De Novo Transcriptome Profiling of Naegleria fowleri Trophozoites and Cysts via RNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of N. fowleri and RNA Isolation
2.2. Sequencing and De Novo Transcriptome Assembly
2.3. Functional Annotation
2.4. Identification of Stage-Specific Genes
2.5. Reverse-Transcription PCR
3. Results
3.1. Sequencing and De Novo Assembly
3.2. Transcriptome Annotation
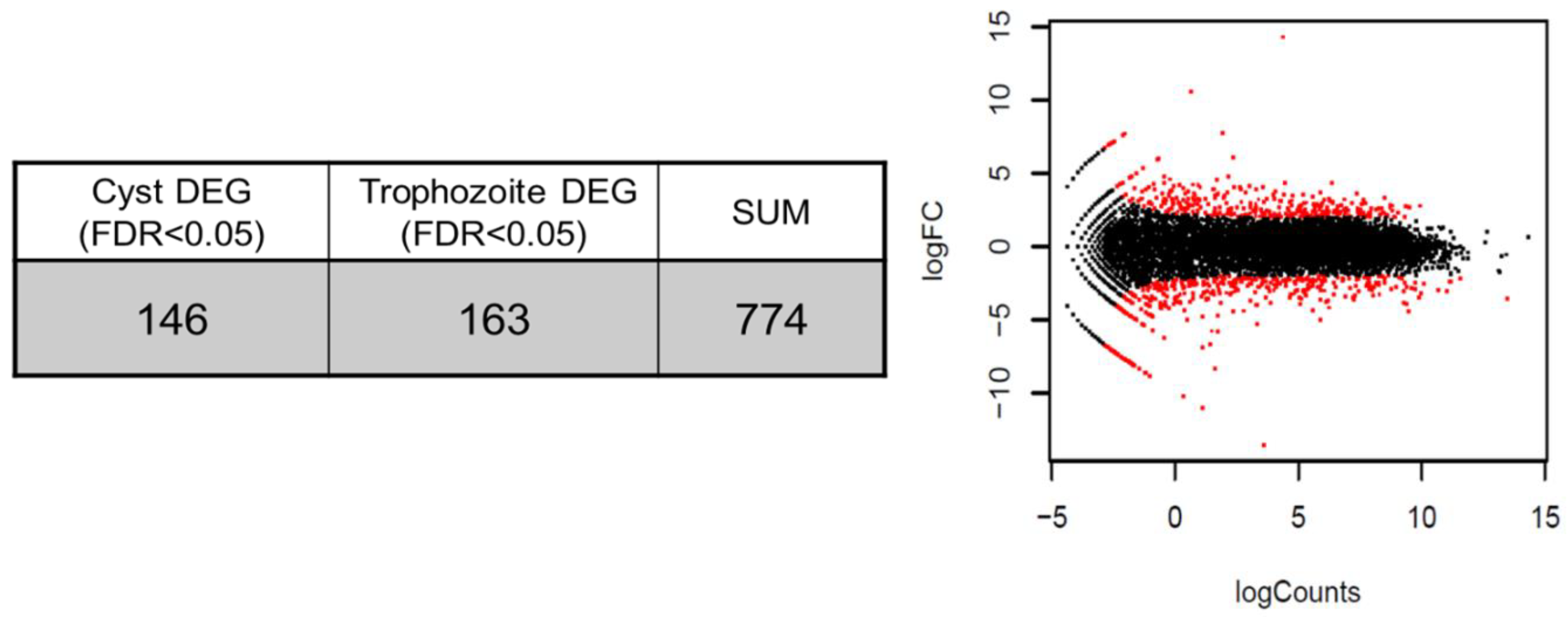
3.3. Differential Expression Genes (DEGs)
3.4. Gene Expression of Profilin in N. fowleri Trohozoites and Cysts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, K.; Jamieson, A. Primary amoebic meningoencephalitis. Lancet 1972, 2, 379. [Google Scholar] [CrossRef]
- Carter, R.F. Primary amoebic meningo-encephalitis. An appraisal of present knowledge. Trans. R Soc. Trop. Med. Hyg. 1972, 66, 193–213. [Google Scholar] [CrossRef]
- Culbertson, C.G. Pathogenic Naegleria and Hartmannella (Acenthamoeba). Ann. N. Y. Acad. Sci. 1970, 174, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Visvesvara, G.S. Amebic meningoencephalitides and keratitis: Challenges in diagnosis and treatment. Curr. Opin. Infect. Dis. 2010, 23, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Marciano-Cabral, F.; Cabral, G.A. The immune response to Naegleria fowleri amebae and pathogenesis of infection. FEMS Immunol. Med. Microbiol. 2007, 51, 243–259. [Google Scholar] [CrossRef] [Green Version]
- Visvesvara, G.S.; Moura, H.; Schuster, F.L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007, 50, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoder, J.S.; Eddy, B.A.; Visvesvara, G.S.; Capewell, L.; Beach, M.J. The epidemiology of primary amoebic meningoencephalitis in the USA, 1962–2008. Epidemiol. Infect. 2010, 138, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Schuster, F.L.; Visvesvara, G.S. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 2004, 34, 1001–1027. [Google Scholar] [CrossRef]
- Maher, C.A.; Kumar-Sinha, C.; Cao, X.; Kalyana-Sundaram, S.; Han, B.; Jing, X.; Sam, L.; Barrette, T.; Palanisamy, N.; Chinnaiyan, A.M. Transcriptome sequencing to detect gene fusions in cancer. Nature 2009, 458, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, E.K.; Chung, D.I.; Hong, Y.C.; Ahn, T.I.; Kong, H.H. Acanthamoeba castellanii: Gene profile of encystation by ESTs analysis and KOG assignment. Exp. Parasitol. 2008, 119, 111–116. [Google Scholar] [CrossRef]
- Moon, E.K.; Kim, M.J.; Lee, H.A.; Quan, F.S.; Kong, H.H. Comparative analysis of differentially expressed genes in Acanthamoeba after ingestion of Legionella pneumophila and Escherichia coli. Exp. Parasitol. 2022, 232, 108188. [Google Scholar] [CrossRef]
