Drosophila melanogaster Systemic Infection Model to Study Altered Virulence during Polymicrobial Infection by Aeromonas
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Collection and Culture Conditions
2.2. Drosophila melanogaster Infection Model
2.3. Quantitative Microbiology
| Strains Characteristics | Context of Strain Isolation | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Strain | Region, Country, Year of Isolation | Pair | Origin | Patient No. | Type of Infection | Age/Sex | Comorbidities and Predisposing Factors | Comments |
| A. rivipollensis * | 76c | Barcelona, Spain, 1992 | 1 | Stool | 1 | Diarrhea with fever and unremarkable presentation | 5/M | Hydrocephalus and ventricular-peritoneal shunt, catheter-related coinfection | Aeromonads considered by the clinician as responsible for the diarrhea after no other pathogen was isolated during the stool culture bacterial screening |
| A. veronii | 77c | Barcelona, Spain, 1992 | 1 | Stool | |||||
| A. hydrophyla | 25a | Saint-Brieuc, France, 2006 | 2 | Respiratory tract | 2 | No infection (colonization) | 62/M | Bronchiectasis Corticosteroid treatment [30] | Clinician decision not to treat [30] |
| A. veronii | 25b | Saint-Brieuc, France, 2006 | 2 | Respiratory tract | |||||
| A. veronii | 186 | Montpellier, France, 2015 | 3 | Blood | 3 | Bloodstream infection with septic shock | 72/M | Cardiopathy Chronic kidney failure History of cholecystectomy following a large cholelithiasis with a resulting blind loop of the bowel | Clinician decision to administer aeromonad-targeted antimicrobial treatment |
| E. coli | 187 | Montpellier, France, 2015 | 3 | Blood | |||||
2.4. Statistical Analysis
3. Results
3.1. Characteristics of the Infections
3.2. Characteristics of Individual Strain Virulence
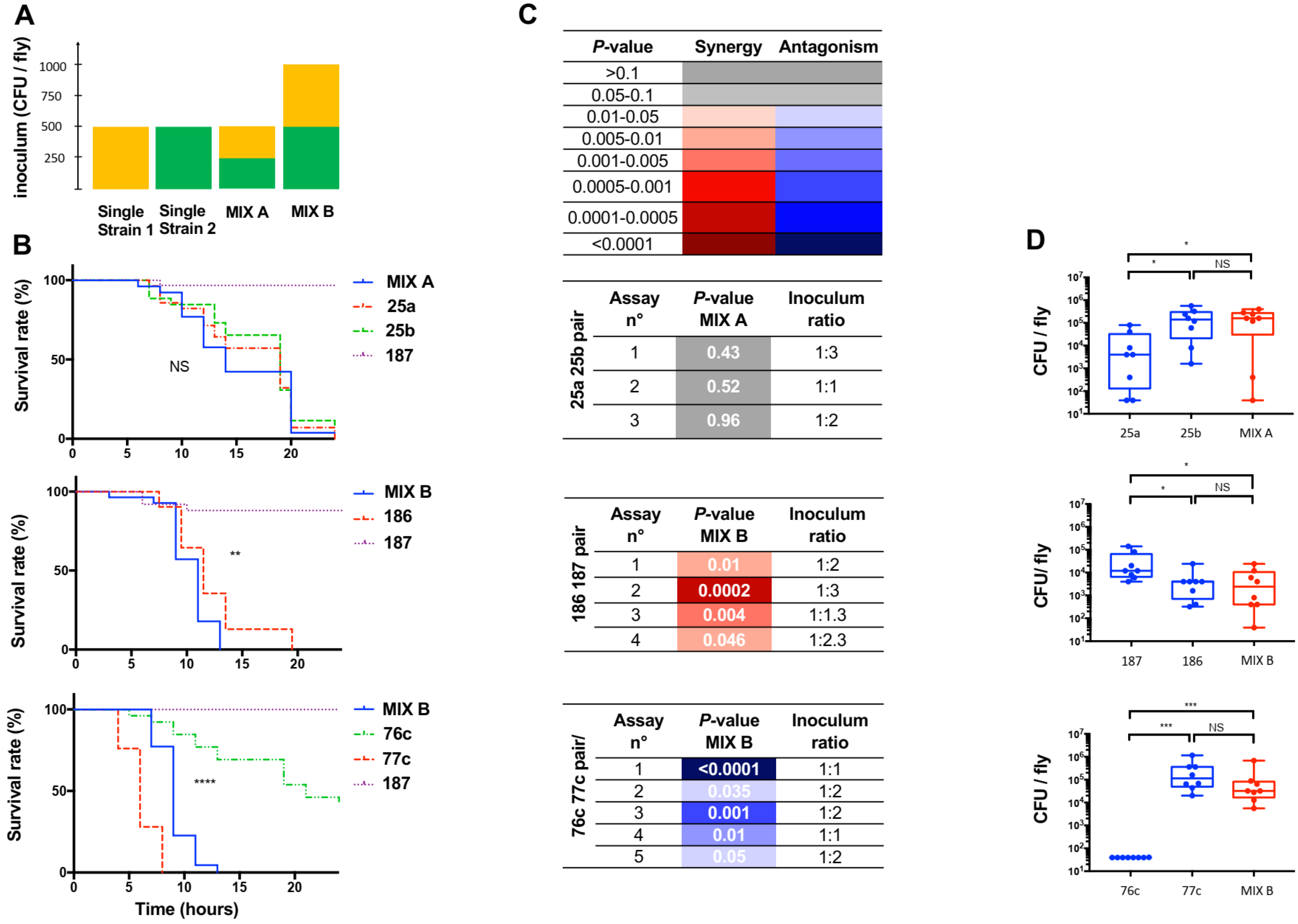
3.3. Assessing the Effect of Polymicrobial Situation on the Infection Outcome
3.4. Screening Bacterial Relationships during Polymicrobial Infection in the Host
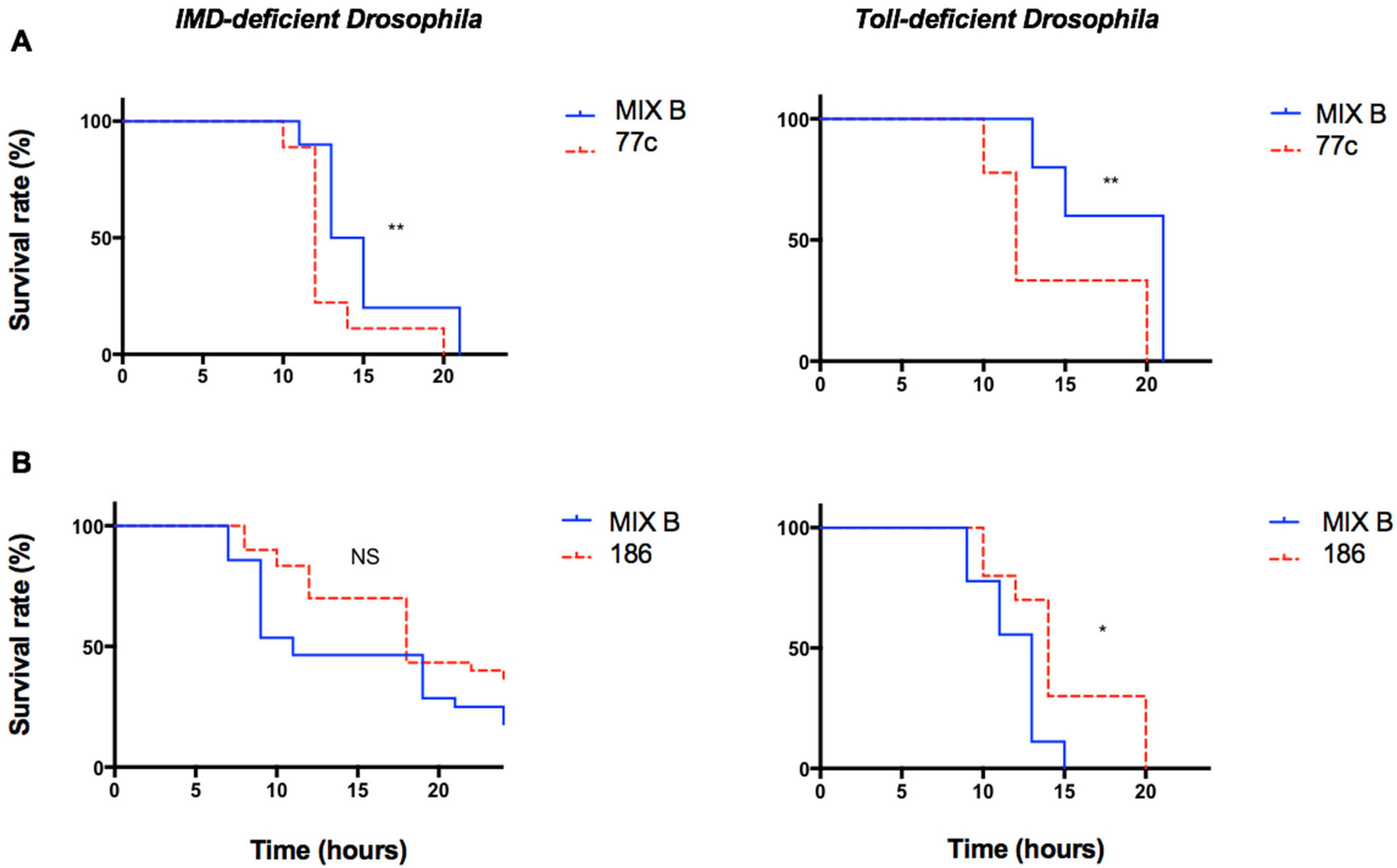
3.5. Exploring the Role of Innate Immunity during Polymicrobial Infection
4. Discussion
4.1. Drosophila Melanogaster, a Model Organism to Study Polymicrobial Infection
4.2. Characteristics, Possibilities, Limitations, and Weaknesses of the Fly Infection Model
