Dynamic Transcriptome Profiling of Mungbean Genotypes Unveil the Genes Respond to the Infection of Mungbean Yellow Mosaic Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genotype Panel and Pathogen Inoculation
2.2. Library Preparation and Illumina Sequencing
2.3. Reads Filtering and Mapping
2.4. Analysis of Differentially Expressed Genes
2.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
3. Results
3.1. Mungbean Genotypes Reaction to MYMV
3.2. Summary of Transcriptome Data Set
3.3. DEGs in Response to MYMV Infection
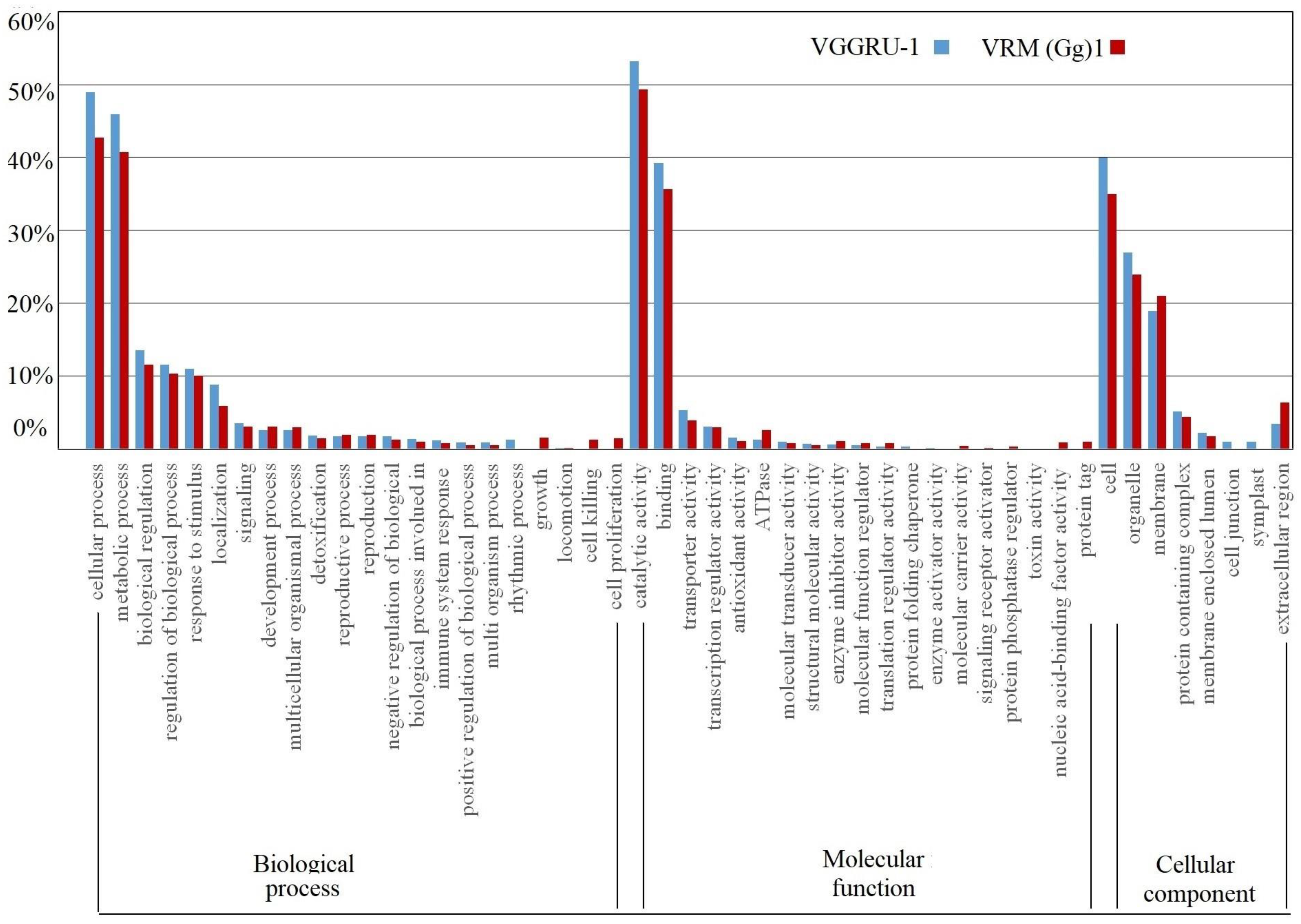
3.4. Gene Ontology Analysis of DEGs
3.5. Analysing DEGs Related to Defense Response to Pathogen Infection
3.6. Validation of Defense Response-Related DEGs
4. Discussion
4.1. Defense and Pathogenesis Related Genes
4.2. LRR-RLK/STK Genes
4.3. Genes Involved in SA, JA, and ET Pathway
4.4. Transcription Factors and Secondary Metabolites
4.5. Comparison of the Major DEGs with the Previous Reports of YMD Resistance in Mungbean
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qazi, J.; Ilyas, M.; Mansoor, S.; Briddon, R.W. Legume yellow mosaic viruses: Genetically isolated begomoviruses. Mol. Plant Pathol. 2007, 8, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.; Vanitharani, R.; Balaji, V.; Anuradha, S.; Thillaichidambaram, P.; Shivaprasad, P.; Parameswari, C.; Balamani, V.; Saminathan, M.; Veluthambi, K. Analysis of an isolate of Mungbean yellow mosaic virus (MYMV) with a highly variable DNA B component. Arch. Virol. 2004, 149, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Usharani, K.; Surendranath, B.; Haq, Q.; Malathi, V. Yellow mosaic virus infecting soybean in northern India is distinct from the species infecting soybean in southern and western India. Curr. Sci. 2004, 845–850. [Google Scholar]
- Sudha, M.; Karthikeyan, A.; Nagarajan, P.; Raveendran, M.; Senthil, N.; Pandiyan, M.; Angappan, K.; Ramalingam, J.; Bharathi, M.; Rabindran, R. Screening of mungbean (Vigna radiata) germplasm for resistance to Mungbean yellow mosaic virus using agroinoculation. Can. J. Plant Pathol. 2013, 35, 424–430. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Shobhana, V.; Sudha, M.; Raveendran, M.; Senthil, N.; Pandiyan, M.; Nagarajan, P. Mungbean yellow mosaic virus (MYMV): A threat to green gram (Vigna radiata) production in Asia. Int. J. Pest Manag. 2014, 60, 314–324. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Sudha, M.; Pandiyan, M.; Senthil, N.; Shobana, V.; Nagarajan, P. Screening of MYMV resistant mungbean (Vigna radiata L. Wilczek) progenies through agroinoculation. Int. J. Plant Pathol. 2011, 2, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Nair, R.; Götz, M.; Winter, S.; Giri, R.; Boddepalli, V.; Sirari, A.; Bains, T.; Taggar, G.; Dikshit, H.; Aski, M. Identification of mungbean lines with tolerance or resistance to yellow mosaic in fields in India where different begomovirus species and different Bemisia tabaci cryptic species predominate. Eur. J. Plant Pathol. 2017, 149, 349–365. [Google Scholar] [CrossRef] [Green Version]
