Differential Gene Expression Pattern of Importin β3 and NS5 in C6/36 Cells Acutely and Persistently Infected with Dengue Virus 2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mosquito Cells
2.2. Dengue Virus
2.3. Amplified Fragment Length Polymorphism (AFLP)
2.4. Bioinformatic Analysis of Importin from Ae. albopictus
2.5. DENV-2 Infection Time-Course Assays
2.6. RNA Purification and Treatment
2.7. cDNA Synthesis
2.8. Verification of the DENV-2 Genome
2.9. Quantification of the DENV-2 Genome
2.10. Importinβ3 Expression Level
2.11. Western Blot Analysis
2.12. Immunofluorescence with Anti-Importin β3 and Anti-DENV NS5
2.13. Confocal Microscopy Analysis
2.14. Statistical Analyses
3. Results
3.1. Identification of Importin β3 as a Dengue Virus-Regulated Gene
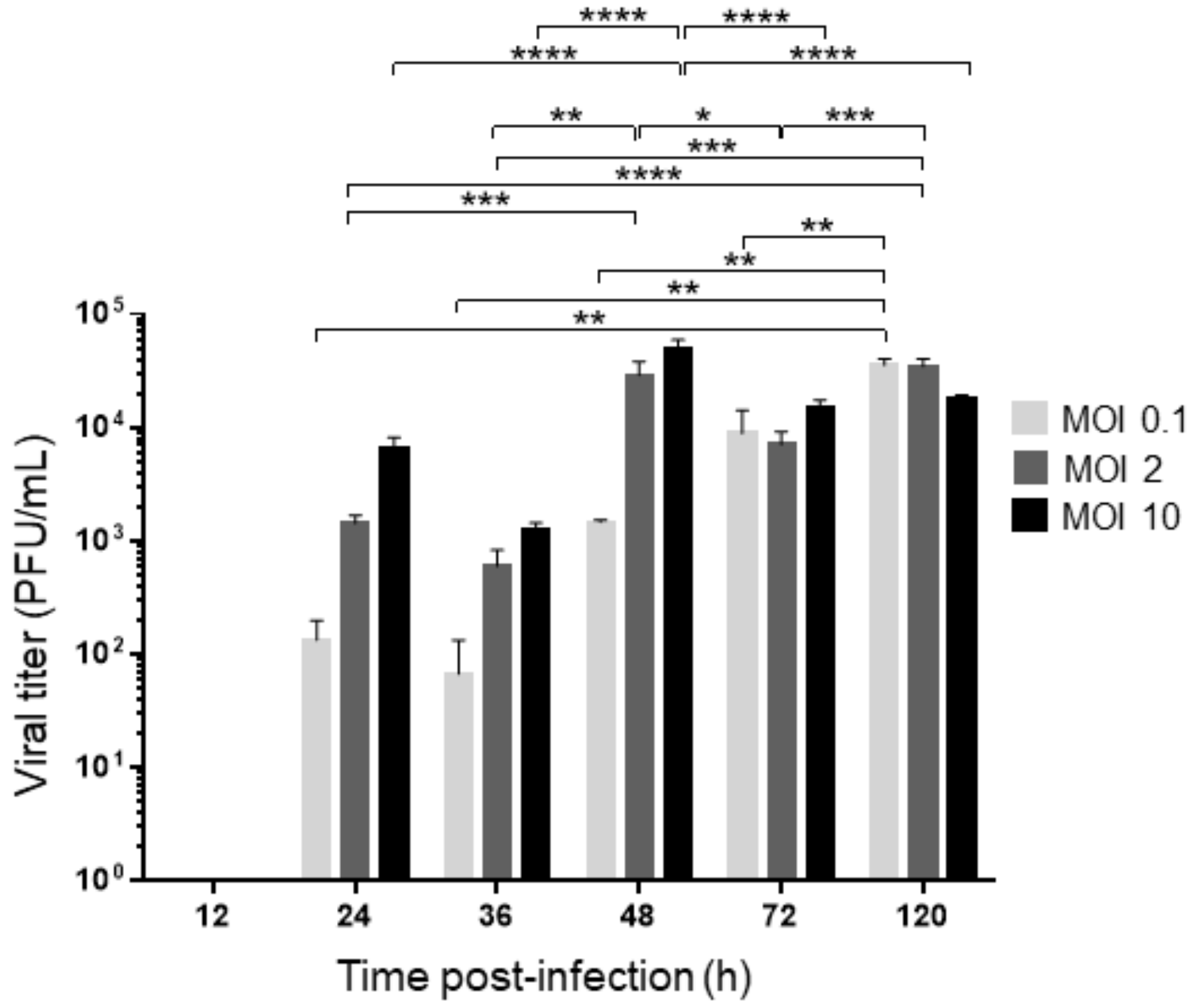
3.2. Models of Viral Infection
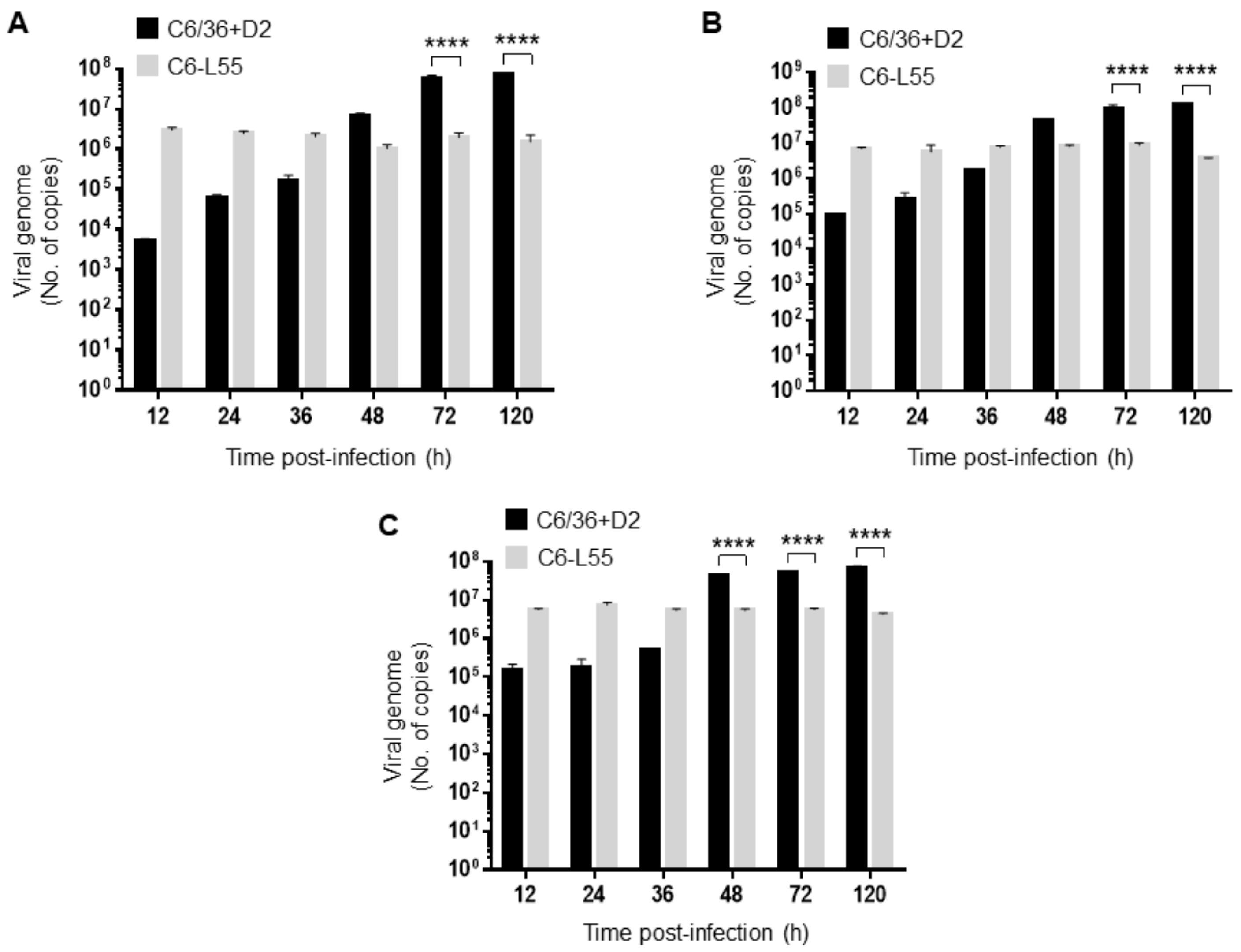
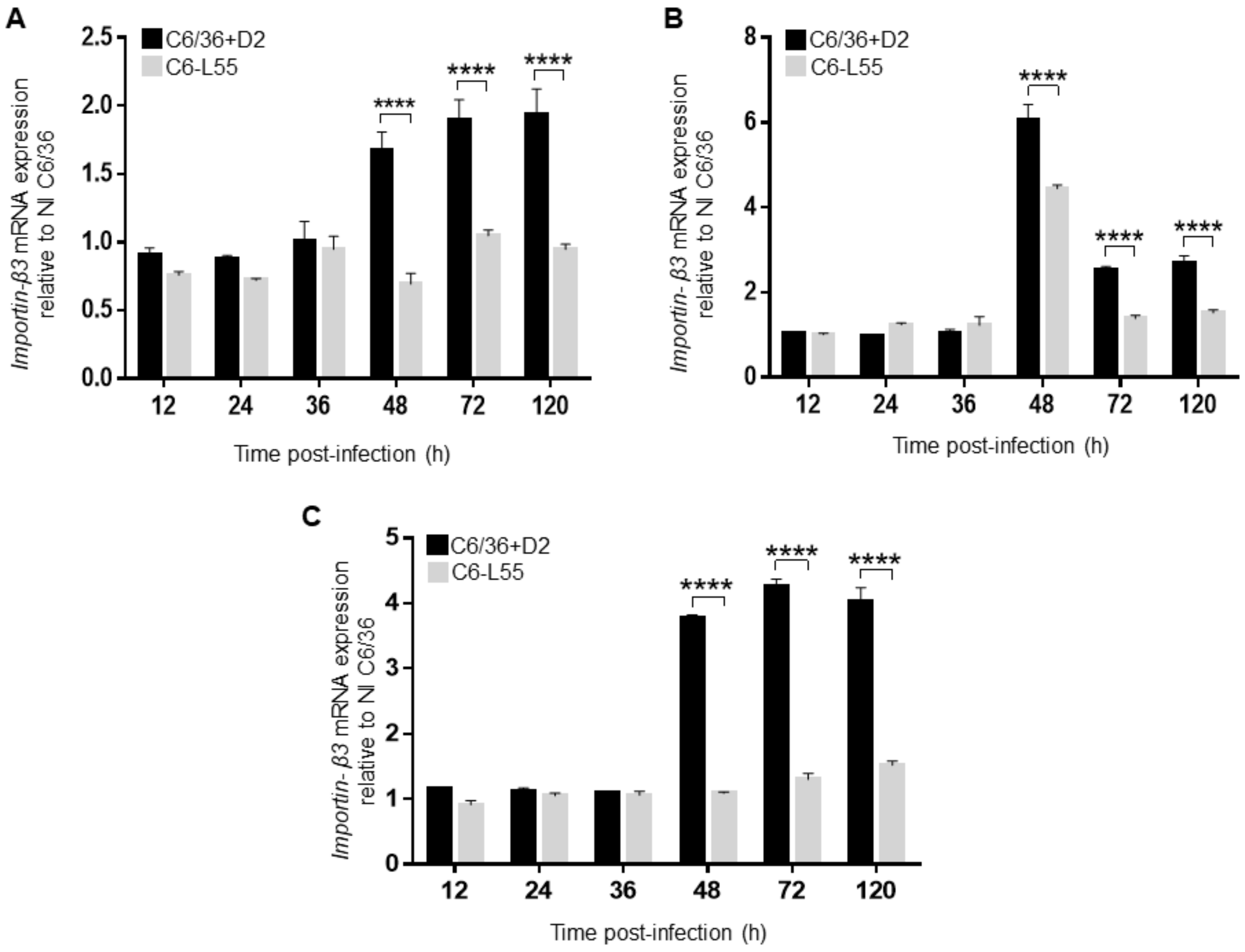
3.3. Importin β3 in Acute and Persistently DENV-2-Infected Cells
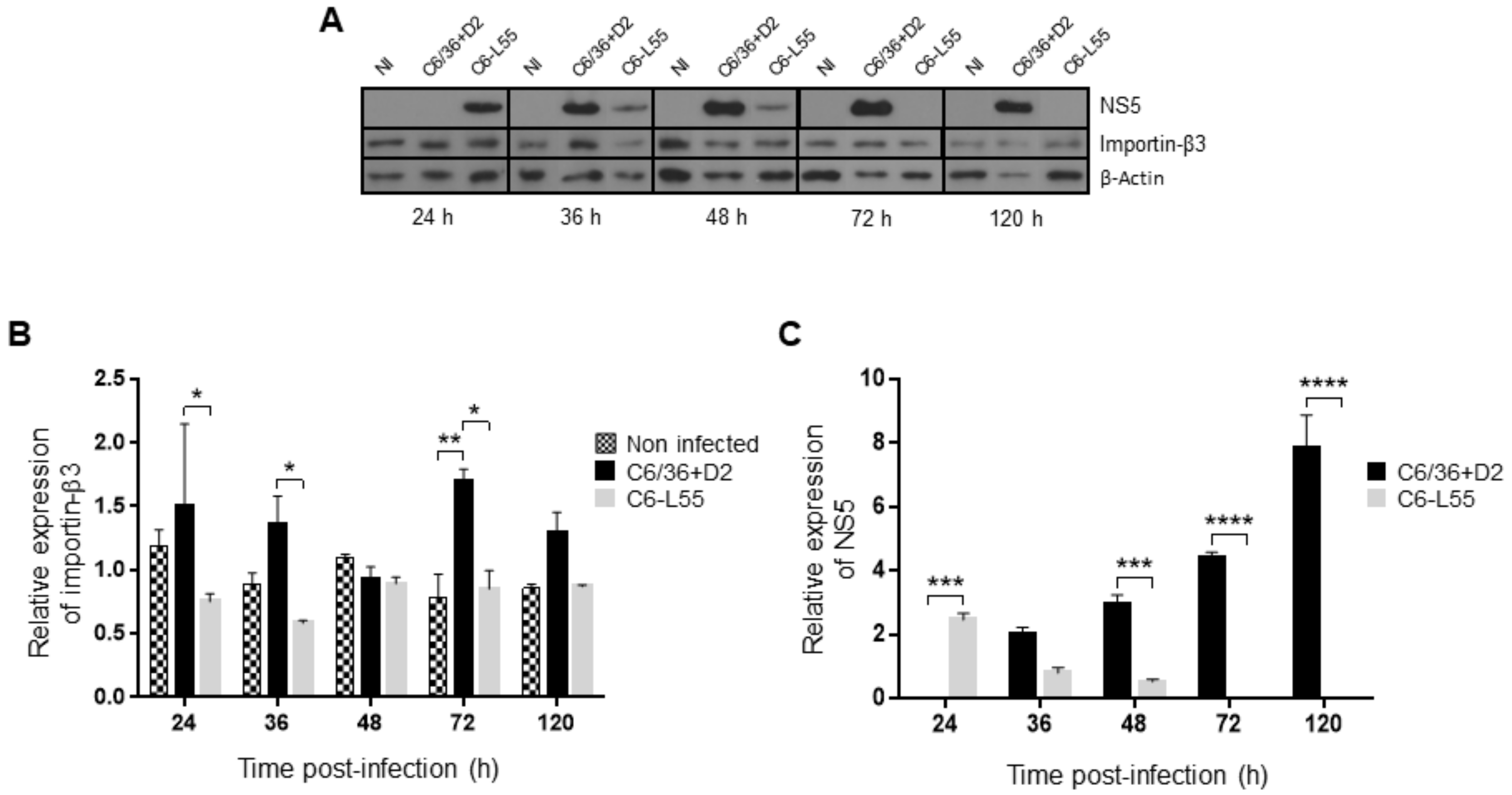
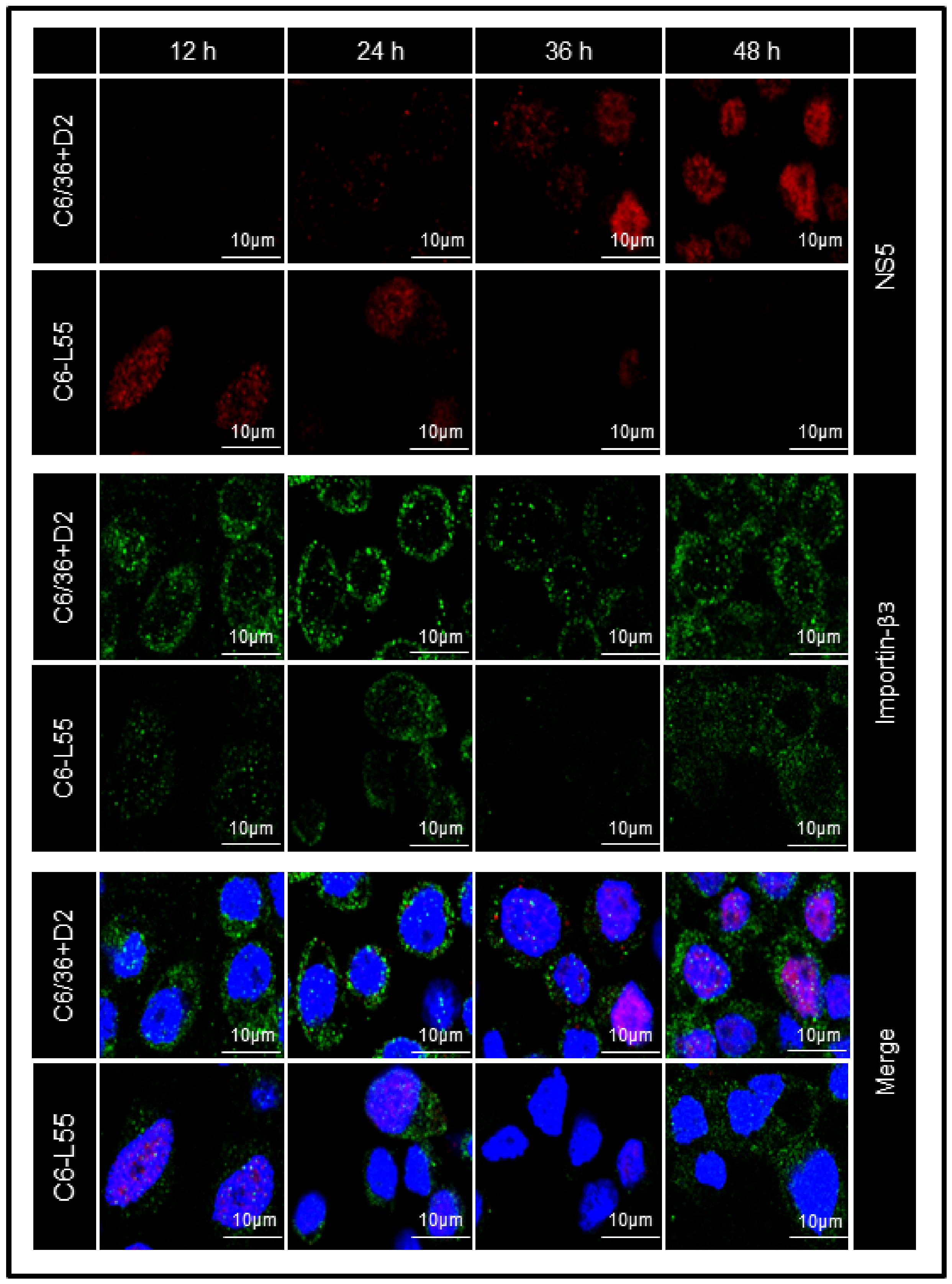
3.4. Level of Expression and Localization of DENV NS5 in Acute and Persistently Infected Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martínez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Oliveira, C.; Freire, J.M.; Conceicao, T.M.; Higa, L.M.; Castanho, M.A.; Da Poian, A.T. Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol. Rev. 2015, 39, 155–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercado-Curiel, R.F.; Esquinca-Avilés, H.A.; Tovar, R.; Díaz-Badillo, A.; Camacho-Nuez, M.; Muñoz, M.d.L. The four serotypes of dengue recognize the same putative receptors in Aedes aegypti midgut and Ae. albopictus cells. BMC Microbiol. 2006, 6, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.S.; Webster, J.A.; Madzokere, E.T.; Stephenson, E.B.; Herrero, L.J. Mosquito antiviral defense mechanisms: A delicate balance between innate immunity and persistent viral infection. Parasit Vectors. 2019, 12, 165. [Google Scholar] [CrossRef] [Green Version]
- Salas-Benito, J.S.; De Nova-Ocampo, M. Viral Interference and Persistence in Mosquito-Borne Flaviviruses. J. Immunol. Res. 2015, 2015, 873404. [Google Scholar] [CrossRef] [Green Version]
- Juárez-Martínez, A.B.; Vega-Almeida, T.O.; Salas-Benito, M.; García-Espitia, M.; De Nova-Ocampo, M.; del Ángel, R.M.; Salas-Benito, J.S. Detection and sequencing of defective viral genomes in C6/36 cells persistently infected with dengue virus 2. Arch. Virol. 2013, 158, 583–599. [Google Scholar] [CrossRef]
- Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Bautista-Carbajal, P.; Jaworski, E.; Soto-Acosta, R.; Cervantes-Salazar, M.; Angel-Ambrocio, A.H.; Castillo-Munguía, J.P.; Chávez-Munguía, B.; De Nova-Ocampo, M.; et al. Mosquito cells persistently infected with dengue virus produce viral particles with host-dependent replication. Virology 2019, 531, 1–18. [Google Scholar] [CrossRef]
- Igarashi, A. Isolation of a singh’s Aedes albopictus cell clone sensitive to dengue and chikungunya viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef]
- Kuno, G.; Oliver, A. Maintaining mosquito cell lines at high temperatures: Effects on the replication of flaviviruses. In Vitro Cell Dev. Biol. 1989, 25, 193–196. [Google Scholar] [CrossRef]
- Gould, E.A.; Clegg, J.C.S. Growth, titration and purification of alphaviruses and flaviviruses. In Virology: A practical Approach; Mahy, B.W.J., Ed.; IRL Press: Oxford, UK, 1991; pp. 43–78. [Google Scholar]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Monroy, V.S.; Flores, M.O.; Villalba-Magdaleno, J.D.; Garcia, C.G.; Ishiwara, D.G. Entamoeba histolytica: Differential gene expression during programmed cell death and identification of early pro- and anti-apoptotic signals. Exp. Parasitol. 2010, 126, 497–505. [Google Scholar] [CrossRef]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; Pérez-Ishiwara, D.G.; Cerecedo-Mercado, D.A.; del Ángel, R.M.; Salas-Benito, J.S. miR-927 has pro-viral effects during acute and persistent infection with dengue virus type 2 in C6/36 mosquito cells. J. Gen. Virol. 2020, 101, 825–839. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mercado-Curiel, R.F.; Black, W.C., 4th; Muñoz, M.D.L. A dengue receptor as possible genetic marker of vector competence in Aedes aegypti. BMC Microbiol. 2008, 8, 118. [Google Scholar] [CrossRef] [Green Version]
- Savelkoul, P.H.M.; Aarts, H.J.M.; De Haas, J.; Dijkshoorn, L.; Duim, B.; Otsen, M.; Rademaker, J.L.W.; Schouls, L.; Lenstra, J.A. Amplified-Fragment Length Polymorphism Analysis: The State of an Art. J. Clin. Microbiol. 1999, 37, 3083–3091. [Google Scholar] [CrossRef] [Green Version]
- Yan, G.; Romero-Severson, J.; Walton, M.; Chadee, D.D.; Severson, D.W. Population genetics of the yellow fever mosquito in Trinidad: Comparisons of amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 1999, 8, 951–963. [Google Scholar] [CrossRef] [Green Version]
- Ravel, S.; Monteny, N.; Velasco Olmos, D.; Escalante Verdugo, J.; Cuny, G. A preliminary study of the population genetics of Aedes aegypti (Diptera: Culicidae) from Mexico using microsatellite and AFLP markers. Acta Trop. 2001, 78, 241–250. [Google Scholar] [CrossRef]
- Paupy, C.; Orsoni, A.; Mousson, L.; Huber, K. Comparisons of amplified fragment length polymorphism (AFLP), microsatellite, and isoenzyme markers: Population genetics of Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 2004, 41, 664–671. [Google Scholar] [CrossRef]
