Protein Levels of Pro-Inflammatory Cytokines and Chemokines as Biomarkers of Mycobacterium bovis Infection and BCG Vaccination in Cattle
Abstract
1. Introduction
2. Results
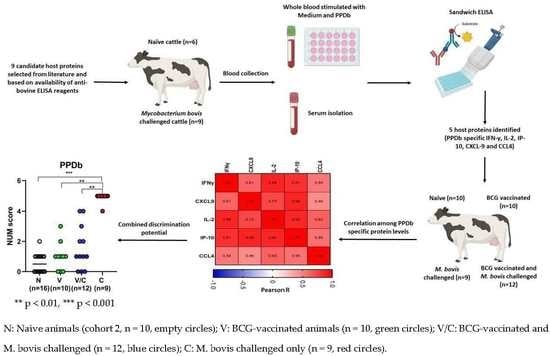
2.1. Cytokine and Chemokines Levels in Naive and M. bovis Challenged Animals
2.2. Comparison of Cytokine and Chemokine Levels between Naive, BCG-Vaccinated, BCG-Vaccinated-M. bovis Challenged and Animals Challenged with M. bovis Alone
2.3. Performance of Host Proteins to Discriminate between Naive and M. bovis Challenged Animals
2.4. Multibiomarker Analysis and Correlation between Cytokine and Chemokine Levels to Accurately Discriminate between Groups of Animals
2.5. Correlations of Host Protein Concentrations with Bacteriology Counts and Total Lesion Scores
3. Discussion
4. Materials and Methods
4.1. Selection of Candidate Biomarkers
4.2. Samples and Animals
Preparation of Samples for Analysis
4.3. ELISA Protocol
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, R.S.; Sweeney, S.J. Mycobacterium bovis (bovine tuberculosis) infection in North American wildlife: Current status and opportunities for mitigation of risks of further infection in wildlife populations. Epidemiol. Infect. 2013, 141, 1357–1370. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.V.; Waters, W.R. Advances in bovine tuberculosis diagnosis and pathogenesis: What policy makers need to know. Vet. Microbiol. 2006, 112, 181–190. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schiller, I.; Oesch, B.; Vordermeier, H.; Palmer, M.; Harris, B.; Orloski, K.; Buddle, B.; Thacker, T.; Lyashchenko, K.; Waters, W. Bovine tuberculosis: A review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound. Emerg. Dis. 2010, 57, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Olea-Popelka, F.; Muwonge, A.; Perera, A.; Dean, A.S.; Mumford, E.; Erlacher-Vindel, E.; Forcella, S.; Silk, B.J.; Ditiu, L.; El Idrissi, A. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—A call for action. Lancet Infect. Dis. 2017, 17, e21–e25. [Google Scholar] [CrossRef]
- Palmer, M.V.; Thacker, T.C.; Waters, W.R. Differential cytokine gene expression in granulomas from lungs and lymph nodes of cattle experimentally infected with aerosolized Mycobacterium bovis. PLoS ONE 2016, 11, e0167471. [Google Scholar] [CrossRef]
- Widdison, S.; Schreuder, L.; Villarreal-Ramos, B.; Howard, C.; Watson, M.; Coffey, T. Cytokine expression profiles of bovine lymph nodes: Effects of Mycobacterium bovis infection and bacille Calmette–Guérin vaccination. Clin. Exp. Immunol. 2006, 144, 281–289. [Google Scholar] [CrossRef]
- Waters, W.R.; Maggioli, M.F.; McGill, J.L.; Lyashchenko, K.P.; Palmer, M.V. Relevance of bovine tuberculosis research to the understanding of human disease: Historical perspectives, approaches, and immunologic mechanisms. Vet. Immunol. Immunopathol. 2014, 159, 113–132. [Google Scholar] [CrossRef]
- De la Rua-Domenech, R.; Goodchild, A.; Vordermeier, H.; Hewinson, R.; Christiansen, K.; Clifton-Hadley, R. Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, γ-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef]
- APHA. Skin Test Day Two. Available online: http://apha.defra.gov.uk/External_OV_Instructions/TB_Instructions/Skin_Test/Skin_Test_Day_Two.html (accessed on 17 April 2022).
