Environmental Monitoring for Enteroviruses in Maputo, Mozambique—2018
Abstract
1. Introduction
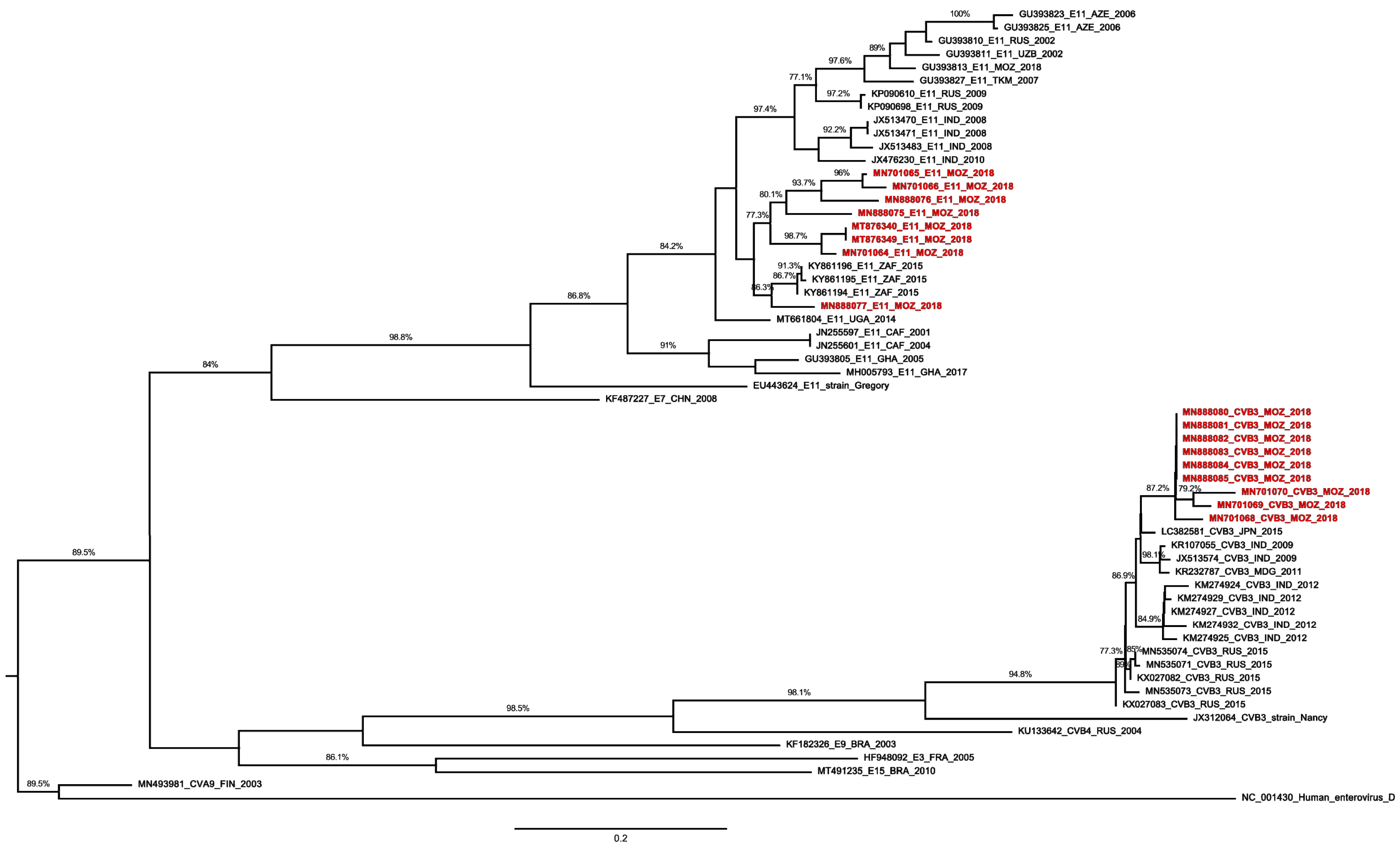
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Gorbalenya, A.E.; Hovi, H.H.T.; Lindberg, N.J.K.A.M. Recommendations for the Nomenclature of Enteroviruses and Rhinoviruses. Arch. Virol. 2020, 165, 793–797. [Google Scholar] [CrossRef] [PubMed]
- WHO. Enterovirus Surveillance Guidelines. Available online: http://www.euro.who.int/__data/assets/pdf_file/0020/272810/EnterovirusSurveillanceGuidelines.pdf (accessed on 13 July 2020).