- Joyce, A.L.; Murillo Torres, M.; Torres, R.; Moreno, M. Genetic variability of the Aedes aegypti (Diptera: Culicidae) mosquito in El Salvador, vector of dengue, yellow fever, chikungunya and Zika. Parasites Vectors 2018, 11, 637. [Google Scholar] [CrossRef] [Green Version]
- Reijans, M.; Lascaris, R.; Groeneger, A.O.; Wittenberg, A.; Wesselink, E.; van Oeveren, J.; de Wit, E.; Boorsma, A.; Voetdijk, B.; van der Spek, H.; et al. Quantitative comparison of cDNA-AFLP, microarrays, and GeneChip expression data in Saccharomyces cerevisiae. Genomics. 2003, 82, 606–618. [Google Scholar] [CrossRef]
- Cheng, S.F.; Huang, Y.P.; Wu, Z.R.; Hu, C.C.; Hsu, Y.H.; Tsai, C.H. Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biol. 2010, 10, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, X.; Yuan, B.; Qiang, S. Differential gene expression for Curvularia eragrostidis pathogenic incidence in crabgrass (Digitaria sanguinalis) revealed by cDNA-AFLP analysis. PLoS ONE 2013, 8, e75430. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Jan, F.J.; Lin, C.W.; Chung, C.H.; Chen, J.C.; Yeh, S.D.; Ku, H.M. Differential gene expression in response to papaya ringspot virus Infection in Cucumis metuliferus using cDNA-Amplified Fragment Length Polymorphism Analysis. PLoS ONE 2013, 8, e68749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLean, J.E.; Hamaguchi, N.; Belenky, P.; Mortimer, S.E.; Stanton, M.; Hedstrom, L. Inosine 5′-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo. Biochem. J. 2004, 379 Pt 2, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Angleró-Rodríguez, Y.I.; MacLeod, H.J.; Kang, S.; Carlson, J.S.; Jupatanakul, N.; Dimopoulos, G. Aedes aegypti molecular responses to Zika virus: Modulation of Infection by the Toll and Jak/Stat Immune Pathways and Virus Host Factors. Front Microbiol. 2017, 8, 2050. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Shields, A.R.; Jupatanakul, N.; Dimopoulos, G. Suppressing dengue-2 infection by chemical inhibition of Aedes aegypti host factors. PLoS Negl. Trop. Dis. 2014, 8, e3084. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.W.; Pietrantonio, P.V. In vitro expression and pharmacology of the 5-HT7-like receptor present in the mosquito Aedes aegypti tracheolar cells and hindgut-associated nerves. Insect Mol. Biol. 2003, 12, 561–569. [Google Scholar] [CrossRef]
- Dunkov, B.C.; Georgieva, T.; Yoshiga, T.; Hall, M.; Law, J.H. Aedes aegypti ferritin heavy chain homologue: Feeding of iron or blood influences message levels, lengths and subunit abundance. J. Insect Sci. 2002, 2, 7. [Google Scholar] [CrossRef]