- Parihar, A.K.; Basandrai, A.K.; Sirari, A.; Dinakaran, D.; Singh, D.; Kannan, K.; Kushawaha, K.P.; Adinarayan, M.; Akram, M.; Latha, T.K.S. Assessment of mungbean genotypes for durable resistance to Yellow Mosaic D isease: Genotype× Environment interactions. Plant Breed. 2017, 136, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Basavaraj, S.; Padmaja, A.; Nagaraju, N.; Ramesh, S. Identification of stable sources of resistance to mungbean yellow mosaic virus (MYMV) disease in mungbean [Vigna radiata (L.) Wilczek]. Plant Genet. Resour. 2019, 17, 362–370. [Google Scholar]
- Bai, T.-T.; Xie, W.-B.; Zhou, P.-P.; Wu, Z.-L.; Xiao, W.-C.; Zhou, L.; Sun, J.; Ruan, X.-L.; Li, H.-P. Transcriptome and expression profile analysis of highly resistant and susceptible banana roots challenged with Fusarium oxysporum f. sp. cubense tropical race 4. PLoS ONE 2013, 8, e73945. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Chittem, K.; Brueggeman, R.; Osorno, J.M.; Richards, J.; Nelson, B.D., Jr. Comparative transcriptome analysis of resistant and susceptible common bean genotypes in response to soybean cyst nematode infection. PLoS ONE 2016, 11, e0159338. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Keller, B.; McDonald, B.A.; Palma-Guerrero, J.; Wicker, T. Comparative transcriptomics reveals how wheat responds to infection by Zymoseptoria tritici. Mol. Plant-Microbe Interact. 2018, 31, 420–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, X.; Zhang, L.; Feng, S.; Zhao, Z.; Wang, X.; Gao, H. Transcriptome analysis of apple leaves in response to powdery mildew (Podosphaera leucotricha) infection. Int. J. Mol. Sci. 2019, 20, 2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Wang, Y.; Yang, J.; Yang, W. Comparative Transcriptome Analysis of Resistant and Susceptible Tomato Lines in Response to Infection by Xanthomonas perforans Race T3. Front. Plant Sci. 2015, 6, 1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lekota, M.; Muzhinji, N.; Van der Waals, J.E. Identification of differentially expressed genes in tolerant and susceptible potato cultivars in response to Spongospora subterranea f. sp. subterranea tuber infection. Plant Pathol. 2019, 68, 1196–1206. [Google Scholar] [CrossRef]
- Chen, T.; Lv, Y.; Zhao, T.; Li, N.; Yang, Y.; Yu, W.; He, X.; Liu, T.; Zhang, B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE 2013, 8, e80816. [Google Scholar]
- Wang, Y.; Zhou, L.; Yu, X.; Stover, E.; Luo, F.; Duan, Y. Transcriptome profiling of Huanglongbing (HLB) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 2016, 7, 933. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, Y.; Li, C.; Song, X.; Lei, J.; Gao, Y.; Liang, Q. Comparative transcriptome profiling of resistant and susceptible sugarcane genotypes in response to the airborne pathogen Fusarium verticillioides. Mol. Biol. Rep. 2019, 46, 3777–3789. [Google Scholar] [CrossRef]
- Dasgupta, U.; Mishra, G.P.; Dikshit, H.K.; Mishra, D.C.; Bosamia, T.; Roy, A.; Bhati, J.; Aski, M.; Kumar, R.R.; Singh, A.K. Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]. PLoS ONE 2021, 16, e0244593. [Google Scholar] [CrossRef]
- Chakraborty, N.; Basak, J. Comparative transcriptome profiling of a resistant vs. susceptible Vigna mungo cultivar in response to Mungbean yellow mosaic India virus infection reveals new insight into MYMIV resistance. Curr. Plant Biol. 2018, 15, 8–24. [Google Scholar] [CrossRef]