- Griffin, D.W.; Donaldson, K.A.; Paul, J.H.; Rose, J.B. Pathogenic Human Viruses in Coastal Waters. Clin. Microbiol. Rev. 2003, 16, 129–143. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines for Environmental Surveillance of Poliovirus Circulation Guidelines for Environmental. Available online: http://apps.who.int/iris/bitstream/10665/67854/1/WHO_V-B_03.03_eng.pdf (accessed on 13 July 2020).
- Bisseux, M.; Colombet, J.; Mirand, A.; Roque-Afonso, A.M.; Abravanel, F.; Izopet, J.; Archimbaud, C.; Peigue-Lafeuille, H.; Debroas, D.; Bailly, J.L.; et al. Monitoring Human Enteric Viruses in Wastewater and Relevance to Infections Encountered in the Clinical Setting: A One-Year Experiment in Central France, 2014 to 2015. Eurosurveillance 2018, 23, 17-00237. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, K.; Oka, T.; Takeda, N.; Hansman, G.S. Norovirus Infections in Symptomatic and Asymptomatic Food Handlers in Japan. J. Clin. Microbiol. 2007, 45, 3996–4005. [Google Scholar] [CrossRef]
- Kotwal, A.; Singh, H.; Verma, A.K.; Gupta, R.M.; Jain, S.; Sinha, S.; Joshi, R.K.; Teli, P.; Khunga, V.; Bhatnagar, A.; et al. A Study of Hepatitis A and E Virus Seropositivity Profile amongst Young Healthy Adults in India. Med. J. Armed Forces India 2014, 70, 225–229. [Google Scholar] [CrossRef][Green Version]
- Pereira, J.d.O.S.; da Silva, L.R.; de Meireles Nunes, A.; de Souza Oliveira, S.; da Costa, E.V.; da Silva, E.E. Environmental Surveillance of Polioviruses in Rio de Janeiro, Brazil, in Support to the Activities of Global Polio Eradication Initiative. Food Environ. Virol. 2016, 8, 27–33. [Google Scholar] [CrossRef]
- Werneck, L.M.C.; Baptista, M.L.; Miagostovich, M.P.; da Silva, E.E. Dissemination of Enteroviruses in the Production Chain of Organic Lettuce in Rio de Janeiro, Brazil. MicrobiologyOpen 2019, 8, e00653. [Google Scholar] [CrossRef]
- CDC Tracking Progress toward Global Polio Eradication—Worldwide. 2009-MMWR Morb. Mortal. Wkly. Rep. 2011, 60, 441–445. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6014a2.htm21-05-2021 (accessed on 21 May 2021).
- WHO. Circulating Vaccine-Derived Poliovirus Type 2—Mozambique. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/25-january-2019-polio-mozambique-en (accessed on 26 February 2020).
- Cassocera, M.; Chissaque, A.; Martins, M.R.O.; De Deus, N. 40 Years of Immunization in Mozambique: A Narrative Review of Literature, Accomplishments, and Perspectives. Cad. Saude Publica 2020, 36, 1–17. [Google Scholar] [CrossRef]
- Polioeradication. Global Circulating Vaccine-Derived Poliovirus (CVDPV)1,2,3. Available online: https://polioeradication.org/wp-content/uploads/2022/03/weekly-polio-analyses-cVDPV-20220322.pdf (accessed on 29 March 2022).