4.3. The Polymicrobial Condition Modulates the Pathogenicity of Opportunistic Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukherjee, S.; Weimer, K.E.; Seok, S.-C.; Ray, W.C.; Jayaprakash, C.; Vieland, V.J.; Swords, W.E.; Das, J. Host-to-host variation of ecological interactions in polymicrobial infections. Phys. Biol. 2014, 12, 016003. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.; De Soyza, A.; Perry, J.D.; Sutcliffe, I.C.; Cummings, S.P. Polymicrobial challenges to Koch’s postulates: Ecological lessons from the bacterial vaginosis and cystic fibrosis microbiomes. Innate Immun. 2012, 18, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Short, F.L.; Murdoch, S.L.; Ryan, R.P. Polybacterial human disease: The ills of social networking. Trends Microbiol. 2014, 22, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef]
- Lin, J.-N.; Lai, C.-H.; Chen, Y.-H.; Chang, L.-L.; Lu, P.-L.; Tsai, S.-S.; Lin, H.-L.; Lin, H.-H. Characteristics and outcomes of polymicrobial bloodstream infections in the emergency department: A matched case-control study. Acad. Emerg. Med. 2010, 17, 1072–1079. [Google Scholar] [CrossRef]
- Learman, B.S.; Brauer, A.L.; Eaton, K.A.; Armbruster, C.E. A Rare Opportunist, Morganella morganii, Decreases Severity of Polymicrobial Catheter-Associated Urinary Tract Infection. Infect. Immun. 2019, 88, e00691-19. [Google Scholar] [CrossRef]
- Aujoulat, F.; Roger, F.; Bourdier, A.; Lotthé, A.; Lamy, B.; Marchandin, H.; Jumas-Bilak, E. From environment to man: Genome evolution and adaptation of human opportunistic bacterial pathogens. Genes 2012, 3, 191–232. [Google Scholar] [CrossRef]
- Korgaonkar, A.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 1059–1064. [Google Scholar] [CrossRef]
- Ponnusamy, D.; Kozlova, E.V.; Sha, J.; Erova, T.E.; Azar, S.R.; Fitts, E.C.; Kirtley, M.L.; Tiner, B.L.; Andersson, J.A.; Grim, C.J.; et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. USA 2016, 113, 722–727. [Google Scholar] [CrossRef]
- Rezzoagli, C.; Granato, E.T.; Kümmerli, R. Harnessing bacterial interactions to manage infections: A review on the opportunistic pathogen Pseudomonas aeruginosa as a case example. J. Med. Microbiol. 2020, 69, 147–161. [Google Scholar] [CrossRef]
