Haplotype Analysis Sheds Light on the Genetic Evolution of the Powdery Mildew Resistance Locus Pm60 in Triticum Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
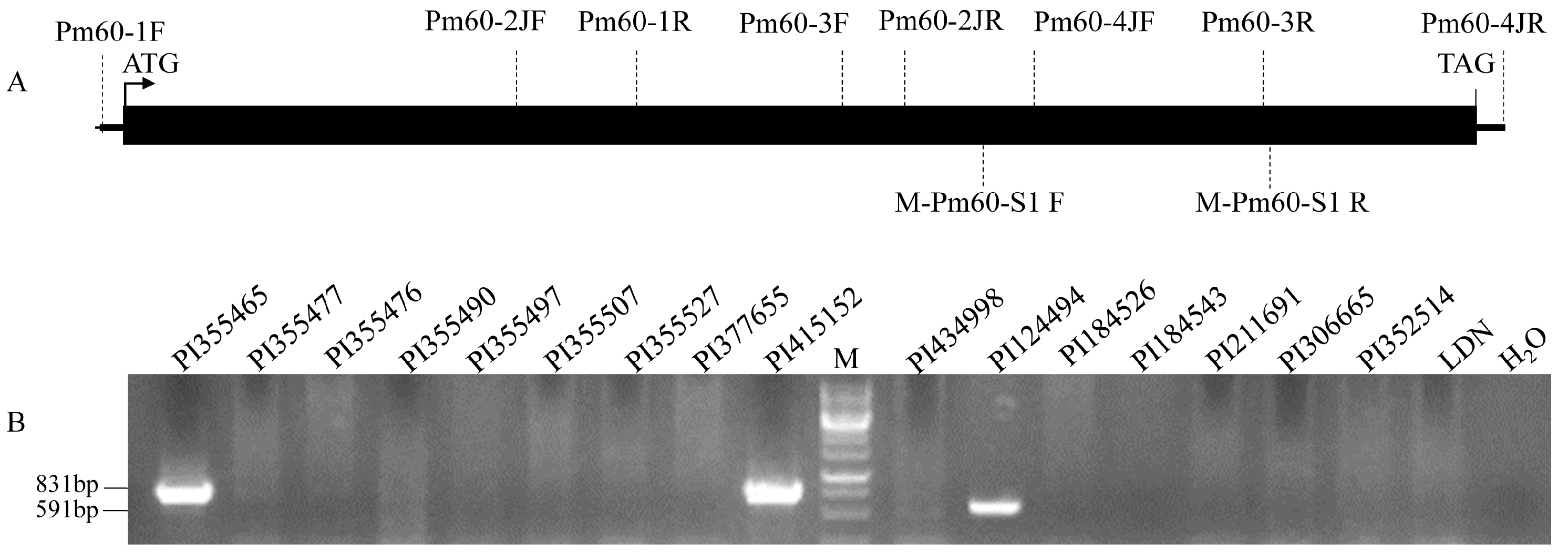
2.2. Genomic DNA Extraction and Marker Analysis
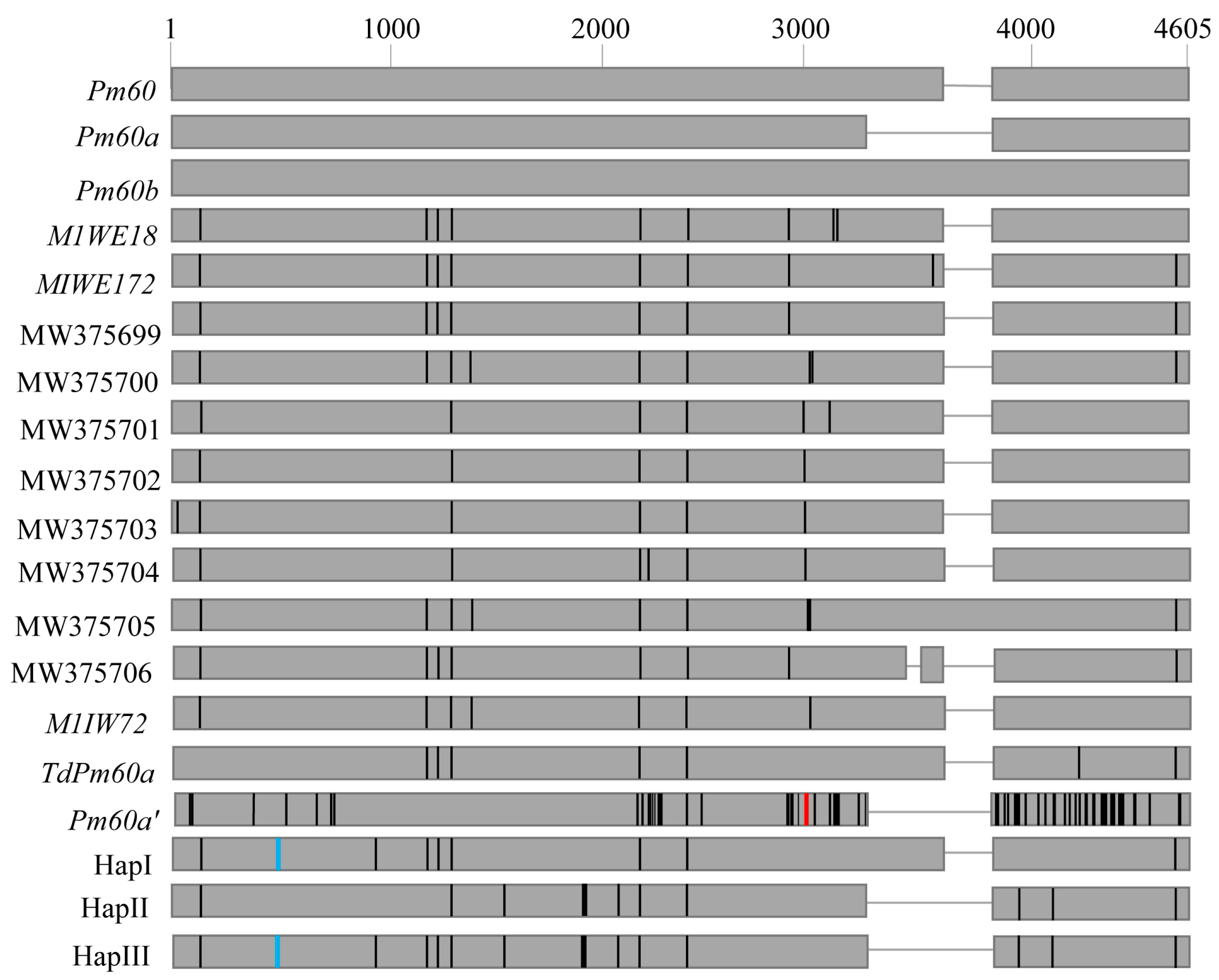
2.3. Sequence Analysis of Pm60
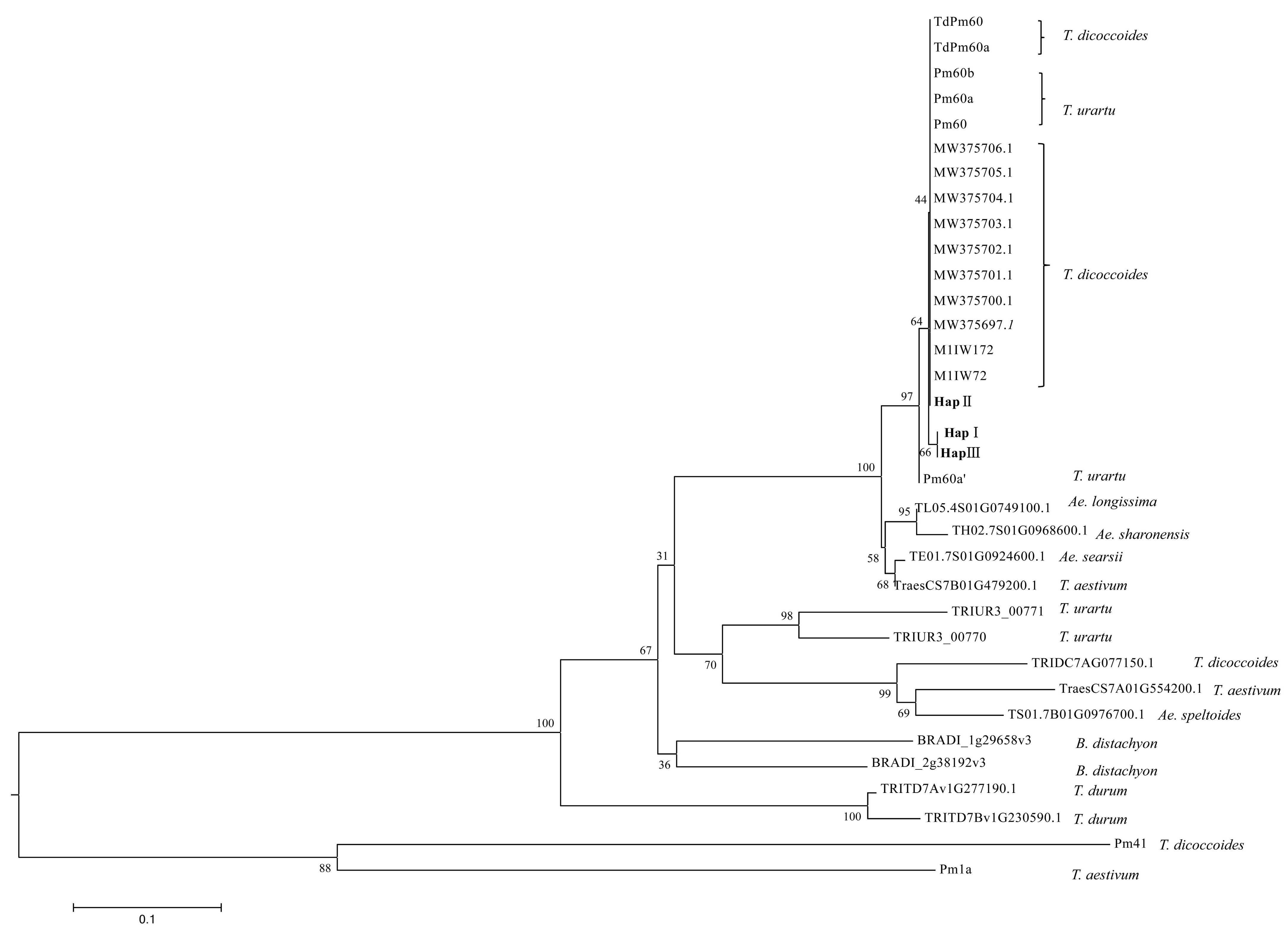
2.4. Phylogenetic Analysis
2.5. Powdery Mildew Resistance Response
3. Results
3.1. Distribution of Pm60 in Wheat Species
3.2. Sequence Variation of Pm60
3.3. Phylogenetic Analysis of Pm60
3.4. Powdery Mildew Responses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, M.; Braun, U.; Takamatsu, S.; Hambleton, S.; Shoukouhi, P.; Bisson, K.R.; Hubbard, K. Taxonomic revision of Blumeria based on multi-gene DNA sequences, host preferences and morphology. Mycoscience 2021, 62, 143–165. [Google Scholar] [CrossRef]
- Singh, R.P.; Singh, P.K.; Rutkoski, J.; Hodson, D.P.; He, X.Y.; Jorgensen, L.N.; Hovmoller, M.S.; Huerta-Espino, J. Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 2016, 54, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, Y.; Fahima, T.; Shen, Q.; Xie, C. Introgression of the powdery mildew resistance genes Pm60 and Pm60b from Triticum urartu to common wheat using durum as a ‘Bridge’. Pathogens 2021, 11, 25. [Google Scholar] [CrossRef]
