Serology as a Tool to Assess Infectious Disease Landscapes and Guide Public Health Policy
Abstract
:1. Introduction and Scope of the Review
2. Case-Based Infectious Disease Surveillance—Current Use and Limitations
2.1. Passive and Active Surveillance
2.2. Implementing Surveillance—Challenges
2.2.1. Definition of Catchment Population
2.2.2. Health Care Seeking Behavior
2.2.3. Case Definitions
2.2.4. Biological Specimen Collection and Diagnostics
3. Serology to Assess Disease Burden
3.1. Rationale for Using Serology to Assess Disease Burden
3.2. Advantages of Using Serology-Based Techniques to Define Disease Burden
3.2.1. Detection of Past Cases Regardless of Symptoms Occurrence
3.2.2. A Variety of Samples Can Be Used
3.2.3. Use of Convenience Sampling and Historical Collections
3.2.4. Multiplexing
3.3. Challenges in Using Serology-Based Techniques to Define Disease Burden
3.3.1. Heterogeneity of Immune Responses
3.3.2. Measurements, Thresholds and Quantifying Immune Responses
3.3.3. Data Analysis and Interpretation
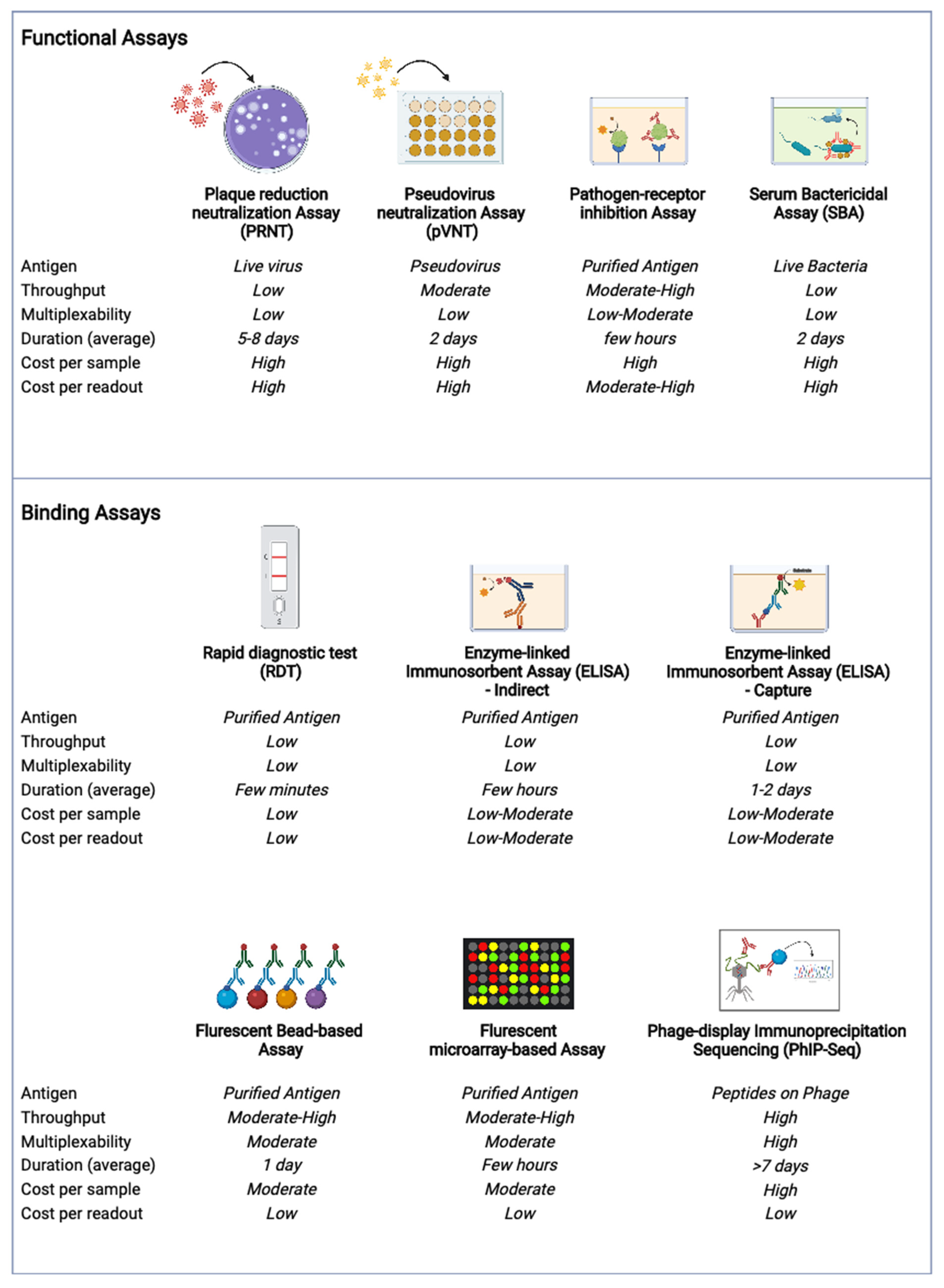
4. Current Methods, New Technologies and Future Directions
5. Real-Life Examples and Public Health Use
5.1. Serology to Guide Child Health Policies and Vaccine Roll-Out
5.2. Serology to Complement Case-Based Clinical Reporting
5.3. Serology to Assess the Burden of Infection beyond Symptomatic Cases
5.4. Serology to Measure Impact of Public Health Interventions
5.5. Serology during the COVID-19 Pandemic
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Strategy on Comprehensive Vaccine-Preventable Disease Surveillance; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Declich, S.; Carter, A.O. Public health surveillance: Historical origins, methods and evaluationi. Bull. World Health Organ. 1994, 72, 285–304. [Google Scholar] [PubMed]
- Jayatilleke, K. Challenges in implementing surveillance tools of high-income countries (HICs) in low middle income countries (LMICs). Curr. Treat. Options Infect. Dis. 2020, 12, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Jamison, D.T.; World Bank; Disease Control Priorities Project. Disease Control Priorities in Developing Countries, 2nd ed.; Oxford University Press: Washington, DC, USA, 2006; Volume xlii, p. 1401. [Google Scholar]