- Wood, P.; Corner, L.; Plackett, P. Development of a simple, rapid in vitro cellular assay for bovine tuberculosis based on the production of γ interferon. Res. Vet. Sci. 1990, 49, 46–49. [Google Scholar] [CrossRef]
- Gormley, E.; Doyle, M.; McGill, K.; Costello, E.; Good, M.; Collins, J. The effect of the tuberculin test and the consequences of a delay in blood culture on the sensitivity of a gamma-interferon assay for the detection of Mycobacterium bovis infection in cattle. Vet. Immunol. Immunopathol. 2004, 102, 413–420. [Google Scholar] [CrossRef]
- Coad, M.; Clifford, D.; Vordermeier, H.; Whelan, A. The consequences of vaccination with the Johne’s disease vaccine, Gudair, on diagnosis of bovine tuberculosis. Vet. Rec. 2013, 172, 266. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.J.; Mannion, C.; Golden, O.; Hacariz, O.; Mulcahy, G. Experimental Fasciola hepatica infection alters responses to tests used for diagnosis of bovine tuberculosis. Infect. Immun. 2007, 75, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, M.; Doherty, M.; Collins, J.; Kazda, J.; Quinn, P. The tuberculin test. Vet. Microbiol. 1994, 40, 111–124. [Google Scholar] [CrossRef]
- Buddle, B.M.; Vordermeier, H.M.; Chambers, M.A.; de Klerk-Lorist, L.-M. Efficacy and safety of BCG vaccine for control of tuberculosis in domestic livestock and wildlife. Front. Vet. Sci. 2018, 5, 259. [Google Scholar] [CrossRef]
- Garnier, T.; Eiglmeier, K.; Camus, J.-C.; Medina, N.; Mansoor, H.; Pryor, M.; Duthoy, S.; Grondin, S.; Lacroix, C.; Monsempe, C. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 2003, 100, 7877–7882. [Google Scholar] [CrossRef]
- Bayissa, B.; Sirak, A.; Zewude, A.; Worku, A.; Gumi, B.; Berg, S.; Hewinson, R.G.; Wood, J.L.; Jones, G.J.; Consortium, E. Field evaluation of specific mycobacterial protein-based skin test for the differentiation of Mycobacterium bovis-infected and Bacillus Calmette Guerin-vaccinated crossbred cattle in Ethiopia. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Srinivasan, S.; Subramanian, S.; Shankar Balakrishnan, S.; Ramaiyan Selvaraju, K.; Manomohan, V.; Selladurai, S.; Jothivelu, M.; Kandasamy, S.; Gopal, D.R.; Kathaperumal, K. A defined antigen skin test that enables implementation of BCG vaccination for control of bovine tuberculosis: Proof of concept. Front. Vet. Sci. 2020, 7, 391. [Google Scholar] [CrossRef]
- Sutherland, J.S.; Mendy, J.; Gindeh, A.; Walzl, G.; Togun, T.; Owolabi, O.; Donkor, S.; Ota, M.O.; Fat, E.T.K.; Ottenhoff, T.H. Use of lateral flow assays to determine IP-10 and CCL4 levels in pleural effusions and whole blood for TB diagnosis. Tuberculosis 2016, 96, 31–36. [Google Scholar] [CrossRef]
- van Hooij, A.; van den Eeden, S.; Richardus, R.; Fat, E.T.K.; Wilson, L.; Franken, K.L.; Faber, R.; Khatun, M.; Alam, K.; Chowdhury, A.S. Application of new host biomarker profiles in quantitative point-of-care tests facilitates leprosy diagnosis in the field. EBioMedicine 2019, 47, 301–308. [Google Scholar] [CrossRef]