- Bero, D.M.; de Deus, N.; da Costa, E.V.; Burlandy, F.M.; Jani, I.V.; da Silva, E.E. Natural Circulation of Human Enterovirus in Maputo City, Mozambique. Afr. J. Microbiol. Res. 2015, 9, 1419–1423. [Google Scholar] [CrossRef][Green Version]
- Rao, C.; Yergolkar, P.; Shankarappa, K. Antigenic Diversity of Enteroviruses Associated with Nonpolio Acute Flaccid Paralysis, India, 2007–2009. Emerg. Infect. Dis. 2012, 18, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.D.; Kebe, O.; Fall, A.D.; Ndiaye, K. Identification and Molecular Characterization of Non-Polio Enteroviruses from Children with Acute Flaccid Paralysis in West Africa, 2013–2014. Sci. Rep. 2017, 7, 3808. [Google Scholar] [CrossRef]
- Laxmivandana, R.; Cherian, S.S.; Yergolkar, P.; Chitambar, S.D. Genomic Characterization of Coxsackievirus Type B3 Strains Associated with Acute Flaccid Paralysis in South-Western India. J. Gen. Virol. 2016, 97, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Odoom, J.; Obodai, E.; Diamenu, S.; Ahove, V.; Addo, J.; Banahene, B.; Taylor, J.; Na, A.; Ko, A.; Dogbe, M.; et al. Environmental Surveillance for Poliovirus in Greater Accra and Eastern Regions of Ghana-2016. Virol. Curr. Res. 2017, 1, 1–6. [Google Scholar]
- Khetsuriani, N.; Kutateladze, T.; Zangaladze, E.; Shutkova, T.; Peñaranda, S.; Nix, W.A.; Pallansch, M.A.; Oberste, M.S. High Degree of Genetic Diversity of Non-Polio Enteroviruses Identified in Georgia by Environmental and Clinical Surveillance, 2002–2005. J. Med. Microbiol. 2010, 59, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Bingjun, T.; Yoshida, H.; Yan, W.; Lin, L.; Tsuji, T.; Shimizu, H.; Miyamura, T. Molecular Typing and Epidemiology of Non-Polio Enteroviruses Isolated From Yunnan Province, the People’s Republic of China. J. Med. Virol. 2008, 4, 670–679. [Google Scholar] [CrossRef]
- Bessaud, M.; Pillet, S.; Ibrahim, W.; Joffret, M.-L.; Pozzetto, B.; Delpeyroux, F.; Gouandjika-Vasilached, I. Molecular Characterization of Human Enteroviruses in the Central African Republic: Uncovering Wide Diversity and Identification of a New Human Enterovirus A71 Genogroup. J. Clin. Microbiol. 2012, 50, 1650–1658. [Google Scholar] [CrossRef]
- Sousa, I.P.; Oliveira, M.d.L.A.; Burlandy, F.M.; Machado, R.S.; Oliveira, S.S.; Tavares, F.N.; Gomes-Neto, F.; da Costa, E.V.; da Silva, E.E. Molecular Characterization and Epidemiological Aspects of Non-Polio Enteroviruses Isolated from Acute Flaccid Paralysis in Brazil: A Historical Series (2005–2017). Emerg. Microbes Infect. 2020, 9, 2536–2546. [Google Scholar] [CrossRef]
- Baicus, A.; Joffret, M.L.; Bessaud, M.; Delpeyroux, F.; Oprisan, G. Reinforced Poliovirus and Enterovirus Surveillance in Romania, 2015–2016. Arch. Virol. 2020, 165, 2627–2632. [Google Scholar] [CrossRef]
- Delogu, R.; Battistone, A.; Buttinelli, G.; Fiore, S.; Fontana, S.; Amato, C.; Cristiano, K.; Gamper, S.; Simeoni, J.; Frate, R.; et al. Poliovirus and Other Enteroviruses from Environmental Surveillance in Italy, 2009–2015. Food Environ. Virol. 2018, 10, 333–342. [Google Scholar] [CrossRef]
- Polioeradication. Global Wild Poliovirus 2016–2022. Available online: https://polioeradication.org/wp-content/uploads/2022/03/weekly-polio-analyses-WPV-20220322.pdf (accessed on 29 March 2022).
- McCarthy, K.; Howard, W.; Yousif, M.; Moonsamy, S.; Suchard, M. The Show Is Not over—Wild-Type Polio in Malawi Is a Wake-up Call and an Opportunity for Elimination Efforts. Int. J. Infect. Dis. 2022, 119, 32–33. [Google Scholar] [CrossRef] [PubMed]
- Odoom, J.K.; Obodai, E.; Boateng, G.; Diamenu, S.; Attiku, K.; Avevor, P.; Duker, E.; Boahene, B.; Eshun, M.; Gberbie, E.; et al. Detection of Vaccine-Derived Poliovirus Circulation by Environmental Surveillance in the Absence of Clinical Cases. Hum. Vaccines Immunother. 2021, 17, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, A.K.; Diop, P.A.M.; Diop, O.M. Environmental Surveillance of Poliovirus and Non-Polio Enterovirus in Urban Sewage in Dakar, Senegal (2007–2013). Pan Afr. Med. J. 2014, 19, 243. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, S.; Paananen, A.; Savolainen-Kopra, C.; Hovi, T.; Roivainen, M. Eight Years of Experience with Molecular Identification of Human Enteroviruses. J. Clin. Microbiol. 2008, 46, 2410–2413. [Google Scholar] [CrossRef] [PubMed]
- INE. Resultados Definitivos Instituto Nacional de Estatistica. Available online: http://www.ine.gov.mz/iv-rgph-2017/mocambique/censo-2017-brochura-dos-resultados-definitivos-do-iv-rgph-nacional.pdf/view (accessed on 12 July 2020).