- Gilbert, R.; Cliff, S.J. Public health surveillance. In Public Health Intelligence; Springer International Publishing: Cham, Switzerland, 2016; Volume 2016, pp. 91–110. [Google Scholar]
- Luby, S.P.; Saha, S.; Andrews, J.R. Towards sustainable public health surveillance for enteric fever. Vaccine 2015, 33 (Suppl. 3), C3–C7. [Google Scholar]
- Groseclose, S.L.; Buckeridge, D.L. Public health surveillance systems: Recent advances in their use and evaluation. Annu. Rev. Public Health 2017, 38, 57–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbst, K.; Juvekar, S.; Jasseh, M.; Berhane, Y.; Chuc, N.T.K.; Seeley, J.; Sankoh, O.; Clark, S.J.; Collinson, M.A. Health and demographic surveillance systems in low- and middle-income countries: History, state of the art and future prospects. Glob. Health Action 2022, 14 (Suppl. 1), 1974676. [Google Scholar] [CrossRef] [PubMed]
- Gerritsen, A.; Bocquier, P.; White, M.; Mbacké, C.; Alam, N.; Béguy, D.; Odhiambo, F.; Sacoor, C.; Phuc, H.D.; Punpuing, S.; et al. Health and demographic surveillance systems: Contributing to an understanding of the dynamics in migration and health. Glob. Health Action 2013, 6, 21496. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.; Cohen, A.L. Infectious disease surveillance. In International Encyclopedia of Public Health; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Bright, T.; Fetlix, L.; Kuper, H.; Polalck, S. A systematic review of strategies to increase access to health services among children in low and middle income countries. BMC Health Serv. Res. 2017, 17, 252. [Google Scholar] [CrossRef] [Green Version]
- Panzner, U.; Pak, G.D.; Aaby, P.; Adu-Sarkodie, Y.; Ali, M.; Aseffa, A.; Baker, S.; Bjerregaard-Andersen, M.; Crump, J.A.; Deerin, J.; et al. Utilization of healthcare in the typhoid fever surveillance in Africa program. Clin. Infect. Dis. 2016, 62 (Suppl. 1), S56–S68. [Google Scholar] [CrossRef] [Green Version]
- CDC. Case definitions for infectious conditions under public health surveillance. Centers for disease control and prevention. MMWR Recomm. Rep. 1997, 46, 1–55.