- Van Hooij, A.; Fat, E.M.T.K.; de Jong, D.; Khatun, M.; Soren, S.; Chowdhury, A.S.; Roy, J.C.; Alam, K.; Kim, J.-P.; Richardus, J.H. Prototype multi-biomarker test for point-of-care leprosy diagnostics. Iscience 2021, 24, 102006. [Google Scholar] [CrossRef]
- Jones, G.J.; Pirson, C.; Hewinson, R.G.; Vordermeier, H.M. Simultaneous measurement of antigen-stimulated interleukin-1β and gamma interferon production enhances test sensitivity for the detection of Mycobacterium bovis infection in cattle. Clin. Vaccine Immunol. 2010, 17, 1946–1951. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coad, M.; Doyle, M.; Steinbach, S.; Gormley, E.; Vordermeier, M.; Jones, G. Simultaneous measurement of antigen-induced CXCL10 and IFN-γ enhances test sensitivity for bovine TB detection in cattle. Vet. Microbiol. 2019, 230, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.H. Revisiting tuberculosis screening: An insight to complementary diagnosis and prospective molecular approaches for the recognition of the dormant TB infection in human and cattle hosts. Microbiol. Res. 2021, 252, 126853. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.; Plain, K.; Purdie, A.; Saunders, B.M.; de Silva, K. Biomarkers for Detecting Resilience against Mycobacterial Disease in Animals. Infect. Immun. 2019, 88, e00401-19. [Google Scholar] [CrossRef]
- Elnaggar, M.M.; Abdellrazeq, G.S.; Elsisy, A.; Mahmoud, A.H.; Shyboub, A.; Sester, M.; Khaliel, S.A.; Singh, M.; Torky, H.A.; Davis, W.C. Evaluation of antigen specific interleukin-1β as a biomarker to detect cattle infected with Mycobacterium bovis. Tuberculosis 2017, 105, 53–59. [Google Scholar] [CrossRef]
- Rhodes, S.G.; McKinna, L.C.; Steinbach, S.; Dean, G.S.; Villarreal-Ramos, B.; Whelan, A.O.; Pirson, C.; Jones, G.J.; Clifford, D.; Vordermeier, H.M. Use of antigen-specific interleukin-2 to differentiate between cattle vaccinated with Mycobacterium bovis BCG and cattle infected with M. bovis. Clin. Vaccine Immunol. 2014, 21, 39–45. [Google Scholar] [CrossRef]
- Thacker, T.C.; Palmer, M.V.; Waters, W.R. Associations between cytokine gene expression and pathology in Mycobacterium bovis infected cattle. Vet. Immunol. Immunopathol. 2007, 119, 204–213. [Google Scholar] [CrossRef]
- Blanco, F.C.; Schierloh, P.; Bianco, M.V.; Caimi, K.; Meikle, V.; Alito, A.E.; Cataldi, A.A.; Sasiain, M.d.C.; Bigi, F. Study of the immunological profile towards Mycobacterium bovis antigens in naturally infected cattle. Microbiol. Immunol. 2009, 53, 460–467. [Google Scholar] [CrossRef]
- Widdison, S.; Watson, M.; Coffey, T.J. Correlation between lymph node pathology and chemokine expression during bovine tuberculosis. Tuberculosis 2009, 89, 417–422. [Google Scholar] [CrossRef]
- Palmer, M.; Thacker, T.; Waters, W. Analysis of cytokine gene expression using a novel chromogenic in-situ hybridization method in pulmonary granulomas of cattle infected experimentally by aerosolized Mycobacterium bovis. J. Comp. Pathol. 2015, 153, 150–159. [Google Scholar] [CrossRef]
