Enolase Inhibitors as Early Lead Therapeutics against Trypanosoma brucei
Abstract
:1. Introduction
2. Materials and Methods
3. Results
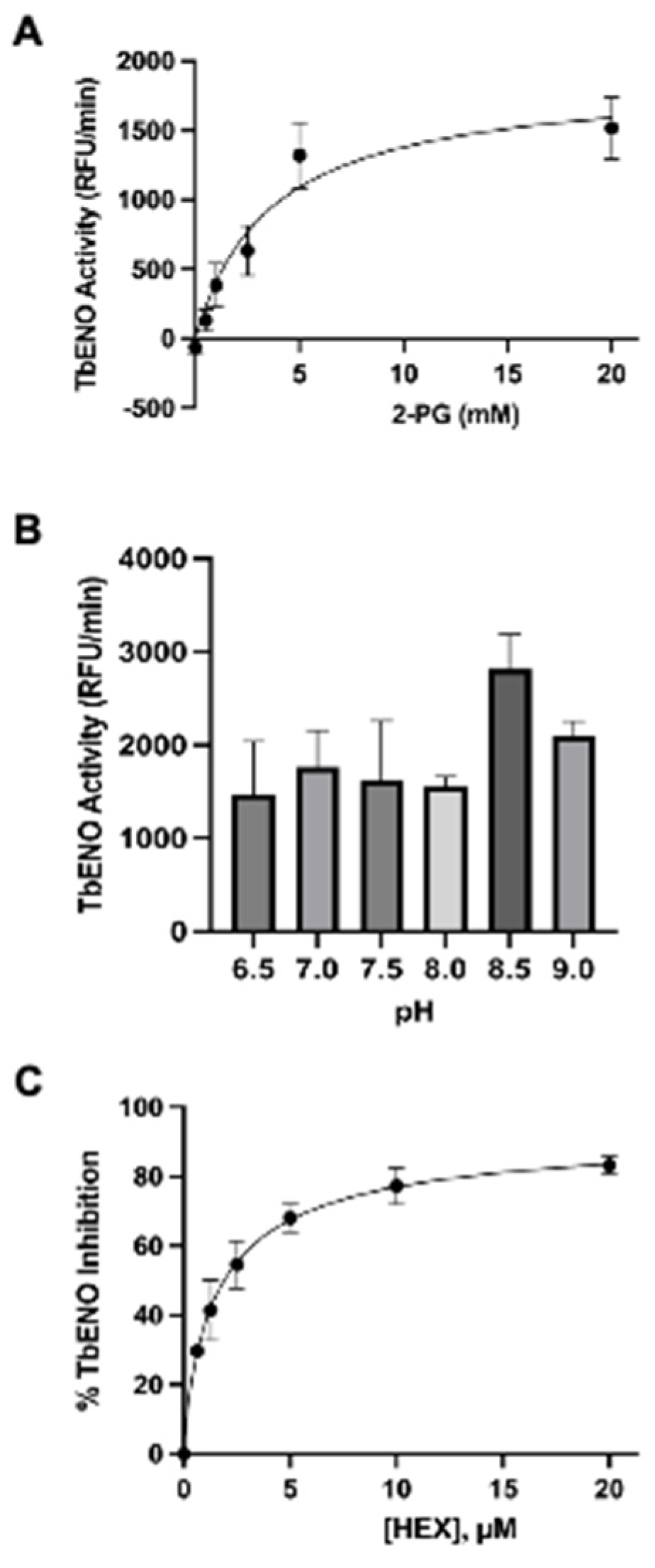
3.1. The Phosphonohydroxamates Inhibit TbENO In Vitro
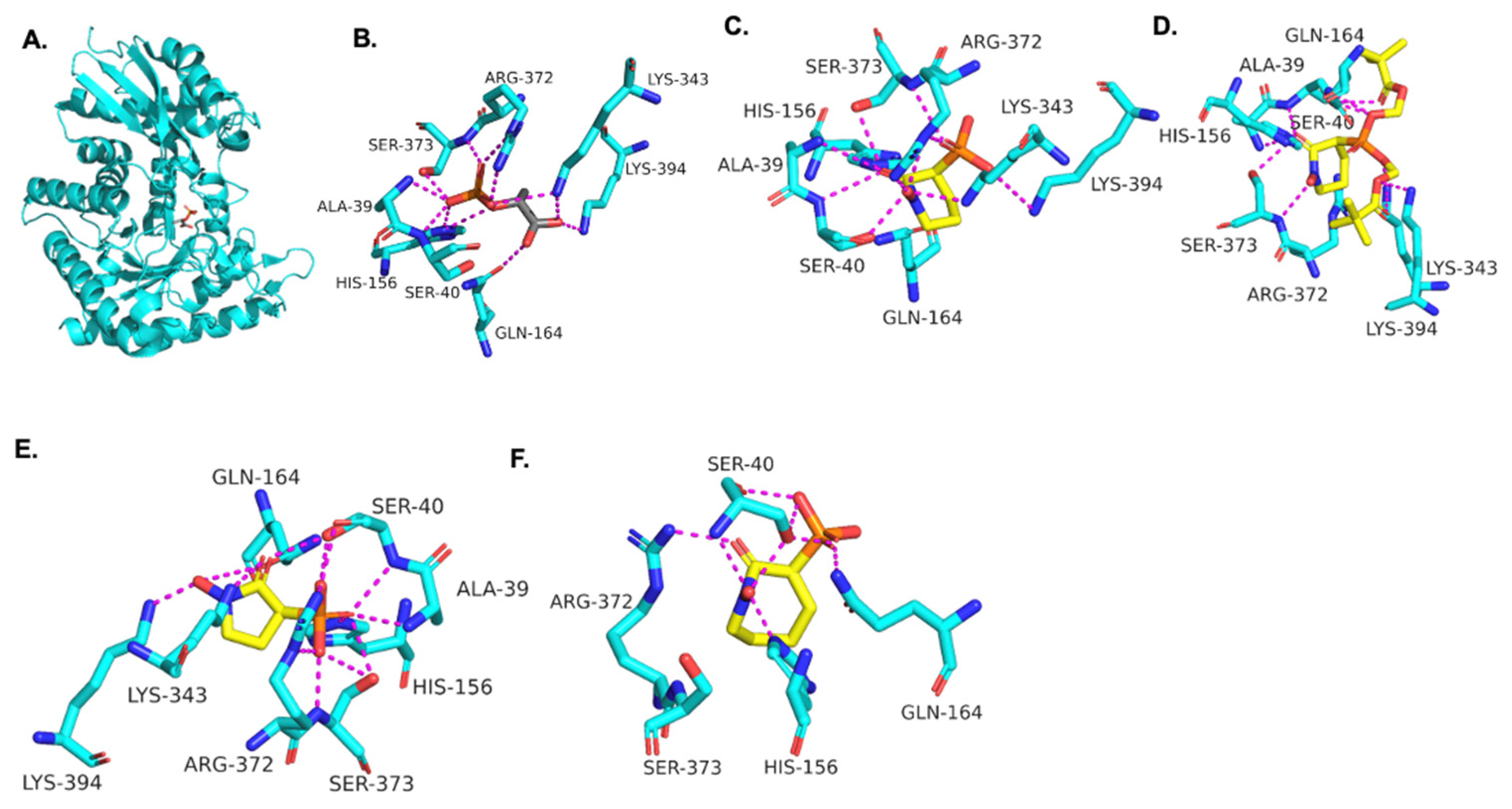
3.2. Molecular Modeling of HEX Compounds Bound to TbENO
3.3. The Phosphonohydroxamates Are Potent Anti-Trypanosomal Compounds
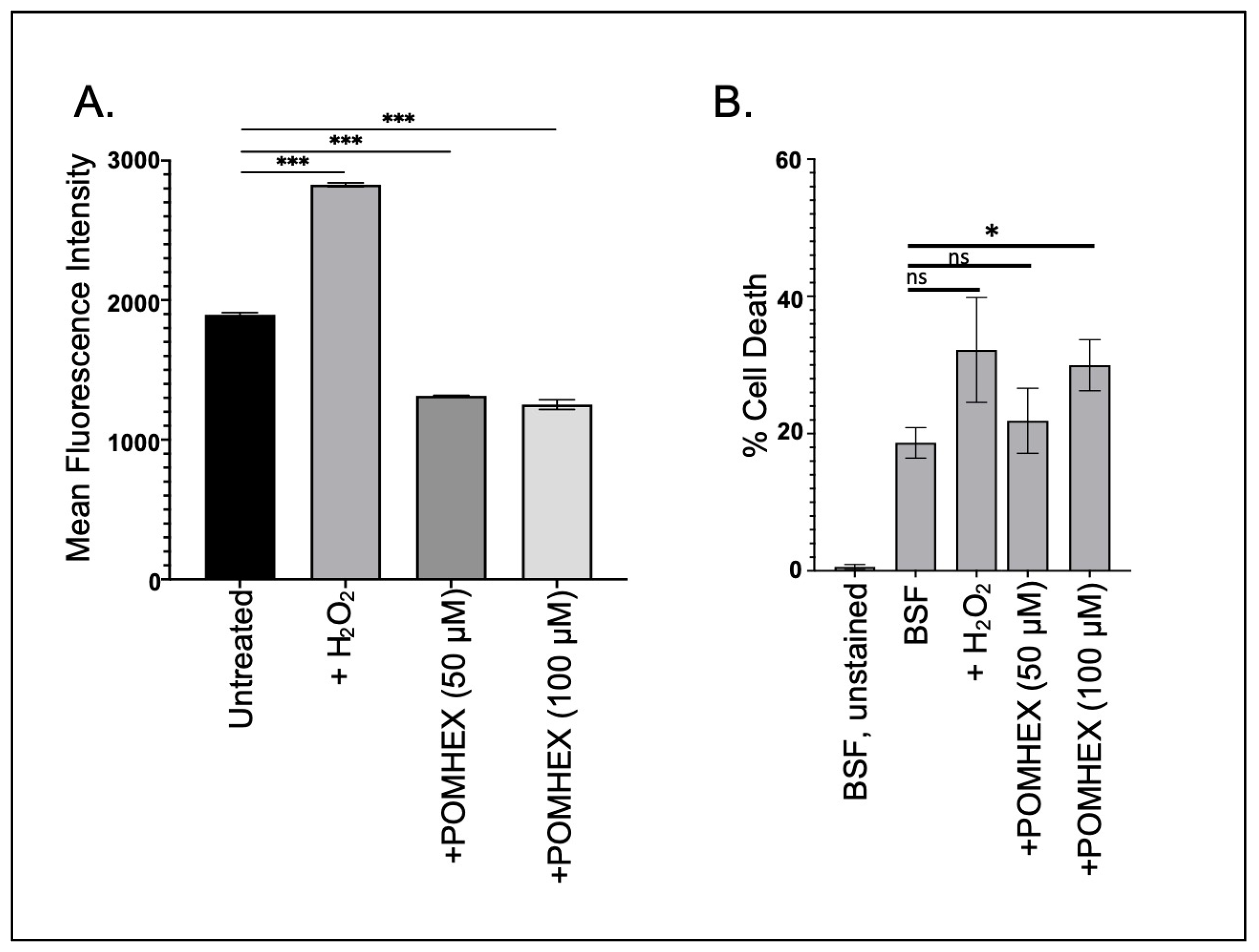
3.4. TbENO Inhibitor Toxicity Is Not Associated with Increased Cellular ROS Levels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharlow, E.; Golden, J.E.; Dodson, H.; Morris, M.; Hesser, M.; Lyda, T.; Leimgruber, S.; Schroeder, C.E.; Flaherty, D.P.; Weiner, W.S.; et al. Identification of Inhibitors of Trypanosoma brucei Hexokinases. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. [Google Scholar]
- Flaherty, D.P.; Harris, M.T.; Schroeder, C.E.; Khan, H.; Kahney, E.W.; Hackler, A.L.; Patrick, S.L.; Weiner, W.S.; Aubé, J.; Sharlow, E.R.; et al. Optimization and Evaluation of Antiparasitic Benzamidobenzoic Acids as Inhibitors of Kinetoplastid Hexokinase 1. ChemMedChem 2017, 12, 1994–2005. [Google Scholar] [CrossRef]
- McNae, I.W.; Kinkead, J.; Malik, D.; Yen, L.-H.; Walker, M.K.; Swain, C.; Webster, S.P.; Gray, N.; Fernandes, P.M.; Myburgh, E.; et al. Fast acting allosteric phosphofructokinase inhibitors block trypanosome glycolysis and cure acute African trypanosomiasis in mice. Nat. Commun. 2021, 12, 1052. [Google Scholar] [CrossRef]
- Albert, M.-A.; Haanstra, J.R.; Hannaert, V.; Van Roy, J.; Opperdoes, F.R.; Bakker, B.M.; Michels, P.A.M. Experimental and in Silico Analyses of Glycolytic Flux Control in Bloodstream Form Trypanosoma brucei. J. Biol. Chem. 2005, 280, 28306–28315. [Google Scholar] [CrossRef] [PubMed]