- Arya, S.; Sharma, G.; Gupta, P.; Tiwari, S. In silico analysis of ubiquitin/ubiquitin-like modifiers and their conjugating enzymes in Entamoeba species. Parasitol. Res. 2012, 111, 37–51. [Google Scholar] [CrossRef]
- Khalil, M.I.; Foda, B.M.; Suresh, S.; Singh, U. Technical advances in trigger-induced RNA interference gene silencing in the parasite Entamoeba histolytica. Int. J. Parasitol. 2016, 46, 205–212. [Google Scholar] [CrossRef] [Green Version]
- MacFarlane, R.C.; Shah, P.H.; Singh, U. Transcriptional profiling of Entamoeba histolytica trophozoites. Int. J. Parasitol. 2005, 35, 533–542. [Google Scholar] [CrossRef]
- Naiyer, S.; Singh, S.S.; Kaur, D.; Mukherjee, A.; Singh, Y.P.; Bhattacharya, A.; Bhattacharya, S. Transcriptomic analysis of ribosome biogenesis and pre-rRNA processing during growth stress in Entamoeba histolytica. Exp. Parasitol. 2022, 239, 108308. [Google Scholar] [CrossRef]
- Toro-Moreno, M.; Sylvester, K.; Srivastava, T.; Posfai, D.; Derbyshire, E.R. RNA-Seq Analysis Illuminates the Early Stages of Plasmodium Liver Infection. mBio 2020, 11, e03234-19. [Google Scholar] [CrossRef]
- Han, S.; Zhang, X.L.; Jiang, X.; Li, X.; Ding, J.; Zuo, L.J.; Duan, S.S.; Chen, R.; Sun, B.B.; Hu, X.Y.; et al. Long Non-Coding RNA and mRNA Expression Analysis in Liver of Mice With Clonorchis sinensis Infection. Front. Cell Infect. Microbiol. 2021, 11, 754224. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Yi, J.; Park, J.; Jeong, J.H.; Kim, J.; Won, J.; Chung, S.; Kim, T.S.; Pak, J.H. Transcriptomic profiling of three-dimensional cholangiocyte spheroids long term exposed to repetitive Clonorchis sinensis excretory-secretory products. Parasit. Vectors. 2021, 14, 213. [Google Scholar] [CrossRef]
- Yoo, W.G.; Kim, D.W.; Ju, J.W.; Cho, P.Y.; Kim, T.I.; Cho, S.H.; Choi, S.H.; Park, H.S.; Kim, T.S.; Hong, S.J. Developmental transcriptomic features of the carcinogenic liver fluke, Clonorchis sinensis. PLoS Negl. Trop. Dis. 2011, 5, e1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, L.; Amaral, M.S.; Beckedorff, F.; Silva, L.F.; Dazzani, B.; Oliveira, K.C.; Almeida, G.T.; Gomes, M.R.; Pires, D.S.; Setubal, J.C.; et al. Schistosoma mansoni Egg, Adult Male and Female Comparative Gene Expression Analysis and Identification of Novel Genes by RNA-Seq. PLoS Negl. Trop. Dis. 2015, 9, e0004334. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Xu, M.J.; Chang, Q.C.; Gao, J.F.; Wang, C.R.; Zhu, X.Q. De novo transcriptomic analysis of the female and male adults of the blood fluke Schistosoma turkestanicum. Parasit. Vectors 2016, 9, 143. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Zhu, B.; Luo, F.; Lin, X.; Sun, C.; You, Y.; Yi, C.; Xu, B.; Wang, J.; Lu, Y.; et al. Comparative transcriptome profiles of Schistosoma japonicum larval stages: Implications for parasite biology and host invasion. PLoS Negl. Trop. Dis. 2022, 16, e0009889. [Google Scholar] [CrossRef] [PubMed]