- Kundu, A.; Singh, P.K.; Dey, A.; Ganguli, S.; Pal, A. Complex molecular mechanisms underlying MYMIV-resistance in Vigna mungo revealed by comparative transcriptome profiling. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.; Shi, S.; Zhang, C.; Zhu, S.; Li, M.; Tan, J.; Yu, Y.; Lin, L.; Jia, S.; Wang, X. Transcriptomic analysis of genes in soybean in response to Peronospora manshurica infection. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soria-Guerra, R.E.; Rosales-Mendoza, S.; Chang, S.; Haudenshield, J.S.; Padmanaban, A.; Rodriguez-Zas, S.; Hartman, G.L.; Ghabrial, S.A.; Korban, S.S. Transcriptome analysis of resistant and susceptible genotypes of Glycine tomentella during Phakopsora pachyrhizi infection reveals novel rust resistance genes. Theor. Appl. Genet. 2010, 120, 1315–1333. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Bagnaresi, P.; Biselli, C.; Carneiro, G.A.; Siciliano, I.; Valé, G.; Gullino, M.L.; Spadaro, D. Comparative transcriptome profiling of resistant and susceptible rice genotypes in response to the seedborne pathogen Fusarium fujikuroi. BMC Genom. 2016, 17, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Ma, L.; Zhao, J.; Li, Z.; Sun, F.; Lu, X. Comparative transcriptome analysis of two rice varieties in response to rice stripe virus and small brown planthoppers during early interaction. PLoS ONE 2013, 8, e82126. [Google Scholar] [CrossRef] [PubMed]
- Balaji, V.; Vanitharani, R.; Karthikeyan, A.; Anbalagan, S.; Veluthambi, K. Infectivity analysis of two variable DNA B components of Mungbean yellow mosaic virus-Vigna in Vigna mungo and Vigna radiata. J. Biosci. 2004, 29, 297–308. [Google Scholar] [CrossRef]
- Sudha, M.; Karthikeyan, A.; Shobhana, V.; Nagarajan, P.; Raveendran, M.; Senthil, N.; Pandiyan, M.; Angappan, K.; Balasubramanian, P.; Rabindran, R. Search for Vigna species conferring resistance to mungbean yellow mosaic virus in mungbean. Plant Genet. Resour. 2015, 13, 162–167. [Google Scholar] [CrossRef]
- Eybishtz, A.; Peretz, Y.; Sade, D.; Akad, F.; Czosnek, H. Silencing of a single gene in tomato plants resistant to Tomato yellow leaf curl virus renders them susceptible to the virus. Plant Mol. Biol. 2009, 71, 157–171. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Luo, X.; Qian, J.; Pang, X.; Song, J.; Qian, G.; Chen, J.; Chen, S. FastUniq: A fast de novo duplicates removal tool for paired short reads. PLoS ONE 2012, 7, e52249. [Google Scholar] [CrossRef] [Green Version]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Sun, F.; Sun, S.; Wang, L.; Wu, J.; Zhu, Z. Transcriptome Analysis of Resistance to Fusarium Wilt in Mung Bean (Vigna radiata L.). Front. Plant Sci. 2021, 12, 1213. [Google Scholar] [CrossRef]
- Ha, J.; Shim, S.; Lee, T.; Lee, E.; Yang, X.; Jeong, H.; Kim, M.Y.; Lee, S.-H. Transcriptomic and biochemical analyses of the accumulation of sucrose in mungbean (Vigna radiata (L.) Wilczek) leaves after pod removal. Theor. Appl. Genet. 2020, 133, 2355–2362. [Google Scholar] [CrossRef]
- Tian, X.; Li, S.; Liu, Y.; Liu, X. Transcriptomic profiling reveals metabolic and regulatory pathways in the desiccation tolerance of Mungbean (Vigna radiata [L.] R. Wilczek). Front. Plant Sci. 2016, 7, 1921. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Adhimoolam, K.; Yuan, Y.; Yin, J.; Ren, R.; Yang, Y.; Zhi, H. Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci. 2017, 68, 156–166. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W. A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef] [Green Version]
