitivity of African Swine Fever Virus to Type I Interferon Is Linked to Genes within Multigene Families 360 and 505. Virology 2016, 493, 154–161. [Google Scholar] [CrossRef] [Green Version]
- Esparza, I.; González, J.C.; Viñuela, E. Effect of Interferon-Alpha, Interferon-Gamma and Tumour Necrosis Factor on African Swine Fever Virus Replication in Porcine Monocytes and Macrophages. J. Gen. Virol. 1988, 69 Pt 12, 2973–2980. [Google Scholar] [CrossRef]
- Paez, E.; Garcia, F.; Gil Fernandez, C. Interferon Cures Cells Lytically and Persistently Infected with African Swine Fever Virus in vitro. Arch. Virol. 1990, 112, 115–127. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; et al. African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J. Virol. 2015, 89, 6048–6056. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Moreno, R.; Cuesta-Geijo, M.Á.; Martínez-Romero, C.; Barrado-Gil, L.; Galindo, I.; García-Sastre, A.; Alonso, C. Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection. PLoS ONE 2016, 11, e0154366. [Google Scholar] [CrossRef]
- Netherton, C.L.; Simpson, J.; Haller, O.; Wileman, T.E.; Takamatsu, H.-H.; Monaghan, P.; Taylor, G. Inhibition of a Large Double-Stranded DNA Virus by MxA Protein. J. Virol. 2009, 83, 2310–2320. [Google Scholar] [CrossRef] [Green Version]
- Razzuoli, E.; Franzoni, G.; Carta, T.; Zinellu, S.; Amadori, M.; Modesto, P.; Oggiano, A. Modulation of Type I Interferon System by African Swine Fever Virus. Pathogens 2020, 9, 361. [Google Scholar] [CrossRef]
- Wang, X.; Wu, J.; Wu, Y.; Chen, H.; Zhang, S.; Li, J.; Xin, T.; Jia, H.; Hou, S.; Jiang, Y.; et al. Inhibition of CGAS-STING-TBK1 Signaling Pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 2018, 506, 437–443. [Google Scholar] [CrossRef]
- Huang, L.; Xu, W.; Liu, H.; Xue, M.; Liu, X.; Zhang, K.; Hu, L.; Li, J.; Liu, X.; Xiang, Z.; et al. African Swine Fever Virus PI215L Negatively Regulates CGAS-STING Signaling Pathway through Recruiting RNF138 to Inhibit K63-Linked Ubiquitination of TBK1. J. Immunol. 2021, 207, 2754–2769. [Google Scholar] [CrossRef]
- Yang, K.; Huang, Q.; Wang, R.; Zeng, Y.; Cheng, M.; Xue, Y.; Shi, C.; Ye, L.; Yang, W.; Jiang, Y.; et al. African Swine Fever Virus MGF505-11R Inhibits Type I Interferon Production by Negatively Regulating the CGAS-STING-Mediated Signaling Pathway. Vet. Microbiol. 2021, 263, 109265. [Google Scholar] [CrossRef]
- Li, D.; Yang, W.; Li, L.; Li, P.; Ma, Z.; Zhang, J.; Qi, X.; Ren, J.; Ru, Y.; Niu, Q.; et al. African Swine Fever Virus MGF-505-7R Negatively Regulates CGAS–STING-Mediated Signaling Pathway. J. Immunol. 2021, 206, 1844–1857. [Google Scholar] [CrossRef]
- Correia, S.; Ventura, S.; Parkhouse, R.M. Identification and Utility of Innate Immune System Evasion Mechanisms of ASFV. Virus Res. 2013, 173, 87–100. [Google Scholar] [CrossRef]
- De Oliveira, V.L.; Almeida, S.C.P.; Soares, H.R.; Crespo, A.; Marshall-Clarke, S.; Parkhouse, R.M.E. A Novel TLR3 Inhibitor Encoded by African Swine Fever Virus (ASFV). Arch. Virol. 2011, 156, 597–609. [Google Scholar] [CrossRef] [Green Version]
- Karalyan, Z.; Zakaryan, H.; Sargsyan, K.H.; Voskanyan, H.; Arzumanyan, H.; Avagyan, H.; Karalova, E. Interferon Status and White Blood Cells during Infection with African Swine Fever Virus in Vivo. Vet. Immunol. Immunopathol. 2012, 145, 551–555. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2021, 7, 1167. [Google Scholar] [CrossRef]
- Fan, W.; Jiao, P.; Zhang, H.; Chen, T.; Zhou, X.; Qi, Y.; Sun, L.; Shang, Y.; Zhu, H.; Hu, R.; et al. Inhibition of African Swine Fever Virus Replication by Porcine Type I and Type II Interferons. Front. Microbiol. 2020, 11, 1203. [Google Scholar] [CrossRef] [PubMed]
| Type I and Type III IFN | Type II IFN | |||
|---|---|---|---|---|
| PRRSV | In vivo early systemic induction of IFN-α response after challenge with PRRSV strains | [30] | In vitro detection of SC-IFN-γ secreting cells at 2–3 weeks P.I. showing an erratic behaviour | [61] |
| In vivodown-regulation of IFN-α production following respiratory infection | [37] | In vivo transient and limited IFN-γ response after PRRSV infection | [66] | |
| Up-regulation of type I IFN gene expression by miR-26a | [44] | Early IFN-γ production in PRRS-infected pigs from NK cells activation | [68] | |
| Down-regulation of type I IFN gene expression by miR-373 and miR-382-5p | [45,46] | |||
| In vivo induction of a “bad IFN-α response” in PRRSV-infected pigs | [52,53] | |||
| Down-regulation of key transcription factor (IRF-1, IRF-3 andNF-κB) after ADE-mediated PRRSV infection | [60] | |||
