Cellular Immune Profiling of Lung and Blood Compartments in Patients with SARS-CoV-2 Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Specimen Processing and Cell Isolation
2.3. Immunophenotyping by Multiparametric Flow Cytometry assay
2.4. Statistical Analysis
3. Results
3.1. Study Population
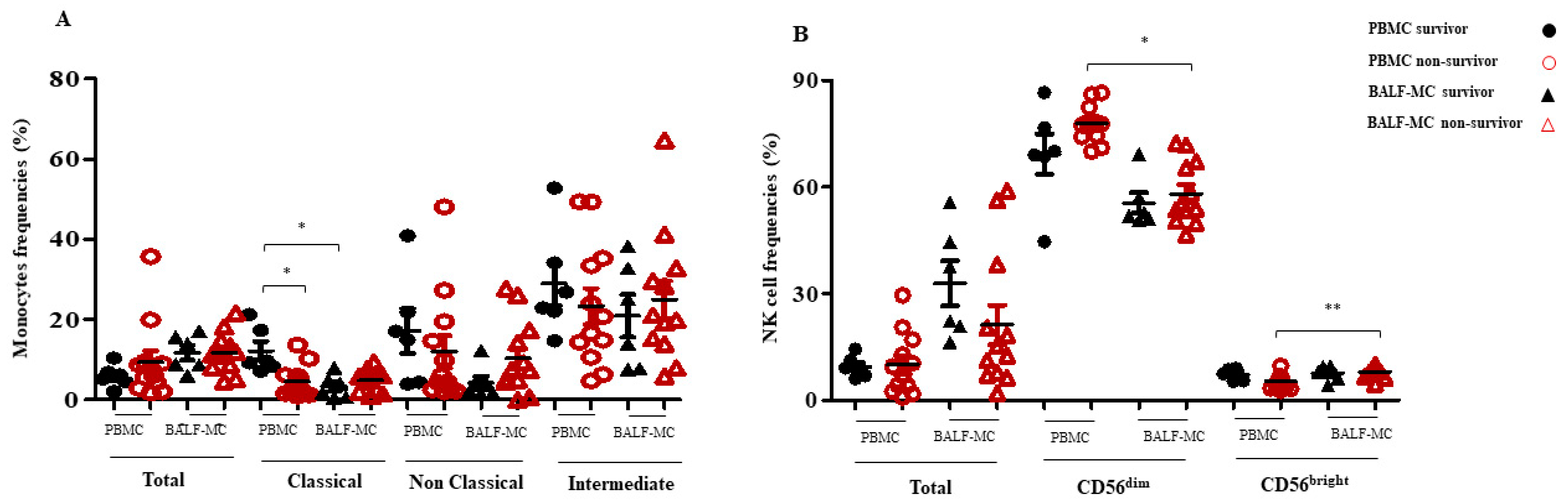
3.2. Monocyte and NK Cell Subset Profiles in BALF-MC and PBMC of SARS-CoV-2-Infected Survivors and Non-Survivors
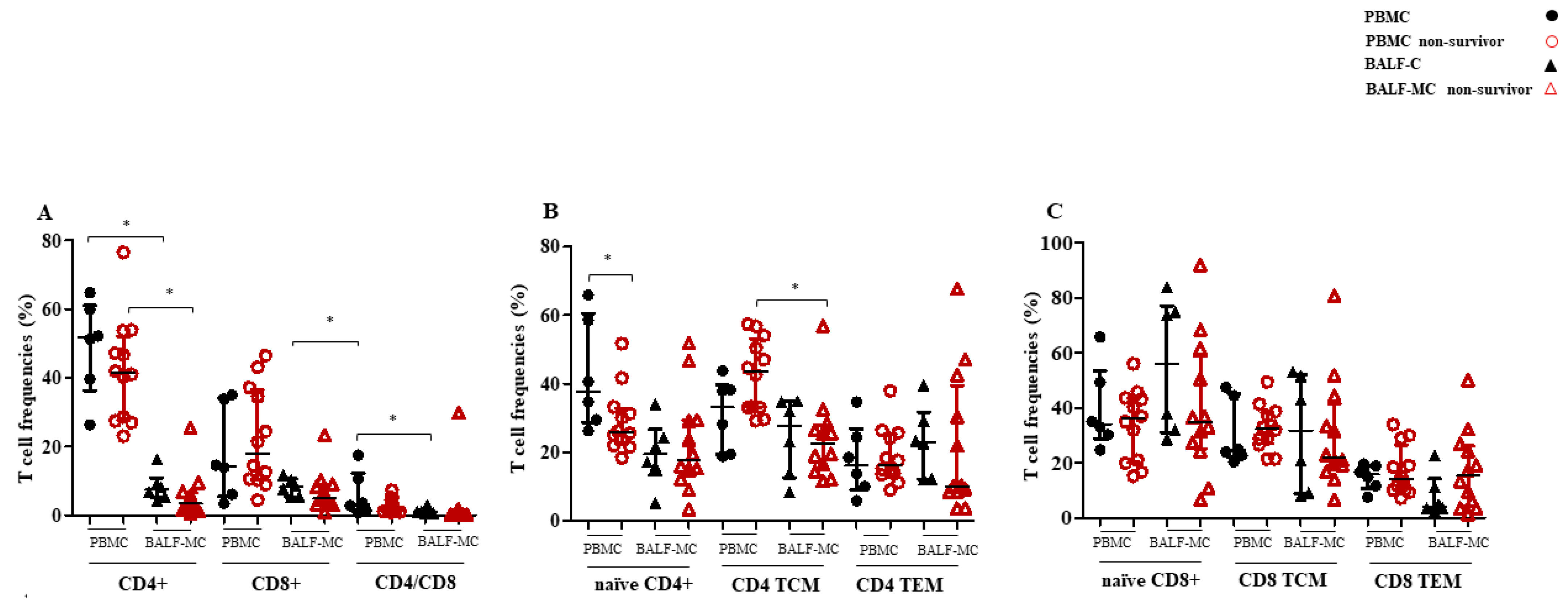
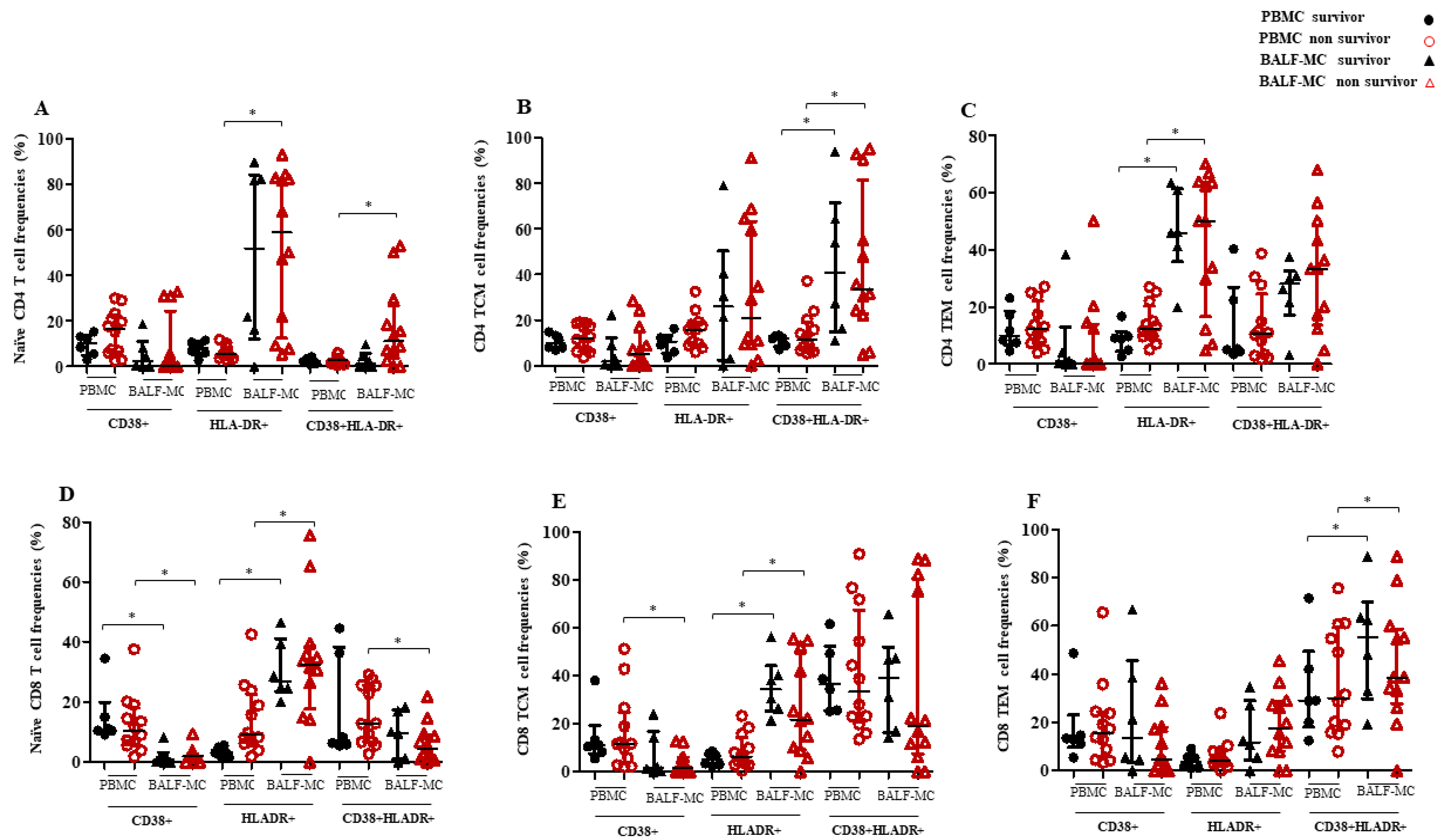