- Wicker, T.; Oberhaensli, S.; Parlange, F.; Buchmann, J.P.; Shatalina, M.; Roffler, S.; Ben-David, R.; Dolezel, J.; Simkova, H.; Schulze-Lefert, P.; et al. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 2013, 45, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Liu, R.; Ma, P.; Du, H.; Zhang, H.; Wu, Q.; Yang, L.; Gong, S.; Liu, T.; Huo, N.; et al. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor Appl Genet 2021, 134, 53–62. [Google Scholar] [CrossRef]
- Li, Y.; Wei, Z.-Z.; Sela, H.; Govta, L.; Klymiuk, V.; Roychowdhury, R.; Singh Chawla, H.; Ens, J.; Wiebe, K.; Bocharova, V.; et al. Long-read genome sequencing accelerated the cloning of Pm69 by resolving the complexity of a rapidly evolving resistance gene cluster in wheat. bioRxiv 2022. bioRxiv: 14.512294. [Google Scholar] [CrossRef]
- Zou, S.H.; Wang, H.; Li, Y.W.; Kong, Z.S.; Tang, D.Z. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2018, 218, 298–309. [Google Scholar] [CrossRef]
- Li, Y.H.; Wei, Z.Z.; Fatiukha, A.; Jaiwar, S.; Wang, H.C.; Hasan, S.; Liu, Z.Y.; Sela, H.; Krugman, T.; Fahima, T. TdPm60 identified in wild emmer wheat is an ortholog of Pm60 and constitutes a strong candidate for PmG16 powdery mildew resistance. Theor. Appl. Genet. 2021, 134, 2777–2793. [Google Scholar] [CrossRef]
