A Loss of Function in LprG−Rv1410c Homologues Attenuates Growth during Biofilm Formation in Mycobacterium smegmatis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Media, and Culture Conditions
2.2. Construction of Mutant and Complement M. smegmatis Strains
2.3. Biofilm Growth and CFU Enumeration
2.4. Lipid Extraction and Analysis
2.5. RNA Extraction and RT-qPCR
3. Results and Discussion
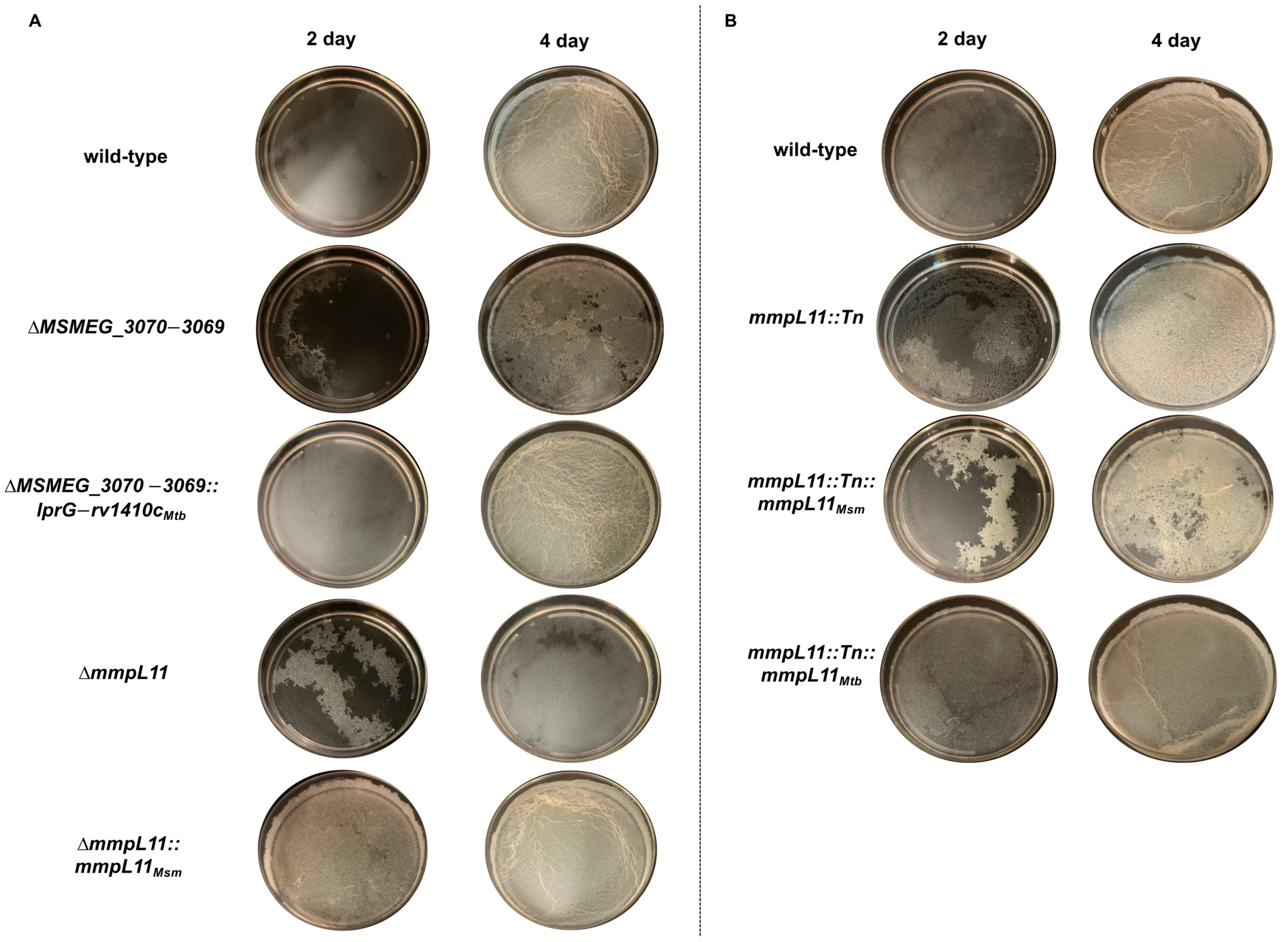
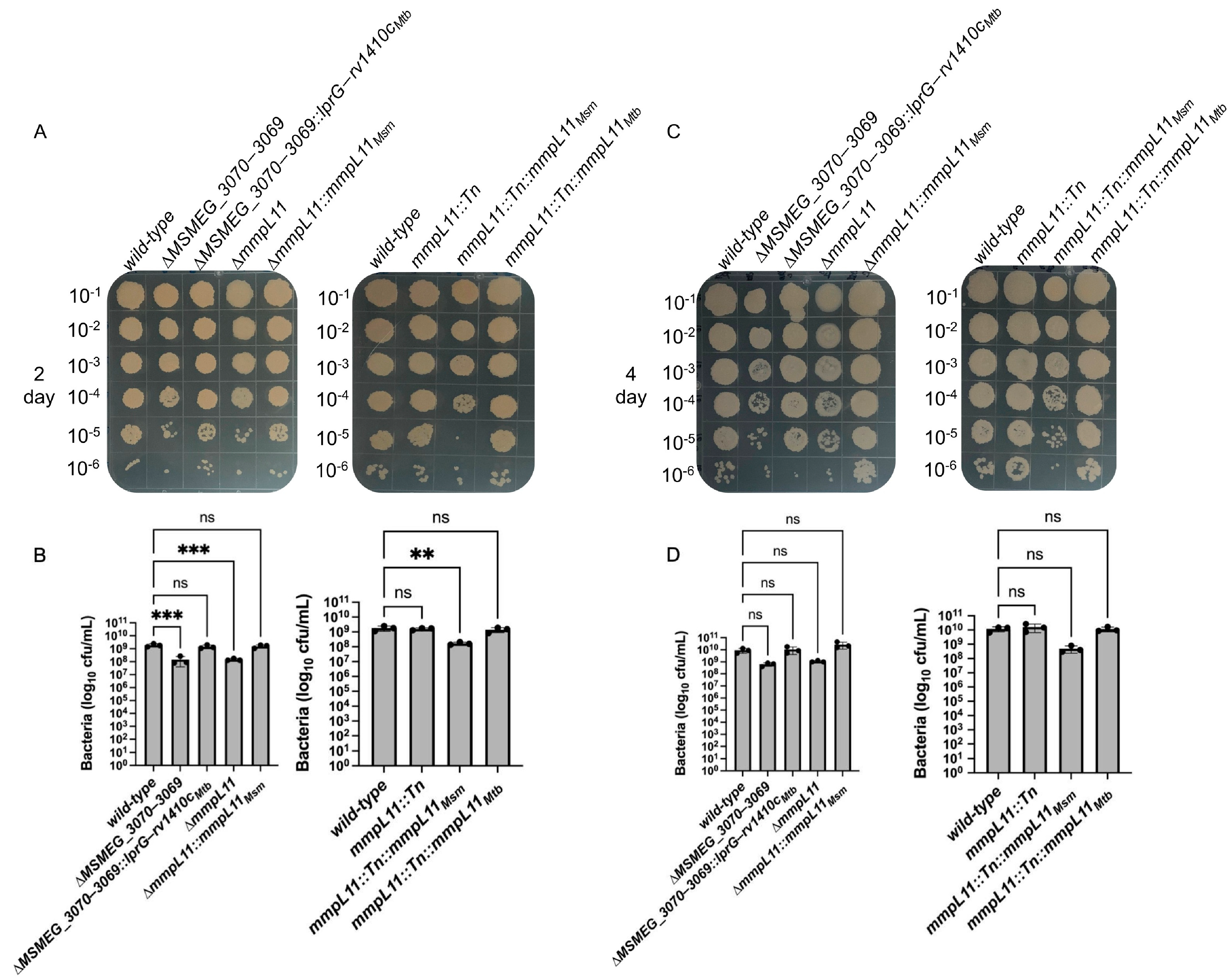
3.1. Loss of MSMEG_3070 3069 or mmpL11 Leads to Similar Defects in Biofilm Formation
3.2. MSMEG_3070−3069 Is Not Required for the Surface Localization of Mycolate Wax Esters and Monomeromycolyl Diacylglycerides
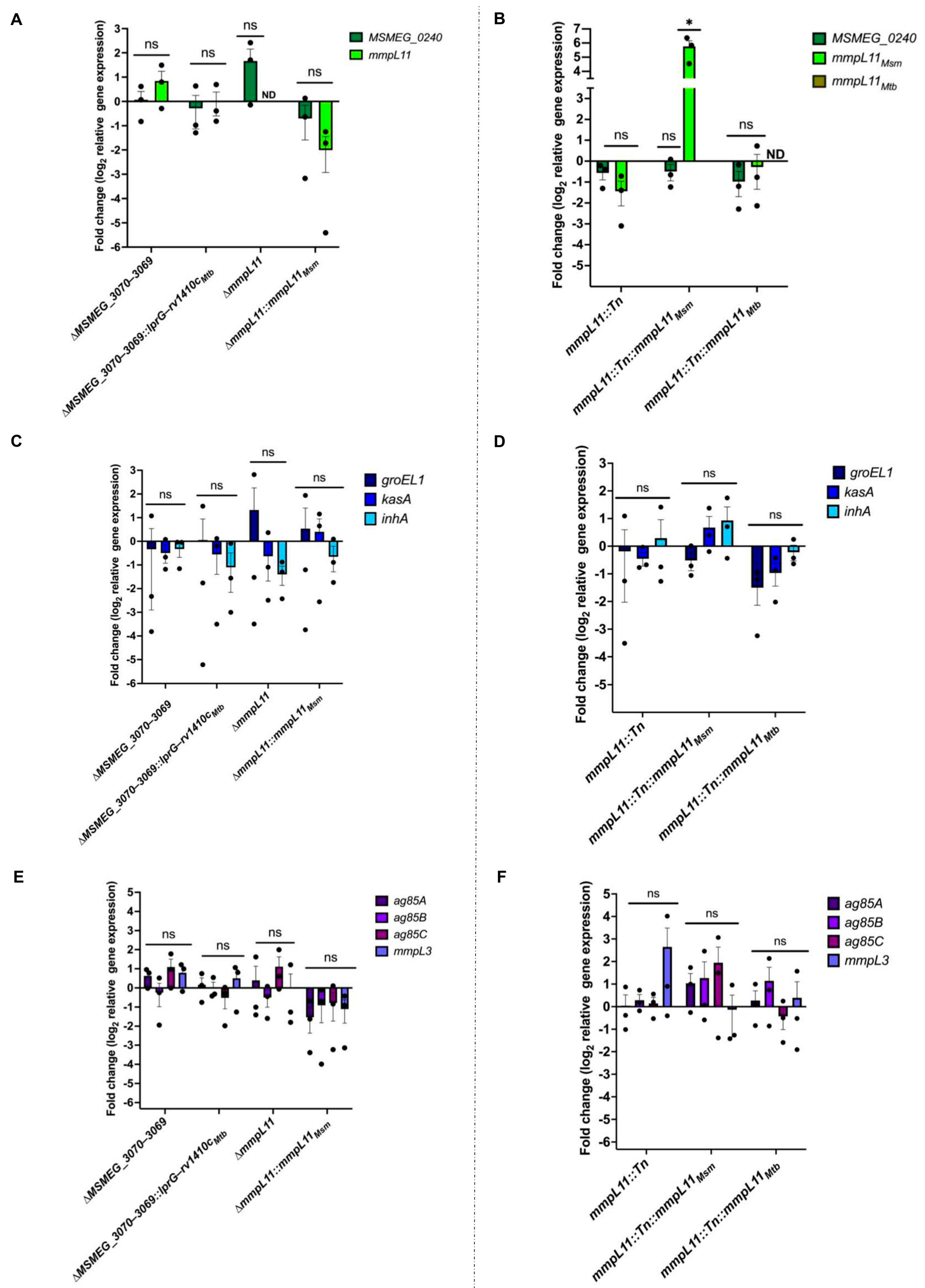
3.3. Loss of MSMEG_3070−3069 or mmpL11 Does Not Correlate with Changes in Expression of Genes Associated with Biofilm Formation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Brennan, P.J.; Nikaido, H. The Envelope of Mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef]
- Hoffmann, C.; Leis, A.; Niederweis, M.; Plitzko, J.M.; Engelhardt, H. Disclosure of the Mycobacterial Outer Membrane: Cryo-Electron Tomography and Vitreous Sections Reveal the Lipid Bilayer Structure. Proc. Natl. Acad. Sci. USA 2008, 105, 3963–3967. [Google Scholar] [CrossRef]
- Jackson, M. The Mycobacterial Cell Envelope-Lipids. Cold Spring Harb. Perspect. Med. 2014, 4, a021105. [Google Scholar] [CrossRef]
- Astarie-Dequeker, C.; Nigou, J.; Passemar, C.; Guilhot, C. The Role of Mycobacterial Lipids in Host Pathogenesis. Drug Discov. Today Dis. Mech. 2010, 7, e33–e41. [Google Scholar] [CrossRef]
- Singh, P.; Rameshwaram, N.R.; Ghosh, S.; Mukhopadhyay, S. Cell Envelope Lipids in the Pathophysiology of Mycobacterium Tuberculosis. Future Microbiol. 2018, 13, 689–710. [Google Scholar] [CrossRef]