- Reboledo, G.; Del Campo, R.; Alvarez, A.; Montesano, M.; Mara, H.; Ponce de León, I. Physcomitrella patens activates defense responses against the pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 2015, 16, 22280–22298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Zhang, X.; Tu, L.; Zeng, F.; Nie, Y.; Guo, X. Isolation and characterization of two novel dirigent-like genes highly induced in cotton (Gossypium barbadense and G. hirsutum) after infection by Verticillium dahliae. J. Plant Pathol. 2007, 89, 41–45. [Google Scholar]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Song, F.; Zheng, Z. Molecular characterization and expression analysis of a rice protein phosphatase 2C gene, OsBIPP2C1, and overexpression in transgenic tobacco conferred enhanced disease resistance and abiotic tolerance. Physiol. Plant. 2006, 127, 225–236. [Google Scholar] [CrossRef]
- Seo, J.-K.; Kwon, S.-J.; Cho, W.K.; Choi, H.-S.; Kim, K.-H. Type 2C protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Sci. Rep. 2014, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-Y.; Nagy, P.D. Blocking tombusvirus replication through the antiviral functions of DDX17-like RH30 DEAD-box helicase. PLoS Pathog. 2019, 15, e1007771. [Google Scholar] [CrossRef]
- Li, D.; Liu, H.; Zhang, H.; Wang, X.; Song, F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 2008, 59, 2133–2146. [Google Scholar] [CrossRef]
- Chae, L.; Sudat, S.; Dudoit, S.; Zhu, T.; Luan, S. Diverse transcriptional programs associated with environmental stress and hormones in the Arabidopsis receptor-like kinase gene family. Mol. Plant 2009, 2, 84–107. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, B.; Xu, Z.; Li, M.; Song, Z.; Li, W.; Li, Y. Tobacco serine/threonine protein kinase gene NrSTK enhances black shank resistance. Genet. Mol. Res. 2015, 14, 16415–16424. [Google Scholar] [CrossRef]
- Armijo, G.; Salinas, P.; Monteoliva, M.I.; Seguel, A.; García, C.; Villarroel-Candia, E.; Song, W.; Van Der Krol, A.R.; Álvarez, M.E.; Holuigue, L. A salicylic acid-induced lectin-like protein plays a positive role in the effector-triggered immunity response of Arabidopsis thaliana to Pseudomonas syringae Avr-Rpm1. Mol. Plant-Microbe Interact. 2013, 26, 1395–1406. [Google Scholar] [CrossRef] [Green Version]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Yang, Q.; Zhi, H. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1998, 1, 316–323. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 2016, 58, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Song, J.T.; Cheong, J.-J.; Lee, Y.-H.; Lee, Y.-W.; Hwang, I.; Lee, J.S.; Do Choi, Y. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivasankar, S.; Sheldrick, B.; Rothstein, S.J. Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 2000, 122, 1335–1342. [Google Scholar] [CrossRef] [Green Version]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Jin, J.-H.; Zhang, H.-X.; Tan, J.-Y.; Yan, M.-J.; Li, D.-W.; Khan, A.; Gong, Z.-H. A new ethylene-responsive factor CaPTI1 gene of pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici. Front. Plant Sci. 2016, 6, 1217. [Google Scholar] [CrossRef]