- Ilgova, J.; Vorel, J.; Roudnicky, P.; Skorpikova, L.; Horn, M.; Kasny, M. Transcriptomic and proteomic profiling of peptidase expression in Fasciola hepatica eggs developing at host’s body temperature. Sci. Rep. 2022, 12, 10308. [Google Scholar] [CrossRef] [PubMed]
- Ricafrente, A.; Cwiklinski, K.; Nguyen, H.; Dalton, J.P.; Tran, N.; Donnelly, S. Stage-specific miRNAs regulate gene expression associated with growth, development and parasite-host interaction during the intra-mammalian migration of the zoonotic helminth parasite Fasciola hepatica. BMC Genom. 2022, 23, 419. [Google Scholar] [CrossRef]
- Xu, J.; Wu, L.; Sun, Y.; Wei, Y.; Zheng, L.; Zhang, J.; Pang, Z.; Yang, Y.; Lu, Y. Proteomics and bioinformatics analysis of Fasciola hepatica somatic proteome in different growth phases. Parasitol. Res. 2020, 119, 2837–2850. [Google Scholar] [CrossRef]
- Willaert, E. Isolation and in vitro culture of the amoeba of the genus Naegleria. Ann. Soc. Belg. Med. Trop. Parasitol. Mycol. 1971, 51, 701–708. [Google Scholar]
- Sohn, H.J.; Kang, H.; Seo, G.E.; Kim, J.H.; Jung, S.Y.; Shin, H.J. Efficient Liquid Media for Encystation of Pathogenic Free-Living Amoebae. Korean J. Parasitol. 2017, 55, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Tseng, P.C.; Runge, M.S.; Cooper, J.A.; Williams, R.C., Jr.; Pollard, T.D. Physical, immunochemical, and functional properties of Acanthamoeba profilin. J. Cell Biol. 1984, 98, 214–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labruyere, E.; Guillen, N. Host tissue invasion by Entamoeba histolytica is powered by motility and phagocytosis. Arch. Med. Res. 2006, 37, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Tavares, P.; Rigothier, M.C.; Khun, H.; Roux, P.; Huerre, M.; Guillen, N. Roles of cell adhesion and cytoskeleton activity in Entamoeba histolytica pathogenesis: A delicate balance. Infect Immun. 2005, 73, 1771–1778. [Google Scholar] [CrossRef] [Green Version]
- Saito-Nakano, Y.; Wahyuni, R.; Nakada-Tsukui, K.; Tomii, K.; Nozaki, T. Rab7D small GTPase is involved in phago-, trogocytosis and cytoskeletal reorganization in the enteric protozoan Entamoeba histolytica. Cell Microbiol. 2021, 23, e13267. [Google Scholar] [CrossRef]
- Heaslip, A.T.; Dzierszinski, F.; Stein, B.; Hu, K. TgMORN1 is a key organizer for the basal complex of Toxoplasma gondii. PLoS Pathog. 2010, 6, e1000754. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Hayes, D.B.; Rebowski, G.; Tardieux, I.; Dominguez, R. Toxofilin from Toxoplasma gondii forms a ternary complex with an antiparallel actin dimer. Proc. Natl. Acad. Sci. USA 2007, 104, 16122–16127. [Google Scholar] [CrossRef] [Green Version]