- Pacheco, S.A.; Hsu, F.-F.; Powers, K.M.; Purdy, G.E. MmpL11 Protein Transports Mycolic Acid-Containing Lipids to the Mycobacterial Cell Wall and Contributes to Biofilm Formation in Mycobacterium Smegmatis. J. Biol. Chem. 2013, 288, 24213–24222. [Google Scholar] [CrossRef]
- Wright, C.C.; Hsu, F.F.; Arnett, E.; Dunaj, J.L.; Davidson, P.M.; Pacheco, S.A.; Harriff, M.J.; Lewinsohn, D.M.; Schlesinger, L.S.; Purdy, G.E. The Mycobacterium Tuberculosis MmpL11 Cell Wall Lipid Transporter Is Important for Biofilm Formation, Intracellular Growth, and Nonreplicating Persistence. Infect. Immun. 2017, 85, e00131-17. [Google Scholar] [CrossRef]
- Recht, J.; Martínez, A.; Torello, S.; Kolter, R. Genetic Analysis of Sliding Motility in Mycobacterium Smegmatis. J. Bacteriol. 2000, 182, 4348–4351. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Danelishvili, L.; Wu, M.; Macnab, M.; Bermudez, L.E. Mycobacterium Avium Genes Associated with the Ability to Form a Biofilm. Appl. Environ. Microbiol. 2006, 72, 819–825. [Google Scholar] [CrossRef]
- Ojha, A.K.; Trivelli, X.; Guerardel, Y.; Kremer, L.; Hatfull, G.F. Enzymatic Hydrolysis of Trehalose Dimycolate Releases Free Mycolic Acids during Mycobacterial Growth in Biofilms. J. Biol. Chem. 2010, 285, 17380–17389. [Google Scholar] [CrossRef]
- Ojha, A.; Anand, M.; Bhatt, A.; Kremer, L.; Jacobs, W.R.J.; Hatfull, G.F. GroEL1: A Dedicated Chaperone Involved in Mycolic Acid Biosynthesis during Biofilm Formation in Mycobacteria. Cell 2005, 123, 861–873. [Google Scholar] [CrossRef]