- Avilán, L.; Gualdrón-López, M.; Quiñones, W.; González-González, L.; Hannaert, V.; Michels, P.A.M.; Concepción, J.-L. Enolase: A Key Player in the Metabolism and a Probable Virulence Factor of Trypanosomatid Parasites—Perspectives for Its Use as a Therapeutic Target. Enzym. Res. 2011, 2011, 932549. [Google Scholar] [CrossRef]
- Hannaert, V.; Albert, M.-A.; Rigden, D.J.; Giotto, M.T.d.S.; Thiemann, O.; Garratt, R.C.; Van Roy, J.; Opperdoes, F.R.; Michels, P.A.M. Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase. Eur. J. Biochem. 2003, 270, 3205–3213. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Satani, N.; Hammoudi, N.; Yan, V.C.; Barekatain, Y.; Khadka, S.; Ackroyd, J.J.; Georgiou, D.K.; Pham, C.-D.; Arthur, K.; et al. An enolase inhibitor for the targeted treatment of ENO1-deleted cancers. Nat. Metab. 2020, 2, 1413–1426. [Google Scholar] [CrossRef]
- Muller, F.L.; Colla, S.; Aquilanti, E.; Manzo, V.E.; Genovese, G.; Lee, J.; Eisenson, D.; Narurkar, R.; Deng, P.; Nezi, L.; et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 2012, 488, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Leonard, P.G.; Satani, N.; Maxwell, D.; Lin, Y.-H.; Hammoudi, N.; Peng, Z.; Pisaneschi, F.; Link, T.M.; Lee, G.R.; Sun, D.; et al. SF2312 is a natural phosphonate inhibitor of enolase. Nat. Chem. Biol. 2016, 12, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Sharlow, E.R.; Lyda, T.A.; Dodson, H.C.; Mustata, G.; Morris, M.T.; Leimgruber, S.S.; Lee, K.