- Lai, Y.; Dang, F.; Lin, J.; Yu, L.; Shi, Y.; Xiao, Y.; Huang, M.; Lin, J.; Chen, C.; Qi, A. Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol. Biochem. 2013, 62, 70–78. [Google Scholar] [CrossRef]
- Tezuka, D.; Kawamata, A.; Kato, H.; Saburi, W.; Mori, H.; Imai, R. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae. Plant Physiol. Biochem. 2019, 135, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Akio Amorim, L.L.; da Fonseca dos Santos, R.; Pacifico Bezerra Neto, J.; Guida-Santos, M.; Crovella, S.; Maria Benko-Iseppon, A. Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, E.; Choi, D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief. Funct. Genom. 2015, 14, 260–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Yu, J.; Ai, G.; Shen, D.; Chai, C.; Jia, Y.; Liu, W.; Dou, D. Bioinformatical analysis and prediction of Nicotiana benthamiana bHLH transcription factors in Phytophthora parasitica resistance. Genomics 2019, 111, 473–482. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Zhang, C.-L.; Wang, G.-L.; Wang, Y.-X.; Qi, C.-H.; Zhao, Q.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.; Van Der Linden, C. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef] [Green Version]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Dong, L.; Gao, T.; Liu, T.; Li, N.; Wang, L.; Chang, X.; Wu, J.; Xu, P.; Zhang, S. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018, 69, 2527–2541. [Google Scholar] [CrossRef]
- Ibraheem, F.; Gaffoor, I.; Tan, Q.; Shyu, C.-R.; Chopra, S. A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize. Molecules 2015, 20, 2388–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Cao, T.; Lahiri, I.; Singh, V.; Louis, J.; Shah, J.; Ayre, B.G. Metabolic engineering of raffinose-family oligosaccharides in the phloem reveals alterations in carbon partitioning and enhances resistance to green peach aphid. Front. Plant Sci. 2013, 4, 263. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chen, H.; Yuan, J.S.; Köllner, T.G.; Chen, Y.; Guo, Y.; Zhuang, X.; Chen, X.; Zhang, Y.J.; Fu, J. The rice terpene synthase gene Os TPS 19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 2018, 16, 1778–1787. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 1–20. [Google Scholar] [CrossRef]
- He, Y.; Wu, L.; Liu, X.; Jiang, P.; Yu, L.; Qiu, J.; Wang, G.; Zhang, X.; Ma, H. TaUGT6, a novel UDP-Glycosyltransferase gene enhances the resistance to FHB and DON accumulation in wheat. Front. Plant Sci. 2020, 11, 1549. [Google Scholar] [CrossRef]
- Li, R.; Yuan, S.; He, Y.; Fan, J.; Zhou, Y.; Qiu, T.; Lin, X.; Yao, Y.; Liu, J.; Fu, S. Genome-wide identification and expression profiling analysis of the galactinol synthase gene family in cassava (Manihot esculenta Crantz). Agronomy 2018, 8, 250. [Google Scholar] [CrossRef] [Green Version]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Schölkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef]
- Shi, H.; Liu, W.; Yao, Y.; Wei, Y.; Chan, Z. Alcohol dehydrogenase 1 (ADH1) confers both abiotic and biotic stress resistance in Arabidopsis. Plant Sci. 2017, 262, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Huot, B.; Foune, C.; Doddapaneni, H.; Enyedi, A. Expression of a β-glucosidase gene results in increased accumulation of salicylic acid in transgenic Nicotiana tabacum cv. Xanthi-nc NN genotype. Plant Cell Rep. 2007, 26, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.S.; Kim, N.H.; Hwang, B.K. Pepper mitochondrial FORMATE DEHYDROGENASE1 regulates cell death and defense responses against bacterial pathogens. Plant Physiol. 2014, 166, 1298–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariola, T.; Brader, G.; Li, J.; Palva, E.T. Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 2005, 17, 282–294. [Google Scholar] [CrossRef] [Green Version]