- Ojha, A.K.; Baughn, A.D.; Sambandan, D.; Hsu, T.; Trivelli, X.; Guerardel, Y.; Alahari, A.; Kremer, L.; Jacobs, W.R.J.; Hatfull, G.F. Growth of Mycobacterium Tuberculosis Biofilms Containing Free Mycolic Acids and Harbouring Drug-Tolerant Bacteria. Mol. Microbiol. 2008, 69, 164–174. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Qvist, T.; Eickhardt, S.; Kragh, K.N.; Andersen, C.B.; Iversen, M.; Høiby, N.; Bjarnsholt, T. Chronic Pulmonary Disease with Mycobacterium Abscessus Complex Is a Biofilm Infection. Eur. Respir. J. 2015, 46, 1823–1826. [Google Scholar] [CrossRef]
- Fennelly, K.P.; Ojano-Dirain, C.; Yang, Q.; Liu, L.; Lu, L.; Progulske-Fox, A.; Wang, G.P.; Antonelli, P.; Schultz, G. Biofilm Formation by Mycobacterium Abscessus in a Lung Cavity. Am. J. Respir. Crit. Care Med. 2016, 193, 692–693. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Danelishvili, L.; Wu, M.; Hidaka, E.; Katsuyama, T.; Stang, B.; Petrofsky, M.; Bildfell, R.; Bermudez, L.E. The Ability to Form Biofilm Influences Mycobacterium Avium Invasion and Translocation of Bronchial Epithelial Cells. Cell. Microbiol. 2006, 8, 806–814. [Google Scholar] [CrossRef]
- Chakraborty, P.; Bajeli, S.; Kaushal, D.; Radotra, B.D.; Kumar, A. Biofilm Formation in the Lung Contributes to Virulence and Drug Tolerance of Mycobacterium Tuberculosis. Nat. Commun. 2021, 12, 1606. [Google Scholar] [CrossRef]
- Greendyke, R.; Byrd, T.F. Differential Antibiotic Susceptibility of Mycobacterium Abscessus Variants in Biofilms and Macrophages Compared to That of Planktonic Bacteria. Antimicrob. Agents Chemother. 2008, 52, 2019–2026. [Google Scholar] [CrossRef]
- Bardouniotis, E.; Ceri, H.; Olson, M.E. Biofilm Formation and Biocide Susceptibility Testing of Mycobacterium Fortuitum and Mycobacterium Marinum. Curr. Microbiol. 2003, 46, 28–32. [Google Scholar] [CrossRef]