- Geiser, D.L.; Chavez, C.A.; Flores-Munguia, R.; Winzerling, J.J.; Pham, D.Q. Aedes aegypti ferritin. Eur. J. Biochem. 2003, 270, 3667–3674. [Google Scholar] [CrossRef]
- Pham, D.Q.; Douglass, P.L.; Chavez, C.A.; Shaffer, J.J. Regulation of the ferritin heavy-chain homologue gene in the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 2005, 14, 223–236. [Google Scholar] [CrossRef]
- Geiser, D.L.; Zhou, G.; Mayo, J.J.; Winzerling, J.J. The effect of bacterial challenge on ferritin regulation in the yellow fever mosquito, Aedes aegypti. Insect Sci. 2013, 20, 601–619. [Google Scholar] [CrossRef] [Green Version]
- Shao, L.; Devenport, M.; Fujioka, H.; Ghosh, A.; Jacobs-Lorena, M. Identification and characterization of a novel peritrophic matrix protein, Ae-Aper50, and the microvillar membrane protein, AEG12, from the mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2005, 35, 947–959. [Google Scholar] [CrossRef]
- Du, X.J.; Wang, J.X.; Liu, N.; Zhao, X.F.; Li, F.H.; Xiang, J.H. Identification and molecular characterization of a peritrophin-like protein from fleshy prawn (Fenneropenaeus chinensis). Mol. Immunol. 2006, 43, 1633–1644. [Google Scholar] [CrossRef]
- Bakovic, M.; Fullerton, M.D.; Michel, V. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: The role of CTP: Phosphoethanolamine cytidylyltransferase (Pcyt2). Biochem. Cell Biol. 2007, 85, 283–300. [Google Scholar] [CrossRef]
- Coleman, M.; Vontas, J.G.; Hemingway, J. Molecular characterization of the amplified aldehyde oxidase from insecticide resistant Culex quinquefasciatus. Eur. J. Biochem. 2002, 269, 768–779. [Google Scholar] [CrossRef]
- Alvisi, G.; Rawlinson, S.M.; Ghildyal, R.; Ripalti, A.; Jans, D.A. Regulated nucleocytoplasmic trafficking of viral gene products: A therapeutic target? Biochim. Biophys. Acta. 2008, 1784, 213–227. [Google Scholar] [CrossRef]
- Pryor, M.J.; Rawlinson, S.M.; Butcher, R.E.; Barton, C.L.; Waterhouse, T.A.; Vasudevan, S.G.; Bardin, P.G.; Wright, P.J.; Jans, D.A.; Davidson, A.D. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic 2007, 8, 795–807. [Google Scholar] [CrossRef]
- Rawlinson, S.M.; Pryor, M.J.; Wright, P.J.; Jans, D.A. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J. Biol. Chem. 2009, 284, 15589–15597. [Google Scholar] [CrossRef] [Green Version]
- Netsawang, J.; Noisakran, S.; Puttikhunt, C.; Kasinrerk, W.; Wongwiwat, W.; Malasit, P.; Yenchitsomanus, P.T.; Limjindaporn, T. Nuclear localization of dengue virus capsid protein is required for DAXX interaction and apoptosis. Virus Res. 2010, 147, 275–283. [Google Scholar] [CrossRef]
- Fraser, J.E.; Rawlinson, S.M.; Heaton, S.M.; Jans, D.A. Dynamic nucleolar targeting of dengue virus polymerase NS5 in response to extracellular pH. J. Virol. 2016, 90, 5797–5807. [Google Scholar] [CrossRef] [Green Version]