| Reduced/unchanged IFN type III expression in PAM-pCD163 cells following nsp2-deletion mutants infection | [72] | |||
| FMD | In vitro limited induction of a type I IFN response | [102] | In vitroinduction of IFN-γ response in whole blood saples from FMD-vaccinated cattle | [111] |
| Up-regulation of IFN-β expression in bovine kidney cells following O1Lif mutant infection | [102,103] | In vivoproductionof IFN-γ after injection of high potency, emergency FMD vaccines in swine | [84] | |
| Down-regulation of IFN-β production after infection with non mutant FMDV strains | [104] | |||
| In vivo constitutive expression of Type I IFNs response | [105,106] | |||
| In vitroup-regulation of type I IFN production following stimulation of bovine pDC with FMDV immune complexes | [108] | |||
| PRCV | Detection of high levels of IFN-α during subclinical course of PRCV infection | [135] | Detection of high IFN-γ levels IN BALs fluid and serum of PRCV infected pigs | [131] |
| Type I IFN production in lung secretions within 24 h PI in experimentally infected pigs | [130] | |||
| Detection of high IFN-α levels in BALs fluids and serum of PRCV infected pigs | [131,136] | |||
| PCV2 | Impairment of pDCs ability to produce IFN-α through TLR-7 and TLR-9 receptors-mediated pathway | [188] | Induction of IFN-γ following vaccination with inactivated PCV2 vaccine and VLPs-based vaccine | [174,195] |
| In vivoidetectionof IFN-α response in PCV2 infected piglets and pigs | [189,191] | Failure in inducing an IFN-γ response after vaccination with Non-Assembled ORF2 Capsid Protein of Porcine Circovirus 2b | [196] | |
| Detection of high levels of IFN-α in PCV2-infected swine alveolar macrophages (AMs) | [192] | |||
| Induction of IFN-β in PK-15 cells through a RIG-1 and MDA-5 signaling pathway | [193] | |||
| PRV | Induction of IFN-α response in purified pDCs | [224] | Induction of a persistent IFN-γ response in PBMCs following PRV infection | [229] |
| increased IFN-α response associated with PRV gE-gI-deleted mutants | [225] | In vitro induction of IFN-γ response in whole blood saples from PRV-vaccinated pigs | [230] | |
| Down-regulation of IFN-β production by PRV gE via CREB-CBP degradation | [226] | |||
| ASFV | In vitro up-regulation of IFN-α subtypes in unactivated and activated moMφ infected with both virulent and attenuated strains | [265] | Suppression of type II IFN response following infection with ASFV-encoded multigene families (MGFs) strains | [270,271] |
| In vitro statistically significant up-regulation of IFN-α10, IFN-α12, IFN-α13, IFN-α15, IFN-α16, IFN-α17 and IFN-β in unactivated moMφ infected with an attenuated strain | [265] | In vivo increased levels of IFN-γ following infection with ASFV virulent strain | [272] | |
| In vitro statistically significant up-regulation of IFN-α1, IFN-α10, IFN-α15, IFN-α16 and IFN-α17 in activated moMφ infected with an attenuated strain | [265] | |||
| Up-regulation of IFN-β expression in infected Mφ with attenuated strain through cGAS-STING-IRF3 signaling pathway during early infection | [248] | |||
| Suppression of type I IFN response following infection with ASFV-encoded multigene families (MGFs) strains | [270,271] | |||
| In vitroinduction of high levels of IFN-β mRNA after infection with deleted-MGF360 and MGF530/505 Benin 97/1 strain | [250] | |||
| In vivo increased levels of type I IFN following infection with ASFV virulent strain | [259,272,273] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razzuoli, E.; Armando, F.; De Paolis, L.; Ciurkiewicz, M.; Amadori, M. The Swine IFN System in Viral Infections: Major Advances and Translational Prospects. Pathogens 2022, 11, 175. https://doi.org/10.3390/pathogens11020175
Razzuoli E, Armando F, De Paolis L, Ciurkiewicz M, Amadori M. The Swine IFN System in Viral Infections: Major Advances and Translational Prospects. Pathogens. 2022; 11(2):175. https://doi.org/10.3390/pathogens11020175
Chicago/Turabian StyleRazzuoli, Elisabetta, Federico Armando, Livia De Paolis, Malgorzata Ciurkiewicz, and Massimo Amadori. 2022. "The Swine IFN System in Viral Infections: Major Advances and Translational Prospects" Pathogens 11, no. 2: 175. https://doi.org/10.3390/pathogens11020175
APA StyleRazzuoli, E., Armando, F., De Paolis, L., Ciurkiewicz, M., & Amadori, M. (2022). The Swine IFN System in Viral Infections: Major Advances and Translational Prospects. Pathogens, 11(2), 175. https://doi.org/10.3390/pathogens11020175