- Viljoen, A.; Dufrêne, Y.F.; Nigou, J. Mycobacterial Adhesion: From Hydrophobic to Receptor-Ligand Interactions. Microorganisms 2022, 10, 454. [Google Scholar] [CrossRef]
- Richards, J.P.; Ojha, A.K. Mycobacterial Biofilms. Microbiol. Spectr. 2014, 2, 1–11. [Google Scholar] [CrossRef]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2018, 8, 2651. [Google Scholar] [CrossRef]
- Chakraborty, P.; Kumar, A. The Extracellular Matrix of Mycobacterial Biofilms: Could We Shorten the Treatment of Mycobacterial Infections? Microb. Cell 2019, 6, 105–122. [Google Scholar] [CrossRef]
- Chen, J.M.; German, G.J.; Alexander, D.C.; Ren, H.; Tan, T.; Liu, J. Roles of Lsr2 in Colony Morphology and Biofilm Formation of Mycobacterium Smegmatis. J. Bacteriol. 2006, 188, 633–641. [Google Scholar] [CrossRef]
- Melly, G.C.; Stokas, H.; Davidson, P.M.; Roma, J.S.; Rhodes, H.L.; Purdy, G.E. Identification of Residues Important for M. Tuberculosis MmpL11 Function Reveals That Function Is Modulated by Phosphorylation in the C-Terminal Domain. Mol. Microbiol. 2021, 115, 208–221. [Google Scholar] [CrossRef]
- Recht, J.; Kolter, R. Glycopeptidolipid Acetylation Affects Sliding Motility and Biofilm Formation in Mycobacterium Smegmatis. J. Bacteriol. 2001, 183, 5718–5724. [Google Scholar] [CrossRef]
- Yang, Y.; Thomas, J.; Li, Y.; Vilchèze, C.; Derbyshire, K.M.; Jacobs, W.R.J.; Ojha, A.K. Defining a Temporal Order of Genetic Requirements for Development of Mycobacterial Biofilms. Mol. Microbiol. 2017, 105, 794–809. [Google Scholar] [CrossRef]
- Gupta, K.R.; Kasetty, S.; Chatterji, D. Novel Functions of (p)ppGpp and Cyclic Di-GMP in Mycobacterial Physiology Revealed by Phenotype Microarray Analysis of Wild-Type and Isogenic Strains of Mycobacterium Smegmatis. Appl. Environ. Microbiol. 2015, 81, 2571–2578. [Google Scholar] [CrossRef]