- Zellweger, R.M.; Yacoub, S.; Chan, Y.F.Z.; Soon, D.; Shafi, H.; Ooi, S.T.; Chan, M.; Jacobson, L.; Sessions, O.M.; Vincent, A.; et al. Disentangling etiologies of CNS infections in Singapore using multiple correspondence analysis and random forest. Sci. Rep. 2020, 10, 18219. [Google Scholar] [CrossRef]
- Turner, P.; Fox-Lewis, A.; Shrestha, P.; Dance, D.A.B.; Wangrangsimakul, T.; Cusack, T.-P.; Ling, C.L.; Hopkins, J.; Roberts, T.; Limmathurotsakul, D.; et al. Microbiology investigation criteria for reporting objectively (MICRO): A framework for the reporting and interpretation of clinical microbiology data. BMC Med. 2019, 17, 70. [Google Scholar] [CrossRef] [PubMed]
- Skodvin, B.; Wathne, J.S.; Lindemann, P.C.; Harthug, S.; Nilsen, R.M.; Charani, E.; Syre, H.; Kittang, B.R.; Kleppe, L.K.S.; Smith, I. Use of microbiology tests in the era of increasing AMR rates—A multicentre hospital cohort study. Antimicrob. Resist. Infect. Control. 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coggon, D.; Martyn, C.; Palmer, K.T.; Evanoff, B.A. Assessing case definitions in the absence of a diagnostic gold standard. Int. J. Epidemiol. 2005, 34, 949–952. [Google Scholar] [CrossRef] [Green Version]
- Bharadwaj, M.; Bengtson, M.; Golverdingen, M.; Waling, L.; Dekker, C. Diagnosing point-of-care diagnostics for neglected tropical diseases. PLoS Negl. Trop. Dis. 2021, 15, e0009405. [Google Scholar] [CrossRef] [PubMed]
- Anh, D.D.; Choisy, M.; Clapham, H.E.; Cuong, H.Q.; Dung, V.T.V.; Duong, T.N.; Hang, N.L.K.; Ha, H.T.T.; Hien, N.T.; Hoa, T.T.N.; et al. Plans for nationwide serosurveillance network in Vietnam. Emerg. Infect. Dis. 2020, 26, e190641. [Google Scholar] [CrossRef]
- Arnold, B.F.; Scobie, H.M.; Priest, J.W.; Lammie, P.J. Integrated serologic surveillance of population immunity and disease transmission. Emerg. Infect. Dis. 2018, 24, 1188–1194. [Google Scholar] [CrossRef] [Green Version]
- Cutts, F.T.; Hanson, M. Seroepidemiology: An underused tool for designing and monitoring vaccination programmes in low- and middle-income countries. Trop. Med. Int. Health 2016, 21, 1086–1098. [Google Scholar] [CrossRef]
- Galipeau, Y.; Greig, M.; Liu, G.; Driedger, M.; Langlois, M.-A. Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020, 11, 610688. [Google Scholar]
- Metcalf, C.J.; Farrar, J.; Cutts, F.T.; Basta, N.E.; Graham, A.L.; Lessler, J.; Ferguson, N.M.; Burke, D.S.; Grenfell, B.T. Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet 2016, 388, 728–730. [Google Scholar] [CrossRef]
- Mina, M.J.; Metcalf, C.J.; McDermott, A.B.; Douek, D.C.; Farrar, J.; Grenfell, B.T. A global lmmunological observatory to meet a time of pandemics. Elife 2020, 9, e58989. [Google Scholar] [CrossRef]
- Anastassopoulou, C.G.; Kafatos, G.; Nardone, A.; Andrews, N.; Pebody, R.G.; Mossong, J.; Davidkin, I.; Gelb, D.; De Ory, F.; Thierfelder, W.; et al. The European Sero-Epidemiology Network 2 (ESEN2): Standardization of assay results for hepatitis A virus (HAV) to enable comparisons of seroprevalence data across 15 countries. Epidemiol. Infect. 2009, 137, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Fischer, C.; Jo, W.K.; Haage, V.; Moreira-Soto, A.; de Oliveira-Filho, E.F.; Drexler, J.F. Challenges towards serologic diagnostics of emerging arboviruses. Clin. Microbiol. Infect. 2021, 27, 1221–1229. [Google Scholar] [PubMed]