- Bouyer, S.; Rodier, M.H.; Guillot, A.; Hechard, Y. Acanthamoeba castellanii: Proteins involved in actin dynamics, glycolysis, and proteolysis are regulated during encystation. Exp. Parasitol. 2009, 123, 90–94. [Google Scholar] [CrossRef]
- Gonzalez-Robles, A.; Salazar-Villatoro, L.; Omana-Molina, M.; Lorenzo-Morales, J.; Martinez-Palomo, A. Acanthamoeba royreba: Morphological features and in vitro cytopathic effect. Exp. Parasitol. 2013, 133, 369–375. [Google Scholar] [CrossRef]
- Phan, I.Q.; Rice, C.A.; Craig, J.; Noorai, R.E.; McDonald, J.R.; Subramanian, S.; Tillery, L.; Barrett, L.K.; Shankar, V.; Morris, J.C.; et al. The transcriptome of Balamuthia mandrillaris trophozoites for structure-guided drug design. Sci. Rep. 2021, 11, 21664. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, R.; Rajendran, K.; Abdella, B.; Ayub, Q.; Lim, S.Y.; Khan, N.A. Naegleria fowleri: Differential genetic expression following treatment with Hesperidin conjugated with silver nanoparticles using RNA-Seq. Parasitol. Res. 2020, 119, 2351–2358. [Google Scholar] [CrossRef] [PubMed]
- Sohn, H.J.; Kim, J.H.; Shin, M.H.; Song, K.J.; Shin, H.J. The Nf-actin gene is an important factor for food-cup formation and cytotoxicity of pathogenic Naegleria fowleri. Parasitol. Res. 2010, 106, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Sohn, H.J.; Song, K.J.; Kang, H.; Ham, A.J.; Lee, J.H.; Chwae, Y.J.; Kim, K.; Park, S.; Kim, J.H.; Shin, H.J. Cellular characterization of actin gene concerned with contact-dependent mechanisms in Naegleria fowleri. Parasite Immunol. 2019, 41, e12631. [Google Scholar] [CrossRef] [PubMed]
- Didry, D.; Carlier, M.F.; Pantaloni, D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 1998, 273, 25602–25611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ressad, F.; Didry, D.; Egile, C.; Pantaloni, D.; Carlier, M.F. Control of actin filament length and turnover by actin depolymerizing factor (ADF/cofilin) in the presence of capping proteins and ARP2/3 complex. J. Biol. Chem. 1999, 274, 20970–20976. [Google Scholar] [CrossRef]


| Contig Count | 42,220 |
|---|---|
| Type | De novo assembly |
| Total read count | 135,733,193 |
| Mean read length (bp) | 1180.5 |
| Total read length (bp) | 135,733,193 |
| Mean contig length (bp) | 2471.5 |
| Total contig length (bp) | 33,118,105 |
| Contig N50 value (bp) | 5360 |
| GC content (%) | 37.21 |
| Pathway | |
|---|---|
| Cellular Processes | 43 |
| Cell motility | 22 |
| Cell growth and death | 10 |
| Cellular community | 4 |
| Transport and catabolism | 7 |
| Environmental Information Processing | 22 |
| Signal transduction | 12 |
| Membrane transport | 7 |
| Signaling molecules and interaction | 3 |
| Genetic Information Processing | 66 |