- Men, W.Q.; Fan, Z.W.; Ma, C.; Zhao, Y.; Wang, C.L.; Tian, X.B.; Chen, Q.F.; Miao, J.N.; He, J.Q.; Qian, J.J.; et al. Mapping of the novel powdery mildew resistance gene Pm2Mb from Aegilops biuncialis based on ph1b-induced homoeologous recombination. Theor. Appl. Genet 2022, 135, 2993–3003. [Google Scholar] [CrossRef]
- Zou, S.H.; Shi, W.Q.; Ji, J.H.; Wang, H.M.; Tang, Y.S.; Yu, D.Z.; Tang, D.Z. Diversity and similarity of wheat powdery mildew resistance among three allelic functional genes at the Pm60 locus. Plant J. 2022, 110, 1781–1790. [Google Scholar] [CrossRef]
- Zhao, F.K.; Li, Y.H.; Yang, B.J.; Yuan, H.B.; Jin, C.; Zhou, L.X.; Pei, H.C.; Zhao, L.F.; Li, Y.W.; Zhou, Y.L.; et al. Powdery mildew disease resistance and marker-assisted screening at the Pm60 locus in wild diploid wheat Triticum urartu. Crop J. 2020, 8, 252–259. [Google Scholar] [CrossRef]
- Wu, Q.H.; Zhao, F.; Chen, Y.X.; Zhang, P.P.; Zhang, H.Z.; Guo, G.H.; Xie, J.Z.; Dong, L.L.; Lu, P.; Li, M.M.; et al. Bulked segregant CGT-Seq-facilitated map-based cloning of a powdery mildew resistance gene originating from wild emmer wheat (Triticum dicoccoides). Plant Biotechnol. J. 2021, 19, 1288–1290. [Google Scholar] [CrossRef]