- Kar, P.; Karna, R. A review of the diagnosis and management of Hepatitis E. Curr. Treat. Options Infect. Dis. 2020, 12, 310–320. [Google Scholar]
- Holroyd, T.A.; Schiaffino, F.; Chang, R.H.; Wanyiri, J.W.; Saldanha, I.J.; Gross, M.; Moss, W.J.; Hayford, K. Diagnostic accuracy of dried blood spots for serology of vaccine-preventable diseases: A systematic review. Expert Rev. Vaccines 2021, 21, 185–200. [Google Scholar]
- Cervia, C.; Nilsson, J.; Zurbuchen, Y.; Valaperti, A.; Schreiner, J.; Wolfensberger, A.; Raeber, M.E.; Adamo, S.; Weigang, S.; Emmenegger, M.; et al. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J. Allergy Clin. Immunol. 2021, 147, 545–557.e9. [Google Scholar] [CrossRef] [PubMed]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claër, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223. [Google Scholar] [PubMed]
- He, T.; Kaplan, S.; Kamboj, M.; Tang, Y.-W. Laboratory diagnosis of central nervous system infection. Curr. Infect. Dis. Rep. 2016, 18, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aro, K.; Wei, F.; Wong, D.T.; Tu, M. Saliva liquid biopsy for point-of-care applications. Front. Public Health 2017, 5, 77. [Google Scholar]
- Hettegger, P.; Huber, J.; Paßecker, K.; Soldo, R.; Kegler, U.; Nöhammer, C.; Weinhäusel, A. High similarity of IgG antibody profiles in blood and saliva opens opportunities for saliva based serology. PLoS ONE 2019, 14, e0218456. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Carreras-Presas, C.M.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasim, H.; Olausson, P.; Hedenberg-Magnusson, B.; Ernberg, M.; Ghafouri, B. The proteomic profile of whole and glandular saliva in healthy pain-free subjects. Sci. Rep. 2016, 6, 39073. [Google Scholar] [CrossRef] [PubMed]
- Osborne, K.; Weinberg, J.; Miller, E. The European sero-epidemiology network. Euro. Surveill. 1997, 2, 29–31. [Google Scholar] [CrossRef] [PubMed]
- Franklin, I.M.; Dow, B.C.; Jordan, A.D. Benefits of a blood donation archive repository: International survey of donor repository procedures and Scottish experiences. Transfusion 2007, 47, 1172–1179. [Google Scholar] [CrossRef]
- Zhang, Y.; Leung, K.; Perera, R.A.P.M.; Lee, C.-K.; Peiris, J.S.M.; Wu, J.T.K. Harnessing the potential of blood donation archives for influenza surveillance and control. PLoS ONE 2020, 15, e0233605. [Google Scholar] [CrossRef] [PubMed]
- Luang-Suarkia, D.; Ernst, T.; Alpers, M.P.; Garruto, R.M.; Smith, D.; Imrie, A. Serological evidence for transmission of multiple dengue virus serotypes in Papua New Guinea and West Papua prior to 1963. PLoS Negl. Trop. Dis. 2017, 11, e0005488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.J.; Kula, T.; Xu, Q.; Li, M.Z.; Vernon, S.D.; Ndung’U, T.; Ruxrungtham, K.; Sanchez, J.; Brander, C.; Chung, R.T.; et al. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348, aaa0698. [Google Scholar] [CrossRef] [Green Version]