- Caly, L.; Wagstaff, K.M.; Jans, D.A. Nuclear trafficking of proteins from RNA viruses: Potential target for antivirals? Antiviral Res. 2012, 95, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Glingston, R.S.; Deb, R.; Kumar, S.; Nagotu, S. Organelle dynamics and viral infections: At cross roads. Microbes Infect. 2019, 21, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Imamoto, N. Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic 2014, 15, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.N.Y.; Atkinson, S.C.; Fraser, J.E.; Wang, C.; Maher, B.; Roman, N.; Forwood, J.K.; Wagstaff, K.M.; Borg, N.A.; Jans, D.A. Novel flavivirus antiviral that targets the host nuclear transport importin α/β1 heterodimer. Cells 2019, 8, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [Green Version]
- Tay, M.Y.; Fraser, J.E.; Chan, W.K.; Moreland, N.J.; Rathore, A.P.; Wang, C.; Vasudevan, S.G.; Jans, D.A. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013, 99, 301–306. [Google Scholar] [CrossRef]
- Fraser, J.E.; Rawlinson, S.M.; Wang, C.; Jans, D.A.; Wagstaff, K.M. Investigating dengue virus nonstructural protein 5 (NS5) nuclear import. Methods Mol. Biol. 2014, 1138, 301–328. [Google Scholar] [CrossRef]
- Fraser, J.E.; Watanabe, S.; Wang, C.; Chan, W.K.; Maher, B.; Lopez-Denman, A.; Hick, C.; Wagstaff, K.M.; Mackenzie, J.M.; Sexton, P.M.; et al. A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J. Infect. Dis. 2014, 210, 1780–1791. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yang, S.N.Y.; Smith, K.; Forwood, J.K.; Jans, D.A. Nuclear import inhibitor N-(4-hydroxyphenyl) retinamide targets Zika virus (ZIKV) nonstructural protein 5 to inhibit ZIKV infection. Biochem. Biophys. Res. Commun. 2017, 493, 1555–1559. [Google Scholar] [CrossRef]
- Chauhan, C.; Behura, S.K.; Debruyn, B.; Lovin, D.D.; Harker, B.W.; Gomez-Machorro, C.; Mori, A.; Romero-Severson, J.; Severson, D.W. Comparative expression profiles of midgut genes in dengue virus refractory and susceptible Aedes aegypti across critical period for virus infection. PLoS ONE 2012, 7, e47350. [Google Scholar] [CrossRef]
- Yang, L.; Wang, R.; Yang, S.; Ma, Z.; Lin, S.; Nan, Y.; Li, Q.; Tang, Q.; Zhang, Y.J. Karyopherin alpha 6 is required for replication of porcine reproductive and respiratory syndrome virus and Zika virus. J. Virol. 2018, 92, e00072-18. [Google Scholar] [CrossRef] [Green Version]
- Tay, M.Y.F.; Vasudevan, S.G. The transactions of NS3 and NS5 in flaviviral RNA replication. Adv. Exp. Med. Biol. 2018, 1062, 147–163. [Google Scholar] [CrossRef]