-H.; Kashiwada, Y.; Close, D.; Lazo, J.S.; et al. A Target-Based High Throughput Screen Yields Trypanosoma brucei Hexokinase Small Molecule Inhibitors with Antiparasitic Activity. PLOS Neglected Trop. Dis. 2010, 4, e659. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision A.02; Expanding the limits of computational chemistry: Wallingford CT, USA, 2016. [Google Scholar]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Kwain, S.; Dominy, B.N.; Whitehead, K.J.; Miller, B.A.; Whitehead, D.C. Exploring the interactive mechanism of acarbose with the amylase SusG in the starch utilization system of the human gut symbiont Bacteroides thetaiotaomicron through molecular modeling. Chem. Biol. Drug Des. 2023, 102, 486–499. [Google Scholar] [CrossRef]
- Huang, J.H.; He, C.; Wu, L.; Tong, H. Thermal degradation reaction mechanism of xylose: A DFT study. Chem. Phys. Lett. 2016, 658, 114–124. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Hirumi, H.; Hirumi, K. Continuous Cultivation of Trypanosoma brucei Blood Stream Forms in a Medium Containing a Low Concentration of Serum Protein without Feeder Cell Layers. J. Parasitol. 1989, 75, 985–989. [Google Scholar] [CrossRef]
- Reed, G.H.; Poyner, R.R.; Larsen, T.M.; E Wedekind, J.; Rayment, I. Structural and mechanistic studies of enolase. Curr. Opin. Struct. Biol. 1996, 6, 736–743. [Google Scholar] [CrossRef]
- Zhang, E.; Brewer, J.M.; Minor, W.; Carreira, L.A.; Lebioda, L. Mechanism of enolase: The crystal structure of asymmetric dimer enolase-2-phospho-d-glycerate/enolase-phosphoenolpyruvate at 2.0 a resolution. Biochemistry 1997, 36, 12526–12534. [Google Scholar] [CrossRef]
- Greene, T.W.; Wuts, P.G.M. Protective Groups in Organic Synthesis, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1999; pp. 391–392. [Google Scholar]