- Mast, E.E.; Margolis, H.S.; Fiore, A.E.; Brink, E.W.; Goldstein, S.T.; Wang, S.A.; Moyer, L.A.; Bell, B.P.; Alter, M.J.; Advisory Committee on Immunization Practices (ACIP). A comprehensive immunization strategy to eliminate transmission of hepatitis B virus infection in the United States: Recommendations of the Advisory Committee on Immunization Practices (ACIP) part 1: Immunization of infants, children, and adolescents. MMWR Recomm. Rep. 2005, 54, 1–31. [Google Scholar]
- Chan, Y.; Fornace, K.; Wu, L.; Arnold, B.F.; Priest, J.W.; Martin, D.L.; Chang, M.A.; Cook, J.; Stresman, G.; Drakeley, C. Determining seropositivity—A review of approaches to define population seroprevalence when using multiplex bead assays to assess burden of tropical diseases. PLoS Negl. Trop. Dis. 2021, 15, e0009457. [Google Scholar] [CrossRef]
- Plotkin, S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef] [Green Version]
- Basile, A.J.; Goodman, C.; Horiuchi, K.; Sloan, A.; Johnson, B.W.; Kosoy, O.; Laven, J.; Panella, A.J.; Sheets, I.; Medina, F.; et al. Multi-laboratory comparison of three commercially available Zika IgM enzyme-linked immunosorbent assays. J. Virol. Methods 2018, 260, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Huzly, D.; Hanselmann, I.; Schmidt-Chanasit, J.; Panning, M. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro. Surveill. 2016, 21, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Kikuti, M.; Cruz, J.S.; Rodrigues, M.; Tavares, A.S.; Paploski, I.; Silva, M.M.O.; Santana, P.M.; Tauro, L.B.; Silva, G.A.O.F.; Campos, G.S.; et al. Accuracy of the SD BIOLINE Dengue Duo for rapid point-of-care diagnosis of dengue. PLoS ONE 2019, 14, e0213301. [Google Scholar] [CrossRef] [PubMed]
- Kikuti, M.; Tauro, L.B.; Moreira, P.S.S.; Campos, G.S.; Paploski, I.; Weaver, S.C.; Reis, M.G.; Kitron, U.; Ribeiro, G.S. Diagnostic performance of commercial IgM and IgG enzyme-linked immunoassays (ELISAs) for diagnosis of Zika virus infection. Virol. J. 2018, 15, 108. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef]
- Hunsperger, E.A.; Yoksan, S.; Buchy, P.; Nguyen, V.C.; Sekaran, S.D.; Enria, D.A.; Vazquez, S.; Cartozian, E.; Pelegrino, J.L.; Artsob, H.; et al. Evaluation of commercially available diagnostic tests for the detection of dengue virus NS1 antigen and anti-dengue virus IgM antibody. PLoS Negl. Trop. Dis. 2014, 8, e3171. [Google Scholar] [CrossRef] [Green Version]
- Yadouleton, A.; Sander, A.-L.; Moreira-Soto, A.; Tchibozo, C.; Hounkanrin, G.; Badou, Y.; Fischer, C.; Krause, N.; Akogbeto, P.; de Oliveira Filho, E.F.; et al. Limited specificity of serologic tests for SARS-CoV-2 antibody detection, Benin. Emerg. Infect. Dis. 2021, 27, 233–237. [Google Scholar] [CrossRef]
- Azman, A.S.; Lessler, J.; Luquero, F.J.; Bhuiyan, T.R.; Khan, A.I.; Chowdhury, F.; Kabir, A.; Gurwith, M.; Weil, A.A.; Harris, J.B.; et al. Estimating cholera incidence with cross-sectional serology. Sci. Transl. Med. 2019, 11, eaau6242. [Google Scholar] [CrossRef] [Green Version]
- Teunis, P.; van Eijkeren, J. Seroincidence: Estimating Infection Rates from Serological Data. R Package 2018. Available online: https://cran.r-project.org/src/contrib/Archive/seroincidence/ (accessed on 10 June 2022).