- Sibley, C.D.; Duan, K.; Fischer, C.; Parkins, M.D.; Storey, D.G.; Rabin, H.R.; Surette, M.G. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008, 4, e1000184. [Google Scholar] [CrossRef] [PubMed]
- Mosser, T.; Talagrand-Reboul, E.; Colston, S.M.; Graf, J.; Figueras, M.J.; Jumas-Bilak, E.; Lamy, B. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front. Microbiol. 2015, 6, 1218. [Google Scholar] [CrossRef] [PubMed]
- Parlet, C.P.; Brown, M.M.; Horswill, A.R. Commensal Staphylococci Influence Staphylococcus aureus Skin Colonization and Disease. Trends Microbiol. 2019, 27, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L.-A. Virulence factors and their mechanisms of action: The view from a damage-response framework. J. Water Health 2009, 7 (Suppl. S1), S2–S18. [Google Scholar] [CrossRef]
- Tzelepis, I.; Kapsetaki, S.-E.; Panayidou, S.; Apidianakis, Y. Drosophila melanogaster: A first step and a stepping-stone to anti-infectives. Curr. Opin. Pharmacol. 2013, 13, 763–768. [Google Scholar] [CrossRef]
- Atilano, M.L.; Pereira, P.M.; Vaz, F.; Catalão, M.J.; Reed, P.; Grilo, I.R.; Sobral, R.G.; Ligoxygakis, P.; Pinho, M.G.; Filipe, S.R. Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. eLife 2014, 3, e02277. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Kilgore, P.B.; Andersson, J.A.; Blears, E.; Figueras, M.J.; Hasan, N.A.; Colwell, R.R.; Sha, J.; Chopra, A.K. T6SS and ExoA of flesh-eating Aeromonas hydrophila in peritonitis and necrotizing fasciitis during mono- and polymicrobial infections. Proc. Natl. Acad. Sci. USA 2019, 116, 24084–24092. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Jang, H.-J.; Chung, I.-Y.; Cho, Y.-H. Drosophila melanogaster as a polymicrobial infection model for Pseudomonas aeruginosa and Staphylococcus aureus. J. Microbiol. 2018, 56, 534–541. [Google Scholar] [CrossRef]
- Galac, M.R.; Lazzaro, B.P. Comparative pathology of bacteria in the genus Providencia to a natural host, Drosophila melanogaster. Microbes Infect. 2011, 13, 673–683. [Google Scholar] [CrossRef]
- Panayidou, S.; Ioannidou, E.; Apidianakis, Y. Human pathogenic bacteria, fungi, and viruses in Drosophila: Disease modeling, lessons, and shortcomings. Virulence 2014, 5, 253–269. [Google Scholar] [CrossRef]