- Hannemann, H.; Sung, P.Y.; Chiu, H.C.; Yousuf, A.; Bird, J.; Lim, S.P.; Davidson, A.D. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J. Biol. Chem. 2013, 288, 22621–22635. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Bühler, S.; Selisko, B.; Davidson, A.; Mulder, K.; Canard, B.; Miller, S.; Bartenschlager, R. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J. Virol. 2013, 87, 4545–4557. [Google Scholar] [CrossRef] [Green Version]
- Tay, M.Y.; Smith, K.; Ng, I.H.; Chan, K.W.; Zhao, Y.; Ooi, E.E.; Lescar, J.; Luo, D.; Jans, D.A.; Forwood, J.K.; et al. The C-terminal 18 amino acid region of dengue virus NS5 regulates its subcellular localization and contains a conserved arginine residue essential for infectious virus production. PLoS Pathog. 2016, 12, e1005886. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Cervantes-Salazar, M.; Lagunes Guillen, A.E.; Chávez-Munguía, B.; Salas-Benito, J.S.; Del Ángel, R.M. Strand-like structures and the nonstructural proteins 5, 3 and 1 are present in the nucleus of mosquito cells infected with dengue virus. Virology 2018, 515, 74–80. [Google Scholar] [CrossRef]
- Lopez-Denman, A.J.; Russo, A.; Wagstaff, K.M.; White, P.A.; Jans, D.A.; Mackenzie, J.M. Nucleocytoplasmic shuttling of the West Nile virus RNA-dependent RNA polymerase NS5 is critical to infection. Cell Microbiol. 2018, 20, e12848. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Chen, Z.; Li, Y.; Zhao, Z.; He, W.; Zohaib, A.; Song, Y.; Deng, C.; Zhang, B.; Chen, H.; et al. Japanese encephalitis virus NS5 inhibits type I interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and NF-κB. J. Virol. 2017, 91, e00039-17. [Google Scholar] [CrossRef] [Green Version]
- Hertzog, J.; Dias Junior, A.G.; Rigby, R.E.; Donald, C.L.; Mayer, A.; Sezgin, E.; Song, C.; Jin, B.; Hublitz, P.; Eggeling, C.; et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur. J. Immunol. 2018, 48, 1120–1136. [Google Scholar] [CrossRef] [Green Version]
- Ng, I.H.W.; Chan, K.W.; Tan, M.J.A.; Gwee, C.P.; Smith, K.M.; Jeffress, S.J.; Saw, W.G.; Swarbrick, C.M.D.; Watanabe, S.; Jans, D.A.; et al. Zika Virus NS5 forms supramolecular nuclear bodies that sequester importin-α and modulate the host immune and pro-inflammatory response in neuronal cells. ACS Infect. Dis. 2019, 5, 932–948. [Google Scholar] [CrossRef]
- Forwood, J.K.; Brooks, A.; Briggs, L.J.; Xiao, C.Y.; Jans, D.A.; Vasudevan, S.G. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun. 1999, 257, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.J.; Johansson, M.; John, A.V.; Xu, Y.; Jans, D.A.; Vasudevan, S.G. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J. Biol. Chem. 2002, 277, 36399–36407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patouret, R. The nuclear transport protein importin-5: A promising target in oncology and virology. Chimia 2021, 75, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Hu, J.; Dai, Y.; Zhang, H.; Xu, K.; Dong, W.; Yan, Y.; Peng, X.; Zhou, J.; Gu, J. Porcine circovirus type 2 hijacks host IPO5 to sustain the intracytoplasmic stability of its capsid protein. J. Virol. 2022, 96, e0152222. [Google Scholar] [CrossRef] [PubMed]