- Teunis, P.F.; van Eijkeren, J.; Ang, C.W.; Van Duijnhoven, Y. Biomarker dynamics: Estimating infection rates from serological data. Stat. Med. 2012, 31, 2240–2248. [Google Scholar] [CrossRef]
- Biggs, J.R.; Sy, A.K.; Sherratt, K.; Brady, O.J.; Kucharski, A.J.; Funk, S.; Reyes, M.A.J.; Quinones, M.A.; Jones-Warner, W.; Avelino, F.L.; et al. Estimating the annual dengue force of infection from the age of reporting primary infections across urban centres in endemic countries. BMC Med. 2021, 19, 217. [Google Scholar] [CrossRef] [PubMed]
- Nealon, J.; Bouckenooghe, A.; Cortes, M.; Coudeville, L.; Frago, C.; Macina, D.; Tam, C.C. Dengue endemicity, force of infection and variation in transmission intensity in 13 endemic countries. J. Infect. Dis. 2022, 225, 75–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setoh, J.W.S.; Ho, C.; Yung, C.F.; Tam, C.; Yelen; Tee, N.W.S. Epstein-barr virus seroprevalence and force of infection in a multiethnic pediatric cohort, Singapore. Pediatr. Infect. Dis. J. 2019, 38, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.C.; Tissera, H.; De Silva, A.M.; De Silva, A.D.; Margolis, H.S.; Amarasinge, A. Estimates of dengue force of infection in children in Colombo, Sri Lanka. PLoS Negl. Trop. Dis. 2013, 7, e2259. [Google Scholar] [CrossRef] [Green Version]
- Bottomley, C.; Otiende, M.; Uyoga, S.; Gallagher, K.; Kagucia, E.W.; Etyang, A.O.; Mugo, D.; Gitonga, J.; Karanja, H.; Nyagwange, J.; et al. Quantifying previous SARS-CoV-2 infection through mixture modelling of antibody levels. Nat. Commun. 2021, 12, 6196. [Google Scholar] [CrossRef]
- Clapham, H.; Hay, J.; Routledge, I.; Takahashi, S.; Choisy, M.; Cummings, D.; Grenfell, B.; Metcalf, C.J.E.; Mina, M.; Barraquer, I.R.; et al. Seroepidemiologic study designs for determining SARS-COV-2 transmission and immunity. Emerg. Infect. Dis. 2020, 26, 1978–1986. [Google Scholar] [CrossRef]
- Arnold, K.B.; Chung, A.W. Prospects from systems serology research. Immunology 2018, 153, 279–289. [Google Scholar] [CrossRef]
- Tsai, W.Y.; Durbin, A.; Tsai, J.-J.; Hsieh, S.-C.; Whitehead, S.; Wang, W.-K. Complexity of neutralizing antibodies against multiple dengue virus serotypes after heterotypic immunization and secondary infection revealed by in-depth analysis of cross-reactive antibodies. J. Virol. 2015, 89, 7348–7362. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Bertoglio, F.; Steinke, S.; Heine, P.A.; Ynga-Durand, M.A.; Maass, H.; Sammartino, J.C.; Cassaniti, I.; Zuo, F.; Du, L.; et al. Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant. BMC Med. 2022, 20, 102. [Google Scholar] [CrossRef]
- Riepler, L.; Rössler, A.; Falch, A.; Volland, A.; Borena, W.; von Laer, D.; Kimpel, J. Comparison of four SARS-CoV-2 neutralization assays. Vaccines 2020, 9, 13. [Google Scholar] [CrossRef]
- Thomas, S.J.; Jarman, R.; Endy, T.P.; Kalayanarooj, S.; Vaughn, D.W.; Nisalak, A.; Putnak, R.; Anderson, K.B.; Gibbons, R.V.; Libraty, D.H. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: How alterations in assay conditions impact performance. Am. J. Trop. Med. Hyg. 2009, 81, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, E.D.G.; Bröker, M.; Wassil, J.; Welsch, J.; Borrow, R. Serum bactericidal antibody assays—The role of complement in infection and immunity. Vaccine 2015, 33, 4414–4421. [Google Scholar] [CrossRef] [PubMed]