- Talagrand-Reboul, E.; Colston, S.M.; Graf, J.; Lamy, B.; Jumas-Bilak, E. Comparative and Evolutionary Genomics of Isolates Provide Insight into the Pathoadaptation of Aeromonas. Genome Biol. Evol. 2020, 12, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Talagrand-Reboul, E.; Latif-Eugenín, F.; Beaz-Hidalgo, R.; Colston, S.; Figueras, M.-J.; Graf, J.; Jumas-Bilak, E.; Lamy, B. Genome-driven evaluation and redesign of PCR tools for improving the detection of virulence-associated genes in aeromonads. PLoS ONE 2018, 13, e0201428. [Google Scholar] [CrossRef] [PubMed]
- Noonin, C.; Jiravanichpaisal, P.; Söderhäll, I.; Merino, S.; Tomás, J.M.; Söderhäll, K. Melanization and pathogenicity in the insect, Tenebrio molitor, and the crustacean, Pacifastacus leniusculus, by Aeromonas hydrophila AH-3. PLoS ONE 2010, 5, e15728. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kozlova, E.V.; Sha, J.; Fitts, E.C.; van Lier, C.J.; Kirtley, M.L.; Joseph, S.J.; Read, T.D.; Burd, E.M.; Tall, B.D.; et al. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. mBio 2013, 4, e00064-13. [Google Scholar] [CrossRef]
- Barraud, O.; Robert, A.; Laval, L.; Ruimy, R.; Morquin, D.; Boyer, L.; Lamy, B. It takes two to tango: Two Aeromonas isolates combine virulence and multidrug resistance in flap infection following leech therapy. Clin. Microbiol. Infect. 2020, 26, 793–794. [Google Scholar] [CrossRef]
- Colston, S.M.; Fullmer, M.S.; Beka, L.; Lamy, B.; Gogarten, J.P.; Graf, J. Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. mBio 2014, 5, e02136. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Jia, Y.; Jin, S.; Hu, K.; Geng, L.; Han, C.; Kang, R.; Pang, Y.; Ling, E.; Tan, E.K.; Pan, Y.; et al. Gut microbiome modulates Drosophila aggression through octopamine signaling. Nat. Commun. 2021, 12, 2698. [Google Scholar] [CrossRef]
- Arias-Rojas, A.; Iatsenko, I. The Role of Microbiota in Drosophila melanogaster Aging. Front. Aging 2022, 3, 57. [Google Scholar] [CrossRef]
- Lamy, B.; Kodjo, A.; colBVH Study Group; Laurent, F. Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 2009, 47, 1234–1237. [Google Scholar] [CrossRef]