- Nelson, L.M.; Rose, R.C.; Moroianu, J. The L1 major capsid protein of human papillomavirus type 11 interacts with Kap beta2 and Kap beta3 nuclear import receptors. Virology 2003, 306, 162–169. [Google Scholar] [CrossRef] [Green Version]
- Darshan, M.S.; Lucchi, J.; Harding, E.; Moroianu, J. The L2 minor capsid protein of human papillomavirus type 16 interacts with a network of nuclear import receptors. J. Virol. 2004, 78, 12179–12188. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.; Nath, A.; Hauber, J.; Kehlenbach, R.H. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 2006, 281, 20883–20890. [Google Scholar] [CrossRef] [Green Version]
- Deng, T.; Engelhardt, O.G.; Thomas, B.; Akoulitchev, A.V.; Brownlee, G.G.; Fodor, E. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 2006, 80, 11911–11919. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, E.C.; Orr, O.E.; Man Liu, S.; Engelhardt, O.G.; Fodor, E. Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5. J. Gen. Virol. 2011, 92 Pt 8, 1859–1869. [Google Scholar] [CrossRef]
- Swale, C.; Monod, A.; Tengo, L.; Labaronne, A.; Garzoni, F.; Bourhis, J.M.; Cusack, S.; Schoehn, G.; Berger, I.; Ruigrok, R.W.; et al. Structural characterization of recombinant IAV polymerase reveals a stable complex between viral PA-PB1 heterodimer and host RanBP5. Sci. Rep. 2016, 6, 24727. [Google Scholar] [CrossRef] [Green Version]


| Gene | Function | Reference |
|---|---|---|
| Inosine-5-monophosphate dehydrogenase | enzyme that participates in the biosynthesis of guanosine nucleotides but is also required for viral RNA synthesis in mosquitoes infected with ZIKV and DENV | [25,26,27] |
| Adenylate cyclase | enzyme that generates cyclic AMP from ATP | [28] |
| Ferritin | iron-binding protein that is expressed in the midgut, fat body, hemolymph, and ovaries, especially after a blood meal. This protein plays a role as a cytotoxic protector against the oxidative challenge during blood meal in mosquitoes | [29,30,31,32] |
| Peritrophin | component of the peritrofic matrix in the midgut of insects and crustaceans that stimulates the digestion of food and blocks the invasion of microorganisms | [33,34] |
| Ethanolamine-phosphate cytidyltransferase | one of the regulatory enzymes implied in the synthesis of ethanolamine-derived phospholipids, structural components of the cell membranes that regulate some cellular functions such as cell division, cell signaling, phagocytosis, and autophagy | [35] |