- Jansen, W.T.; Väkeväinen-Anttila, M.; Käyhty, H.; Nahm, M.; Bakker, N.; Verhoef, J.; Snippe, H.; Verheul, A.F.M. Comparison of a classical phagocytosis assay and a flow cytometry assay for assessment of the phagocytic capacity of sera from adults vaccinated with a pneumococcal conjugate vaccine. Clin. Diagn. Lab. Immunol. 2001, 8, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Machado, B.A.S.; Hodel, K.V.S.; Barbosa-Júnior, V.G.; Soares, M.B.P.; Badaró, R. The Main Molecular and Serological Methods for Diagnosing COVID-19: An overview based on the literature. Viruses 2020, 13, 40. [Google Scholar] [CrossRef]
- Maple, P.A.; Gunn, A.; Sellwood, J.; Brown, D.W.; Gray, J.J. Comparison of fifteen commercial assays for detecting Varicella Zoster virus IgG with reference to a time resolved fluorescence immunoassay (TRFIA) and the performance of two commercial assays for screening sera from immunocompromised individuals. J. Virol. Methods 2009, 155, 143–149. [Google Scholar] [CrossRef]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef]
- Rosado, J.; Pelleau, S.; Cockram, C.; Merkling, S.H.; Nekkab, N.; Demeret, C.; Meola, A.; Kerneis, S.; Terrier, B.; Fafi-Kremer, S.; et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: An antibody-based diagnostic and machine learning study. Lancet Microbe 2021, 2, e60–e69. [Google Scholar] [CrossRef]
- Davies, D.H.; Liang, X.; Hernandez, J.E.; Randall, A.; Hirst, S.; Mu, Y.; Romero, K.M.; Nguyen, T.T.; Kalantari-Dehaghi, M.; Crotty, S.; et al. Profiling the humoral immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA 2005, 102, 547–552. [Google Scholar] [CrossRef] [Green Version]
- Bruckner, T.A.; Parker, D.M.; Bartell, S.M.; Vieira, V.M.; Khan, S.; Noymer, A.; Drum, E.; Albala, B.; Zahn, M.; Boden-Albala, B. Estimated seroprevalence of SARS-CoV-2 antibodies among adults in Orange County, California. Sci. Rep. 2021, 11, 3081. [Google Scholar] [CrossRef]
- Tiu, C.K.; Zhu, F.; Wang, L.-F.; de Alwis, R. Phage immunoprecipitation sequencing (PhIP-Seq): The promise of high throughput serology. Pathogens 2022, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Klompus, S.; Leviatan, S.; Kalka, I.N.; Weinberger, A.; Wijmenga, C.; Fu, J.; Zhernakova, A.; Weersma, R.K.; Segal, E. Population-wide diversity and stability of serum antibody epitope repertoires against human microbiota. Nat. Med. 2021, 27, 1442–1450. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Rahman, M.; Al Ali, F.; Huang, S.S.Y.; Ata, M.; Zhang, Q.; Bastard, P.; Liu, Z.; Jouanguy, E.; Beziat, V.; et al. Distinct antibody repertoires against endemic human coronaviruses in children and adults. JCI Insight 2021, 6, e144499. [Google Scholar] [CrossRef] [PubMed]
- Andeweg, S.P.; Schepp, R.M.; van de Kassteele, J.; Mollema, L.; Berbers, G.A.M.; van Boven, M. Population-based serology reveals risk factors for RSV infection in children younger than 5 years. Sci. Rep. 2021, 11, 8953. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, K.; Falconi-Agapito, F.; van Esbroeck, M.; Talledo, M.; Ariën, K.K. Reliable serological diagnostic tests for arboviruses: Feasible or utopia? Trends Microbiol. 2020, 28, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Russell, T.L.; Horwood, P.F.; Harrington, H.; Apairamo, A.; Kama, N.J.; Bobogare, A.; MacLaren, D.; Burkot, T.R. Seroprevalence of dengue, Zika, chikungunya and Ross River viruses across the Solomon Islands. PLoS Negl. Trop. Dis. 2022, 16, e0009848. [Google Scholar] [CrossRef]
- Bosch, Q.A.T.; Clapham, H.E.; Lambrechts, L.; Duong, V.; Buchy, P.; Althouse, B.M.; Lloyd, A.; Waller, L.A.; Morrison, A.C.; Kitron, U.; et al. Contributions from the silent majority dominate dengue virus transmission. PLoS Pathog. 2018, 14, e1006965. [Google Scholar]
- Vermund, S.H.; Pitzer, V.E. Asymptomatic transmission and the infection fatality risk for COVID-19: Implications for school reopening. Clin. Infect. Dis. 2021, 72, 1493–1496. [Google Scholar] [CrossRef]