- Boom, R.; Sol, J.A.; Salimans, M.M.M.; Jansen’, C.L.; Wertheim-Van Dillen, P.M.E.; Van Der Noordaa’, J. Rapid and Simple Method for Purification of Nucleic Acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Kilpatrick, D.R.; Iber, J.C.; Chen, Q.; Ching, K.; Yang, S.J.; De, L.; Mandelbaum, M.D.; Emery, B.; Campagnoli, R.; Burns, C.C.; et al. Poliovirus Serotype-Specific VP1 Sequencing Primers. J. Virol. Methods 2011, 174, 128–130. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability Article Fast Track. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 2010–2011. [Google Scholar] [CrossRef]
| Month | Location | No of Samples Collected by Site | Isolation by RD or L20B (Positive Specimens) | Intratypic Differentiation * | Typing |
|---|---|---|---|---|---|
| January | ETAR (2) Marginal Avenue (1) | 2 | RD | Negative | CVB5, E7 CVB5 |
| February | ETAR (1) | 2 | RD and L20B | Positive | PV1 |
| March | ETAR (1) | 2 | E11 | ||
| Marginal Avenue (1) November 10th Avenue (1) | RD | Negative | EV-B75 CVB5 | ||
| April | ETAR (1) November 10th Avenue (1) | 2 | RD | Negative | E7 E11 |
| May | Marginal Avenue (1) November 10th Avenue (2) | 2 | RD | Negative | E11 E11 (2) |
| June | ETAR (1) | 2 | RD | Negative | CVB3 |
| July | ETAR (2) | 2 | RD | Negative | CVB3, E6 |
| August | ETAR (1) November 10th Avenue (1) | 2 | RD | Negative | CVB3 E11 |
| September | ETAR (2) November 10th Avenue (1) | 2 | RD | Negative | CVB3, E27 CVB3 |
| October | ETAR (1) Marginal Avenue (2) | 2 | RD | Negative | E11 CVB3 (2) |
| November | ETAR (1) Marginal Avenue (1) | 1 | RD | Negative | CVB3 CVB3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bero, D.M.; Nhassengo, S.A.; Sousa, I.P., Jr.; de Sousa, S.O.; Machado, R.S.; Dias, A.M.N.; de Sousa Ferreira, C.; Burlandy, F.M.; de Deus, N.; da Silva, E.E. Environmental Monitoring for Enteroviruses in Maputo, Mozambique—2018. Pathogens 2022, 11, 527. https://doi.org/10.3390/pathogens11050527
Bero DM, Nhassengo SA, Sousa IP Jr., de Sousa SO, Machado RS, Dias AMN, de Sousa Ferreira C, Burlandy FM, de Deus N, da Silva EE. Environmental Monitoring for Enteroviruses in Maputo, Mozambique—2018. Pathogens. 2022; 11(5):527. https://doi.org/10.3390/pathogens11050527
Chicago/Turabian StyleBero, Diocreciano Matias, Sheila António Nhassengo, Ivanildo Pedro Sousa, Jr., Silas Oliveira de Sousa, Raiana Scerni Machado, Amanda Meireles Nunes Dias, Cristiane de Sousa Ferreira, Fernanda Marcicano Burlandy, Nilsa de Deus, and Edson Elias da Silva. 2022. "Environmental Monitoring for Enteroviruses in Maputo, Mozambique—2018" Pathogens 11, no. 5: 527. https://doi.org/10.3390/pathogens11050527
APA StyleBero, D. M., Nhassengo, S. A., Sousa, I. P., Jr., de Sousa, S. O., Machado, R. S., Dias, A. M. N., de Sousa Ferreira, C., Burlandy, F. M., de Deus, N., & da Silva, E. E. (2022). Environmental Monitoring for Enteroviruses in Maputo, Mozambique—2018. Pathogens, 11(5), 527. https://doi.org/10.3390/pathogens11050527