- Welsh, M.D.; Cunningham, R.T.; Corbett, D.M.; Girvin, R.M.; McNair, J.; Skuce, R.A.; Bryson, D.G.; Pollock, J.M. Influence of pathological progression on the balance between cellular and humoral immune responses in bovine tuberculosis. Immunology 2005, 114, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, S.; Vordermeier, H.M.; Jones, G.J. CD4+ and γδ T Cells are the main Producers of IL-22 and IL-17A in Lymphocytes from Mycobacterium bovis-infected Cattle. Sci. Rep. 2016, 6, 1–10. [Google Scholar]
- Blanco, F.C.; Bianco, M.V.; Meikle, V.; Garbaccio, S.; Vagnoni, L.; Forrellad, M.; Klepp, L.I.; Cataldi, A.A.; Bigi, F. Increased IL-17 expression is associated with pathology in a bovine model of tuberculosis. Tuberculosis 2011, 91, 57–63. [Google Scholar] [CrossRef]
- Aranday-Cortes, E.; Hogarth, P.J.; Kaveh, D.A.; Whelan, A.O.; Villarreal-Ramos, B.; Lalvani, A.; Vordermeier, H.M. Transcriptional profiling of disease-induced host responses in bovine tuberculosis and the identification of potential diagnostic biomarkers. PLoS ONE 2012, 7, e30626. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Waters, W.; Palmer, M.; Whipple, D.; Carlson, M.; Nonnecke, B. Diagnostic implications of antigen-induced gamma interferon, nitric oxide, and tumor necrosis factor alpha production by peripheral blood mononuclear cells from Mycobacterium bovis-infected cattle. Clin. Vaccine Immunol. 2003, 10, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Aranday-Cortes, E.; Bull, N.; Villarreal-Ramos, B.; Gough, J.; Hicks, D.; Ortiz-Peláez, A.; Vordermeier, H.; Salguero, F. Upregulation of IL-17 A, CXCL 9 and CXCL 10 in Early-Stage Granulomas Induced by M ycobacterium bovis in Cattle. Transbound. Emerg. Dis. 2013, 60, 525–537. [Google Scholar] [CrossRef]
- Klepp, L.I.; Eirin, M.E.; Garbaccio, S.; Soria, M.; Bigi, F.; Blanco, F.C. Identification of bovine tuberculosis biomarkers to detect tuberculin skin test and IFNγ release assay false negative cattle. Res. Vet. Sci. 2019, 122, 7–14. [Google Scholar] [CrossRef]
- Parsons, S.D.; McGill, K.; Doyle, M.B.; Goosen, W.J.; Van Helden, P.D.; Gormley, E. Antigen-specific IP-10 release is a sensitive biomarker of Mycobacterium bovis infection in cattle. PLoS ONE 2016, 11, e0155440. [Google Scholar] [CrossRef]
- Chegou, N.N.; Sutherland, J.S.; Malherbe, S.; Crampin, A.C.; Corstjens, P.L.A.M.; Geluk, A.; Mayanja-Kizza, H.; Loxton, A.G.; van der Spuy, G.; Stanley, K.; et al. Diagnostic performance of a seven-marker serum protein biosignature for the diagnosis of active TB disease in African primary healthcare clinic attendees with signs and symptoms suggestive of TB. Thorax 2016, 71, 785–794. [Google Scholar] [CrossRef]
- Hope, J.; Thom, M.; McAulay, M.; Mead, E.; Vordermeier, H.; Clifford, D.; Hewinson, R.; Villarreal-Ramos, B. Identification of surrogates and correlates of protection in protective immunity against Mycobacterium bovis infection induced in neonatal calves by vaccination with M. bovis BCG Pasteur and M. bovis BCG Danish. Clin. Vaccine Immunol. 2011, 18, 373–379. [Google Scholar] [CrossRef]