| Molybdopterin cofactor synthesis protein | Molybdenum cofactor–dependent enzymes play important roles in several biological processes in mammals, such as purine and sulfur catabolism. They participate in the oxidation of xenobiotics, and it has been associated with insecticide resistance in mosquitoes | [36] |
| Importin 5 | Protein that participates in nuclear import | [37] |
| AALF002645-RA Importin-5-like [Ae. albopictus] | T-COFFEE Structural Alignment | Clustal Omega | |||
|---|---|---|---|---|---|
| Organism | Accession Number | Protein | Identity (%) | Identity (%) | E-Value |
| Ae. aegypti | AAEL010159-PA | importin beta-3 | 96% | 99.09% | 2.4 × 10−58 |
| Cx quinquefasciatus | CPIJ010329-RA | Importin beta-3 | 96% | 94.18% | 1.4 × 10−21 |
| An. gambiae | AGAP003769-PA | importin beta-3 | 95% | 84.35% | 7.7 × 10−47 |
| D. melanogaster | Q9VN44_DROME | Karybeta3 | 95% | 71.91% | 2.9 × 10−52 |
| Homo sapiens | NP_002262.4 | importin-5 | 94% | 55.45% | 4.5 × 10−59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ávila-Ramírez, M.L.; Reyes-Reyes, A.L.; Avila-Bonilla, R.G.; Salas-Benito, M.; Cerecedo, D.; Ramírez-Moreno, M.E.; Villagrán-Herrera, M.E.; Mercado-Curiel, R.F.; Salas-Benito, J.S. Differential Gene Expression Pattern of Importin β3 and NS5 in C6/36 Cells Acutely and Persistently Infected with Dengue Virus 2. Pathogens 2023, 12, 191. https://doi.org/10.3390/pathogens12020191
Ávila-Ramírez ML, Reyes-Reyes AL, Avila-Bonilla RG, Salas-Benito M, Cerecedo D, Ramírez-Moreno ME, Villagrán-Herrera ME, Mercado-Curiel RF, Salas-Benito JS. Differential Gene Expression Pattern of Importin β3 and NS5 in C6/36 Cells Acutely and Persistently Infected with Dengue Virus 2. Pathogens. 2023; 12(2):191. https://doi.org/10.3390/pathogens12020191
Chicago/Turabian StyleÁvila-Ramírez, María Leticia, Ana Laura Reyes-Reyes, Rodolfo Gamaliel Avila-Bonilla, Mariana Salas-Benito, Doris Cerecedo, María Esther Ramírez-Moreno, María Elena Villagrán-Herrera, Ricardo Francisco Mercado-Curiel, and Juan Santiago Salas-Benito. 2023. "Differential Gene Expression Pattern of Importin β3 and NS5 in C6/36 Cells Acutely and Persistently Infected with Dengue Virus 2" Pathogens 12, no. 2: 191. https://doi.org/10.3390/pathogens12020191
APA StyleÁvila-Ramírez, M. L., Reyes-Reyes, A. L., Avila-Bonilla, R. G., Salas-Benito, M., Cerecedo, D., Ramírez-Moreno, M. E., Villagrán-Herrera, M. E., Mercado-Curiel, R. F., & Salas-Benito, J. S. (2023). Differential Gene Expression Pattern of Importin β3 and NS5 in C6/36 Cells Acutely and Persistently Infected with Dengue Virus 2. Pathogens, 12(2), 191. https://doi.org/10.3390/pathogens12020191