- Talagrand-Reboul, E.; Jumas-Bilak, E.; Lamy, B. The Social Life of Aeromonas through Biofilm and Quorum Sensing Systems. Front. Microbiol. 2017, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Balcázar, J.L. Aeromonas rivipollensis sp. nov., a novel species isolated from aquatic samples. J. Basic Microbiol. 2015, 55, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Ngba Essebe, C.; Visvikis, O.; Fines-Guyon, M.; Vergne, A.; Cattoir, V.; Lecoustumier, A.; Lemichez, E.; Sotto, A.; Lavigne, J.-P.; Dunyach-Remy, C. Decrease of Staphylococcus aureus Virulence by Helcococcus kunzii in a Caenorhabditis elegans Model. Front. Cell. Infect. Microbiol. 2017, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Kinnula, H.; Mappes, J.; Sundberg, L.-R. Coinfection outcome in an opportunistic pathogen depends on the inter-strain interactions. BMC Evol. Biol. 2017, 17, 77. [Google Scholar] [CrossRef]
- Sheehan, G.; Tully, L.; Kavanagh, K.A. Candida albicans increases the pathogenicity of Staphylococcus aureus during polymicrobial infection of Galleria mellonella larvae. Microbiology 2020, 166, 375–385. [Google Scholar] [CrossRef]
- Brunke, S.; Quintin, J.; Kasper, L.; Jacobsen, I.D.; Richter, M.E.; Hiller, E.; Schwarzmüller, T.; d’Enfert, C.; Kuchler, K.; Rupp, S.; et al. Of mice, flies--and men? Comparing fungal infection models for large-scale screening efforts. Dis. Model. Mech. 2015, 8, 473–486. [Google Scholar] [CrossRef]
- Yung, D.B.Y.; Sircombe, K.J.; Pletzer, D. Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol. 2021, 116, 1–15. [Google Scholar] [CrossRef]
- Troha, K.; Buchon, N. Methods for the study of innate immunity in Drosophila melanogaster. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e344. [Google Scholar] [CrossRef]
- Fauvarque, M.-O.; Bergeret, E.; Chabert, J.; Dacheux, D.; Satre, M.; Attree, I. Role and activation of type III secretion system genes in Pseudomonas aeruginosa—Induced Drosophila killing. Microb. Pathog. 2002, 32, 287–295. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Rahme, L.G. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 2009, 4, 1285–1294. [Google Scholar] [CrossRef]