- He, S.; Dayton, A.; Kuppusamy, P.; Werbovetz, K.A.; Drew, M.E. Induction of Oxidative Stress in Trypanosoma brucei by the Antitrypanosomal Dihydroquinoline OSU-40. Antimicrob. Agents Chemother. 2012, 56, 2428–2434. [Google Scholar] [CrossRef] [PubMed]
- Figarella, K.; Uzcategui, N.L.; Beck, A.; Schoenfeld, C.; Kubata, B.K.; Lang, F.; Duszenko, M. Prostaglandin-induced programmed cell death in Trypanosoma brucei involves oxidative stress. Cell Death Differ. 2006, 13, 1802–1814. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, M.; Larrea, S.C.V.; Haikarainen, T.; Narwal, M.; Venkannagari, H.; Flawiá, M.M.; Lehtiö, L.; Villamil, S.H.F. Disrupted ADP-ribose metabolism with nuclear Poly (ADP-ribose) accumulation leads to different cell death pathways in presence of hydrogen peroxide in procyclic Trypanosoma brucei. Parasites Vectors 2016, 9, 173. [Google Scholar] [CrossRef] [PubMed]
- Lamour, N.; Rivière, L.; Coustou, V.; Coombs, G.H.; Barrett, M.P.; Bringaud, F. Proline Metabolism in Procyclic Trypanosoma brucei Is Down-regulated in the Presence of Glucose. J. Biol. Chem. 2005, 280, 11902–11910. [Google Scholar] [CrossRef]
- Wargnies, M.; Bertiaux, E.; Cahoreau, E.; Ziebart, N.; Crouzols, A.; Morand, P.; Biran, M.; Allmann, S.; Hubert, J.; Villafraz, O.; et al. Gluconeogenesis is essential for trypanosome development in the tsetse fly vector. PLOS Pathog. 2018, 14, e1007502. [Google Scholar] [CrossRef]
- Kaberdin, V.R.; Lin-Chao, S. Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biol. 2009, 6, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V. Multifunctional α-enolase: Its role in diseases. Cell. Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef] [PubMed]