- Ji, X.L.; Xie, C.J.; Ni, Z.F.; Yang, T.M.; Nevo, E.; Fahima, T.; Liu, Z.Y.; Sun, Q.X. Identification and genetic mapping of a powdery mildew resistance gene in wild emmer (Triticum dicoccoides) accession IW72 from Israel. Euphytica 2008, 159, 385–390. [Google Scholar] [CrossRef]
- Wu, Q.H.; Chen, Y.X.; Li, B.B.; Li, J.; Zhang, P.P.; Xie, J.Z.; Zhang, H.Z.; Guo, G.H.; Lu, P.; Li, M.M.; et al. Functional characterization of powdery mildew resistance gene MlIW172, a new Pm60 allele and its allelic variation in wild emmer wheat. J. Genet. Genomics 2022, 49, 787–795. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Liu, D.C.; Yan, Z.H.; Lan, X.J.; Zheng, Y.L.; Zhou, Y.H. Rapid changes of microsatellite flanking sequence in the allopolyploidization of new synthesized hexaploid wheat. Sci. China C. Life Sci. 2004, 47, 553–561. [Google Scholar] [CrossRef]
- Huang, L.; Sela, H.; Feng, L.H.; Chen, Q.J.; Krugman, T.; Yan, J.; Dubcovsky, J.; Fahima, T. Distribution and haplotype diversity of WKS resistance genes in wild emmer wheat natural populations. Theor. Appl. Genet. 2016, 129, 921–934. [Google Scholar] [CrossRef]
- Hall, T. BioEdit, Version 7.0. 9. Computer program and documentation. Lbis Biosciences: Carlsbad, CA, USA, 2007.