- Roger, F.; Lamy, B.; Jumas-Bilak, E.; Kodjo, A.; colBVH Study Group; Marchandin, H. Ribosomal multi-operon diversity: An original perspective on the genus Aeromonas. PLoS ONE 2012, 7, e46268. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, M.S.; Schoeb, T.; Swords, W.E. Cooperativity between Stenotrophomonas maltophilia and Pseudomonas aeruginosa during Polymicrobial Airway Infections. Infect. Immun. 2020, 88, e00855-19. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.-W.; Lai, Y.-K.; Liu, Y.-T.; Gallo, R.L.; Huang, C.-M. Staphylococcus aureus hijacks a skin commensal to intensify its virulence: Immunization targeting β-hemolysin and CAMP factor. J. Investig. Dermatol. 2011, 131, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Todd, O.A.; Peters, B.M. Candida albicans and Staphylococcus aureus Pathogenicity and Polymicrobial Interactions: Lessons beyond Koch’s Postulates. J. Fungi 2019, 5, 81. [Google Scholar] [CrossRef]
- Rumbaugh, K.P.; Trivedi, U.; Watters, C.; Burton-Chellew, M.N.; Diggle, S.P.; West, S.A. Kin selection, quorum sensing and virulence in pathogenic bacteria. Proc. Biol. Sci. 2012, 279, 3584–3588. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robert, A.; Talagrand-Reboul, E.; Figueras, M.-J.; Ruimy, R.; Boyer, L.; Lamy, B. Drosophila melanogaster Systemic Infection Model to Study Altered Virulence during Polymicrobial Infection by Aeromonas. Pathogens 2023, 12, 405. https://doi.org/10.3390/pathogens12030405
Robert A, Talagrand-Reboul E, Figueras M-J, Ruimy R, Boyer L, Lamy B. Drosophila melanogaster Systemic Infection Model to Study Altered Virulence during Polymicrobial Infection by Aeromonas. Pathogens. 2023; 12(3):405. https://doi.org/10.3390/pathogens12030405
Chicago/Turabian StyleRobert, Alexandre, Emilie Talagrand-Reboul, Maria-Jose Figueras, Raymond Ruimy, Laurent Boyer, and Brigitte Lamy. 2023. "Drosophila melanogaster Systemic Infection Model to Study Altered Virulence during Polymicrobial Infection by Aeromonas" Pathogens 12, no. 3: 405. https://doi.org/10.3390/pathogens12030405
APA StyleRobert, A., Talagrand-Reboul, E., Figueras, M.-J., Ruimy, R., Boyer, L., & Lamy, B. (2023). Drosophila melanogaster Systemic Infection Model to Study Altered Virulence during Polymicrobial Infection by Aeromonas. Pathogens, 12(3), 405. https://doi.org/10.3390/pathogens12030405