- Gao, X.; Guo, X.; Li, M.; Jia, H.; Lin, W.; Fang, L.; Jiang, Y.; Zhu, H.; Zhang, Z.; Ding, J. Interleukin 8 and pentaxin (C-reactive protein) as potential new biomarkers of bovine tuberculosis. J. Clin. Microbiol. 2019, 57, e00274-19. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.V.; Thacker, T.C.; Rabideau, M.M.; Jones, G.J.; Kanipe, C.; Vordermeier, H.M.; Waters, W.R. Biomarkers of cell-mediated immunity to bovine tuberculosis. Vet. Immunol. Immunopathol. 2020, 220, 109988. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, S.; Jalili-Firoozinezhad, S.; Srinivasan, S.; Melo, M.B.; Middleton, S.; Konold, T.; Coad, M.; Hammond, P.T.; Irvine, D.J.; Vordermeier, M. Temporal dynamics of intradermal cytokine response to tuberculin in Mycobacterium bovis BCG-vaccinated cattle using sampling microneedles. Sci. Rep. 2021, 11, 1–9. [Google Scholar]
- Bernitz, N.; Kerr, T.J.; Goosen, W.J.; Clarke, C.; Higgitt, R.; Roos, E.O.; Cooper, D.V.; Warren, R.M.; van Helden, P.D.; Parsons, S.D. Parallel measurement of IFN-γ and IP-10 in QuantiFERON®-TB Gold (QFT) plasma improves the detection of Mycobacterium bovis infection in African buffaloes (Syncerus caffer). Prev. Vet. Med. 2019, 169, 104700. [Google Scholar] [CrossRef] [PubMed]
- O’Halloran, C.; McCulloch, L.; Rentoul, L.; Alexander, J.; Hope, J.C.; Gunn-Moore, D.A. Cytokine and Chemokine Concentrations as Biomarkers of Feline Mycobacteriosis. Sci. Rep. 2018, 8, 17314. [Google Scholar] [CrossRef] [PubMed]
- Roupie, V.; Alonso-Velasco, E.; Van Der Heyden, S.; Holbert, S.; Duytschaever, L.; Berthon, P.; Van Dosselaer, I.; Van Campe, W.; Mostin, L.; Biet, F. Evaluation of mycobacteria-specific gamma interferon and antibody responses before and after a single intradermal skin test in cattle naturally exposed to M. avium subsp. paratuberculosis and experimentally infected with M. bovis. Vet. Immunol. Immunopathol. 2018, 196, 35–47. [Google Scholar] [CrossRef]
- Biet, F.; Boschiroli, M.L. Non-tuberculous mycobacterial infections of veterinary relevance. Res. Vet. Sci. 2014, 97, S69–S77. [Google Scholar] [CrossRef]
- Mazorra-Carrillo, J.L.; Alcaraz-López, O.A.; López-Rincón, G.; Villarreal-Ramos, B.; Gutiérrez-Pabello, J.A.; Esquivel-Solís, H. Host Serum Proteins as Potential Biomarkers of Bovine Tuberculosis Resistance Phenotype. Front. Vet. Sci. 2021, 8, 734087. [Google Scholar] [CrossRef]
- Corstjens, P.; Van Hooij, A.; Fat, E.T.K.; Herdigein, S.; Namuganga, A.R.; Diergaardt, A.; Mutavhatsindi, H.; Gindeh, A.; Mihret, A.; Van De Spuy, G. OC 8435 multi-biomarker test strip for point-of-care screening for active tuberculosis: A five-country multi-centre test evaluation. BMJ Glob. Health 2019, 4, A6. [Google Scholar] [CrossRef]
- Vordermeier, H.M.; Villarreal-Ramos, B.; Cockle, P.J.; McAulay, M.; Rhodes, S.G.; Thacker, T.; Gilbert, S.C.; McShane, H.; Hill, A.V.; Xing, Z. Viral booster vaccines improve Mycobacterium bovis BCG-induced protection against bovine tuberculosis. Infect. Immun. 2009, 77, 3364–3373. [Google Scholar] [CrossRef]
- Dean, G.S.; Rhodes, S.G.; Coad, M.; Whelan, A.O.; Cockle, P.J.; Clifford, D.J.; Hewinson, R.G.; Vordermeier, H.M. Minimum infective dose of Mycobacterium bovis in cattle. Infect. Immun. 2005, 73, 6467–6471. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abcam. Available online: https://www.abcam.com/kits/elisa-terms-glossary (accessed on 28 April 2022).