- Plucinski, M.M.; Candrinho, B.; Chambe, G.; Muchanga, J.; Muguande, O.; Matsinhe, G.; Mathe, G.; Rogier, E.; Doyle, T.; Zulliger, R.; et al. Multiplex serology for impact evaluation of bed net distribution on burden of lymphatic filariasis and four species of human malaria in northern Mozambique. PLoS Negl. Trop. Dis. 2018, 12, e0006278. [Google Scholar] [CrossRef] [Green Version]
- Bousema, T.; Youssef, R.M.; Cook, J.; Cox, J.; Alegana, V.; Amran, J.; Noor, A.M.; Snow, R.; Drakeley, C. Serologic markers for detecting malaria in areas of low endemicity, Somalia, 2008. Emerg. Infect. Dis. 2010, 16, 392–399. [Google Scholar] [CrossRef]
- Li, H.; Mendelsohn, E.; Zong, C.; Zhang, W.; Hagan, E.; Wang, N.; Li, S.; Yan, H.; Huang, H.; Zhu, G.; et al. Human-animal interactions and bat coronavirus spillover potential among rural residents in Southern China. Biosaf. Health 2019, 1, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Daszak, P.; Olival, K.J.; Li, H. A strategy to prevent future epidemics similar to the 2019-nCoV outbreak. Biosaf. Health 2020, 2, 6–8. [Google Scholar] [CrossRef]
- Arora, R.K.; Joseph, A.; van Wyk, J.; Rocco, S.; Atmaja, A.; May, E.; Yan, T.; Bobrovitz, N.; Chevrier, J.; Cheng, M.P.; et al. SeroTracker: A global SARS-CoV-2 seroprevalence dashboard. Lancet Infect. Dis. 2021, 21, e75–e76. [Google Scholar] [CrossRef]
- Halili, R.; Bunjaku, J.; Gashi, B.; Hoxha, T.; Kamberi, A.; Hoti, N.; Agahi, R.; Basha, V.; Berisha, V.; Hoxha, I. Seroprevalence of anti-SARS-CoV-2 antibodies among staff at primary healthcare institutions in Prishtina. BMC Infect. Dis. 2022, 22, 57. [Google Scholar] [CrossRef]
- Krsak, M.; Johnson, S.C.; Poeschla, E.M. COVID-19 serosurveillance may facilitate return-to-work decisions. Am. J. Trop. Med. Hyg. 2020, 102, 1189–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Population-Based Age-Stratified Seroepidemiological Investigation Protocol for Coronavirus 2019 (COVID-19) Infection; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]

| Case-Based | Serology-Based | |
|---|---|---|
| Cases detected | Current/ongoing Symptomatic (mainly) | Past Symptomatic/asymptomatic |
| Health system presentation | Necessary (*) | Not necessary |
| Time window for detection | Short (days) | Medium-Long (month-years) |
| Possible on stored samples | Rarely | Yes |
| Possibility of multiplexing | Yes | Yes |
| Challenges to implementation in low resource settings | Healthcare access barriers Limitations in use of confirmatory diagnostics | Financial and infrastructure constraints depending on test chosen |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haselbeck, A.H.; Im, J.; Prifti, K.; Marks, F.; Holm, M.; Zellweger, R.M. Serology as a Tool to Assess Infectious Disease Landscapes and Guide Public Health Policy. Pathogens 2022, 11, 732. https://doi.org/10.3390/pathogens11070732
Haselbeck AH, Im J, Prifti K, Marks F, Holm M, Zellweger RM. Serology as a Tool to Assess Infectious Disease Landscapes and Guide Public Health Policy. Pathogens. 2022; 11(7):732. https://doi.org/10.3390/pathogens11070732
Chicago/Turabian StyleHaselbeck, Andrea H., Justin Im, Kristi Prifti, Florian Marks, Marianne Holm, and Raphaël M. Zellweger. 2022. "Serology as a Tool to Assess Infectious Disease Landscapes and Guide Public Health Policy" Pathogens 11, no. 7: 732. https://doi.org/10.3390/pathogens11070732
APA StyleHaselbeck, A. H., Im, J., Prifti, K., Marks, F., Holm, M., & Zellweger, R. M. (2022). Serology as a Tool to Assess Infectious Disease Landscapes and Guide Public Health Policy. Pathogens, 11(7), 732. https://doi.org/10.3390/pathogens11070732