| Translation | 31 |
| Folding, sorting, and degradation | 23 |
| Replication and repair | 12 |
| Metabolism | 131 |
| Lipid metabolism | 22 |
| Energy metabolism | 11 |
| Metabolism of other amino acids | 9 |
| Nucleotide metabolism | 19 |
| Carbohydrate metabolism | 37 |
| Amino acid metabolism | 8 |
| Metabolism of terpenoids and polyketides | 3 |
| Xenobiotic biodegradation and metabolism | 6 |
| Metabolism of cofactors and vitamins | 8 |
| Glycan biosynthesis and metabolism | 2 |
| Biosynthesis of other secondary metabolites | 6 |
| Organismal Systems | 20 |
| Immune system | 5 |
| Digestive system | 5 |
| Nervous system | 4 |
| Development | 1 |
| Endocrine system | 5 |
| Upregulated Proteins in N. fowleri Trophozoites | log FC |
|---|---|
| Luminal-binding protein | 14.309 |
| Lysosomal Pro-X carboxypeptidase | 7.017 |
| Chaperone protein | 4.349 |
| 12-oxophytodienoate reductase 1 | 4.344 |
| Probable alpha-L-glutamate ligase | 4.181 |
| fatty acid desaturase | 4.120 |
| Cytoskeleton-associated protein | 3.280 |
| Tubulin alpha-6 chain | 2.659 |
| Actin | 2.115 |
| Microtubule-associated protein | 1.996 |
| Upregulated Proteins in N. fowleri Cysts | log FC |
| Uridine kinase | 13.526 |
| Calpain-5 | 8.641 |
| Translin-associated factor X-interacting protein | 7.394 |
| Gag-Pol polyprotein | 6.912 |
| Profilin | 6.813 |
| Probable E3 ubiquitin-protein ligase | 6.707 |
| Serine/threonine-protein kinase | 6.683 |
| LisH domain-containing protein | 5.032 |
| Phospholipid-transporting ATPase | 4.970 |
| EF-hand domain-containing family member C2 | 4.579 |
| Kinesin-like calmodulin-binding protein | 4.429 |
| Probable glycerol-3-phosphate dehydrogenase(mt) | 4.327 |
| Sphingosine-1-phosphate lyase | 4.285 |
| Nitrile-specifier protein | 4.174 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohn, H.-J.; Kim, J.-H.; Kim, K.; Park, S.; Shin, H.-J. De Novo Transcriptome Profiling of Naegleria fowleri Trophozoites and Cysts via RNA Sequencing. Pathogens 2023, 12, 174. https://doi.org/10.3390/pathogens12020174
Sohn H-J, Kim J-H, Kim K, Park S, Shin H-J. De Novo Transcriptome Profiling of Naegleria fowleri Trophozoites and Cysts via RNA Sequencing. Pathogens. 2023; 12(2):174. https://doi.org/10.3390/pathogens12020174
Chicago/Turabian StyleSohn, Hae-Jin, Jong-Hyun Kim, Kyongmin Kim, Sun Park, and Ho-Joon Shin. 2023. "De Novo Transcriptome Profiling of Naegleria fowleri Trophozoites and Cysts via RNA Sequencing" Pathogens 12, no. 2: 174. https://doi.org/10.3390/pathogens12020174
APA StyleSohn, H.-J., Kim, J.-H., Kim, K., Park, S., & Shin, H.-J. (2023). De Novo Transcriptome Profiling of Naegleria fowleri Trophozoites and Cysts via RNA Sequencing. Pathogens, 12(2), 174. https://doi.org/10.3390/pathogens12020174