- Fluss, R.; Faraggi, D.; Reiser, B. Estimation of the Youden Index and its associated cutoff point. Biom. J. J. Math. Methods Biosci. 2005, 47, 458–472. [Google Scholar] [CrossRef] [PubMed]







| (a) Analysis by comparing levels in bovine tuberculin (PPDb)-stimulated whole blood supernatants | |||||
| Analyte | Cut-off | Sensitivity (95% CI) | Specificity (95% CI) | AUC-ROC | p-value |
| IFNγ | >1943 | 100% (70.09 to 100.0) | 100% (80.64 to 100.0) | 1 | <0.001 |
| IP-10 | >25,846 | 100% (70.09 to 100.0) | 94% (71.67 to 99.68) | 0.9931 | <0.001 |
| IL-2 | >2666 | 100% (70.09 to 100.0) | 100% (80.64 to 100.0) | 1 | <0.001 |
| CXCL9 | >1651 | 100% (70.09 to 100.0) | 81% (56.99 to 93.41) | 0.9167 | <0.001 |
| CCL4 | >21,840 | 89% (56.50 to 99.43) | 69% (44.40 to 85.84) | 0.84 | 0.006 |
| (b) Analysis by comparing serum concentrations | |||||
| Analyte | Cut-off | Sensitivity (95% CI) | Specificity | AUC-ROC | p-value |
| IFNγ | >76.00 | 44% (18.88 to 73.33) | 100% (80.64 to 100.0) | 0.7 | 0.09 |
| IP-10 | >4448 | 100% (70.09 to 100.0) | 50% (28.0 to 72.0) | 0.72 | 0.07 |
| IL-2 | >170.1 | 100% (70.09 to 100.0) | 63% (38.64 to 81.52) | 0.81 | 0.01 |
| CXCL9 | >6377 | 78% (45.26 to 96.05) | 100% (80.64 to 100.0) | 0.96 | <0.001 |
| CCL4 | >15.00 | 100% (70.09 to 100.0) | 63% (38.64 to 81.52) | 0.88 | 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, H.; van Hooij, A.; Connelley, T.K.; Geluk, A.; Hope, J.C. Protein Levels of Pro-Inflammatory Cytokines and Chemokines as Biomarkers of Mycobacterium bovis Infection and BCG Vaccination in Cattle. Pathogens 2022, 11, 738. https://doi.org/10.3390/pathogens11070738
Khalid H, van Hooij A, Connelley TK, Geluk A, Hope JC. Protein Levels of Pro-Inflammatory Cytokines and Chemokines as Biomarkers of Mycobacterium bovis Infection and BCG Vaccination in Cattle. Pathogens. 2022; 11(7):738. https://doi.org/10.3390/pathogens11070738
Chicago/Turabian StyleKhalid, Hamza, Anouk van Hooij, Timothy K. Connelley, Annemieke Geluk, and Jayne C. Hope. 2022. "Protein Levels of Pro-Inflammatory Cytokines and Chemokines as Biomarkers of Mycobacterium bovis Infection and BCG Vaccination in Cattle" Pathogens 11, no. 7: 738. https://doi.org/10.3390/pathogens11070738
APA StyleKhalid, H., van Hooij, A., Connelley, T. K., Geluk, A., & Hope, J. C. (2022). Protein Levels of Pro-Inflammatory Cytokines and Chemokines as Biomarkers of Mycobacterium bovis Infection and BCG Vaccination in Cattle. Pathogens, 11(7), 738. https://doi.org/10.3390/pathogens11070738