- Peng, J.H.; Sun, D.F.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281–301. [Google Scholar] [CrossRef]
- Levy, A.A.; Feldman, M. Evolution and origin of bread wheat. Plant Cell 2022, 34, 2549–2567. [Google Scholar] [CrossRef]
- Luo, M.C.; Yang, Z.L.; You, F.M.; Kawahara, T.; Waines, J.G.; Dvorak, J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 2007, 114, 947–959. [Google Scholar] [CrossRef]
- Koller, T.; Brunner, S.; Herren, G.; Hurni, S.; Keller, B. Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor. Appl. Genet. 2018, 131, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhang, M.H.; Hu, Y.L.; Gan, M.J.; Jiang, B.; Hao, M.; Ning, S.Z.; Yuan, Z.W.; Chen, X.J.; Chen, X.; et al. Pyramiding of adult-plant resistance genes enhances all-stage resistance to wheat stripe rust. Plant Dis. 2023, in press. [Google Scholar] [CrossRef] [PubMed]
| □ | □ | HapI | HapII | □ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bgt Isolates | G18-16 (TdPm60) | PI352335 | PI355465 | PI415152 | CItr7685 | PI190949 | PI113392 | PI124494 | AS285 | Chinese Spring |
| Bgt#1-25 | ND | 4 | 4 | 4 | 3 | 4 | 4 | 4 | 3 | 4 |
| Bgt#15-9-4 | ND | 4 | 4 | 4 | 4 | 3 | 4 | 4 | 3 | 4 |
| Bgt#SZS | 0 | 3 | 4 | 4 | 4 | 4 | 3 | 4 | 4 | 4 |
| Bgt#WJ | 0 | 3 | 4 | 4 | 4 | 3 | 4 | 4 | 4 | 4 |
| Bgt#GH | 0 | 4 | 4 | 3 | 4 | 2 | 2 | 2 | 2 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Jin, X.; Ren, X.; Wu, W.; Ji, W.; Feng, L.; Jiang, B.; Hao, M.; Ning, S.; Yuan, Z.; et al. Haplotype Analysis Sheds Light on the Genetic Evolution of the Powdery Mildew Resistance Locus Pm60 in Triticum Species. Pathogens 2023, 12, 241. https://doi.org/10.3390/pathogens12020241
Huang X, Jin X, Ren X, Wu W, Ji W, Feng L, Jiang B, Hao M, Ning S, Yuan Z, et al. Haplotype Analysis Sheds Light on the Genetic Evolution of the Powdery Mildew Resistance Locus Pm60 in Triticum Species. Pathogens. 2023; 12(2):241. https://doi.org/10.3390/pathogens12020241
Chicago/Turabian StyleHuang, Xuhui, Xueli Jin, Xiaojie Ren, Wenxuan Wu, Wenjun Ji, Lihua Feng, Bo Jiang, Ming Hao, Shunzong Ning, Zhongwei Yuan, and et al. 2023. "Haplotype Analysis Sheds Light on the Genetic Evolution of the Powdery Mildew Resistance Locus Pm60 in Triticum Species" Pathogens 12, no. 2: 241. https://doi.org/10.3390/pathogens12020241