- Richards, J.P.; Cai, W.; Zill, N.A.; Zhang, W.; Ojha, A.K. Adaptation of Mycobacterium Tuberculosis to Biofilm Growth Is Genetically Linked to Drug Tolerance. Antimicrob. Agents Chemother. 2019, 63, e01213-19. [Google Scholar] [CrossRef]
- Pang, J.M.; Layre, E.; Sweet, L.; Sherrid, A.; Moody, D.B.; Ojha, A.; Sherman, D.R. The Polyketide Pks1 Contributes to Biofilm Formation in Mycobacterium Tuberculosis. J. Bacteriol. 2012, 194, 715–721. [Google Scholar] [CrossRef]
- Dokic, A.; Peterson, E.; Arrieta-Ortiz, M.L.; Pan, M.; Di Maio, A.; Baliga, N.; Bhatt, A. Mycobacterium Abscessus Biofilms Produce an Extracellular Matrix and Have a Distinct Mycolic Acid Profile. Cell Surf. 2021, 7, 100051. [Google Scholar] [CrossRef]
- Belardinelli, J.M.; Li, W.; Avanzi, C.; Angala, S.K.; Lian, E.; Wiersma, C.J.; Palčeková, Z.; Martin, K.H.; Angala, B.; de Moura, V.C.N.; et al. Unique Features of Mycobacterium Abscessus Biofilms Formed in Synthetic Cystic Fibrosis Medium. Front. Microbiol. 2021, 12, 743126. [Google Scholar] [CrossRef]
- Savijoki, K.; Myllymäki, H.; Luukinen, H.; Paulamäki, L.; Vanha-Aho, L.-M.; Svorjova, A.; Miettinen, I.; Fallarero, A.; Ihalainen, T.O.; Yli-Kauhaluoma, J.; et al. Surface-Shaving Proteomics of Mycobacterium Marinum Identifies Biofilm Subtype-Specific Changes Affecting Virulence, Tolerance, and Persistence. mSystems 2021, 6, e0050021. [Google Scholar] [CrossRef]
- Sharma, A.; Bansal, S.; Kumari, N.; Vashistt, J.; Shrivastava, R. Comparative Proteomic Investigation Unravels the Pathobiology of Mycobacterium Fortuitum Biofilm. Appl. Microbiol. Biotechnol. 2023, 107, 6029–6046. [Google Scholar] [CrossRef]