- Stefanowicz, K.; Lannoo, N.; Van Damme, E.J. Plant F-box proteins–Judges between life and death. Crit. Rev. Plant Sci. 2015, 34, 523–552. [Google Scholar] [CrossRef]
- Oh, S.-K.; Baek, K.-H.; Seong, E.S.; Joung, Y.H.; Choi, G.-J.; Park, J.M.; Cho, H.S.; Kim, E.A.; Lee, S.; Choi, D. CaMsrB2, pepper methionine sulfoxide reductase B2, is a novel defense regulator against oxidative stress and pathogen attack. Plant Physiol. 2010, 154, 245–261. [Google Scholar] [CrossRef] [Green Version]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [Green Version]
- Chittoor, J.M.; Leach, J.E.; White, F.F. Induction of peroxidase during defense against pathogens. In Pathogenesis-Related Proteins in Plants; CRC Press LLC: Boca Raton, FL, USA, 1999; pp. 171–193. [Google Scholar]
- Mathivathana, M.K.; Murukarthick, J.; Karthikeyan, A.; Jang, W.; Dhasarathan, M.; Jagadeeshselvam, N.; Sudha, M.; Vanniarajan, C.; Karthikeyan, G.; Yang, T.-J. Detection of QTLs associated with mungbean yellow mosaic virus (MYMV) resistance using the interspecific cross of Vigna radiata × Vigna umbellata. J. Appl. Genet. 2019, 60, 255–268. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sudha, M.; Karthikeyan, A.; Madhumitha, B.; Veera Ranjani, R.; Kanimoli Mathivathana, M.; Dhasarathan, M.; Murukarthick, J.; Samu Shihabdeen, M.N.; Eraivan Arutkani Aiyanathan, K.; Pandiyan, M.; et al. Dynamic Transcriptome Profiling of Mungbean Genotypes Unveil the Genes Respond to the Infection of Mungbean Yellow Mosaic Virus. Pathogens 2022, 11, 190. https://doi.org/10.3390/pathogens11020190
Sudha M, Karthikeyan A, Madhumitha B, Veera Ranjani R, Kanimoli Mathivathana M, Dhasarathan M, Murukarthick J, Samu Shihabdeen MN, Eraivan Arutkani Aiyanathan K, Pandiyan M, et al. Dynamic Transcriptome Profiling of Mungbean Genotypes Unveil the Genes Respond to the Infection of Mungbean Yellow Mosaic Virus. Pathogens. 2022; 11(2):190. https://doi.org/10.3390/pathogens11020190
Chicago/Turabian StyleSudha, Manickam, Adhimoolam Karthikeyan, Balasubramaniam Madhumitha, Rajagopalan Veera Ranjani, Mayalagu Kanimoli Mathivathana, Manickam Dhasarathan, Jayakodi Murukarthick, Madiha Natchi Samu Shihabdeen, Karuppiah Eraivan Arutkani Aiyanathan, Muthaiyan Pandiyan, and et al. 2022. "Dynamic Transcriptome Profiling of Mungbean Genotypes Unveil the Genes Respond to the Infection of Mungbean Yellow Mosaic Virus" Pathogens 11, no. 2: 190. https://doi.org/10.3390/pathogens11020190
APA StyleSudha, M., Karthikeyan, A., Madhumitha, B., Veera Ranjani, R., Kanimoli Mathivathana, M., Dhasarathan, M., Murukarthick, J., Samu Shihabdeen, M. N., Eraivan Arutkani Aiyanathan, K., Pandiyan, M., Senthil, N., & Raveendran, M. (2022). Dynamic Transcriptome Profiling of Mungbean Genotypes Unveil the Genes Respond to the Infection of Mungbean Yellow Mosaic Virus. Pathogens, 11(2), 190. https://doi.org/10.3390/pathogens11020190