| TbENO | |
|---|---|
| NfENO | 46 (62) |
| HsENO2 | 63 (78) |
| Compound | Structure | TbENO IC50 (µM) | HsENO2 IC50 (µM) 1 | T. brucei BSF EC50 (µM) |
|---|---|---|---|---|
| HEX | 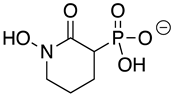 | 2.1 ± 1.1 | 0.36 ± 0.04 | >10 |
| BenzylHEX 2 | 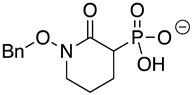 | >10 | >10 | >10 |
| POMHEX |  | 2.8 ± 0.64 | >10 | 0.61 ± 0.08 |
| Deoxy-SF2312 | 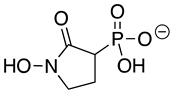 | 0.60 ± 0.23 | 0.10 ± 0.04 | >10 |
| Benzyl-deoxy-SF2312 | 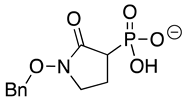 | >10 | >10 | >10 |
| HEPTA |  | >10 | 5.8 ± 0.88 | >10 |
| Benzyl-HEPTA | 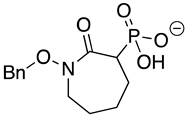 | >10 | >10 | >10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roster, C.P.; LaVigne, D.; Milanes, J.E.; Knight, E.; Anderson, H.D.; Pizarro, S.; Harding, E.M.; Morris, M.T.; Yan, V.C.; Pham, C.-D.; et al. Enolase Inhibitors as Early Lead Therapeutics against Trypanosoma brucei. Pathogens 2023, 12, 1290. https://doi.org/10.3390/pathogens12111290
Roster CP, LaVigne D, Milanes JE, Knight E, Anderson HD, Pizarro S, Harding EM, Morris MT, Yan VC, Pham C-D, et al. Enolase Inhibitors as Early Lead Therapeutics against Trypanosoma brucei. Pathogens. 2023; 12(11):1290. https://doi.org/10.3390/pathogens12111290
Chicago/Turabian StyleRoster, Colm P., Danielle LaVigne, Jillian E. Milanes, Emily Knight, Heidi D. Anderson, Sabrina Pizarro, Elijah M. Harding, Meredith T. Morris, Victoria C. Yan, Cong-Dat Pham, and et al. 2023. "Enolase Inhibitors as Early Lead Therapeutics against Trypanosoma brucei" Pathogens 12, no. 11: 1290. https://doi.org/10.3390/pathogens12111290