- Shukla, S.; Richardson, E.T.; Athman, J.J.; Shi, L.; Wearsch, P.A.; McDonald, D.; Banaei, N.; Boom, W.H.; Jackson, M.; Harding, C.V. Mycobacterium Tuberculosis Lipoprotein LprG Binds Lipoarabinomannan and Determines Its Cell Envelope Localization to Control Phagolysosomal Fusion. PLoS Pathog. 2014, 10, e1004471. [Google Scholar] [CrossRef]
- Gaur, R.L.; Ren, K.; Blumenthal, A.; Bhamidi, S.; González-Nilo, F.D.; Jackson, M.; Zare, R.N.; Ehrt, S.; Ernst, J.D.; Banaei, N. LprG-Mediated Surface Expression of Lipoarabinomannan Is Essential for Virulence of Mycobacterium Tuberculosis. PLoS Pathog. 2014, 10, e1004376. [Google Scholar] [CrossRef]
- Martinot, A.J.; Farrow, M.; Bai, L.; Layre, E.; Cheng, T.-Y.; Tsai, J.H.; Iqbal, J.; Annand, J.W.; Sullivan, Z.A.; Hussain, M.M.; et al. Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium Tuberculosis. PLoS Pathog. 2016, 12, e1005351. [Google Scholar] [CrossRef]
- Melly, G.C.; Stokas, H.; Dunaj, J.L.; Hsu, F.-F.; Rajavel, M.; Su, C.-C.; Yu, E.W.; Purdy, G.E. Structural and Functional Evidence That Lipoprotein LpqN Supports Cell Envelope Biogenesis in M. Tuberculosis. J. Biol. Chem. 2019, 294, 15711–15723. [Google Scholar] [CrossRef]
- Farrow, M.F.; Rubin, E.J. Function of a Mycobacterial Major Facilitator Superfamily Pump Requires a Membrane-Associated Lipoprotein. J. Bacteriol. 2008, 190, 1783–1791. [Google Scholar] [CrossRef]
- Murphy, K.C.; Papavinasasundaram, K.; Sassetti, C.M. Mycobacterial Recombineering. Methods Mol. Biol. 2015, 1285, 177–199. [Google Scholar] [CrossRef]
- Bansal-Mutalik, R.; Nikaido, H. Mycobacterial Outer Membrane Is a Lipid Bilayer and the Inner Membrane Is Unusually Rich in Diacyl Phosphatidylinositol Dimannosides. Proc. Natl. Acad. Sci. USA 2014, 111, 4958–4963. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Touchette, M.H.; Holsclaw, C.M.; Previti, M.L.; Solomon, V.C.; Leary, J.A.; Bertozzi, C.R.; Seeliger, J.C. The Rv1184c Locus Encodes Chp2, an Acyltransferase in Mycobacterium Tuberculosis Polyacyltrehalose Lipid Biosynthesis. J. Bacteriol. 2015, 197, 201–210. [Google Scholar] [CrossRef]
- Converse, S.E.; Mougous, J.D.; Leavell, M.D.; Leary, J.A.; Bertozzi, C.R.; Cox, J.S. MmpL8 Is Required for Sulfolipid-1 Biosynthesis and Mycobacterium Tuberculosis Virulence. Proc. Natl. Acad. Sci. USA 2003, 100, 6121–6126. [Google Scholar] [CrossRef]
- Seeliger, J.C.; Holsclaw, C.M.; Schelle, M.W.; Botyanszki, Z.; Gilmore, S.A.; Tully, S.E.; Niederweis, M.; Cravatt, B.F.; Leary, J.A.; Bertozzi, C.R. Elucidation and Chemical Modulation of Sulfolipid-1 Biosynthesis in Mycobacterium Tuberculosis. J. Biol. Chem. 2012, 287, 7990–8000. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisbett, L.-M.; Previti, M.L.; Seeliger, J.C. A Loss of Function in LprG−Rv1410c Homologues Attenuates Growth during Biofilm Formation in Mycobacterium smegmatis. Pathogens 2023, 12, 1375. https://doi.org/10.3390/pathogens12121375
Nisbett L-M, Previti ML, Seeliger JC. A Loss of Function in LprG−Rv1410c Homologues Attenuates Growth during Biofilm Formation in Mycobacterium smegmatis. Pathogens. 2023; 12(12):1375. https://doi.org/10.3390/pathogens12121375
Chicago/Turabian StyleNisbett, Lisa-Marie, Mary L. Previti, and Jessica C. Seeliger. 2023. "A Loss of Function in LprG−Rv1410c Homologues Attenuates Growth during Biofilm Formation in Mycobacterium smegmatis" Pathogens 12, no. 12: 1375. https://doi.org/10.3390/pathogens12121375