3.3. T Cell Subset Profiles and Immune Activation in BALF-MC and PBMC of SARS-CoV-2-Infected Survivors and Non-Survivors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef] [PubMed]
- Lim, Z.J.; Subramaniam, A.; Reddy, M.P.; Blecher, G.; Kadam, U.; Afroz, A.; Billah, B.; Ashwin, S.; Kubicki, M.; Bilotta, F.; et al. Case Fatality Rates for Patients with COVID-19 Requiring Invasive Mechanical Ventilation. A Meta-analysis. Am. J. Respir. Crit. Care Med. 2021, 203, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, C.J.; Wooster, L.; Sigurslid, H.H.; Li, R.H.; Jiang, W.; Tian, W.; Cardenas, C.L.L.; Malhotra, R. Estimating risk of mechanical ventilation and in-hospital mortality among adult COVID-19 patients admitted to Mass General Brigham: The VICE and DICE scores. Eclinicalmedicine 2021, 33, 100765. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Annunziato, F.; Cosmi, L. Hallmarks of immune response in COVID-19: Exploring dysregulation and exhaustion. Semin. Immunol. 2021, 55, 101508. [Google Scholar] [CrossRef]
- Pierangeli, A.; Gentile, M.; Oliveto, G.; Frasca, F.; Sorrentino, L.; Matera, L.; Nenna, R.; Viscido, A.; Fracella, M.; Petrarca, L.; et al. Comparison by Age of the Local Interferon Response to SARS-CoV-2 Suggests a Role for IFN-ε and -ω. Front. Immunol. 2022, 13, 873232. [Google Scholar] [CrossRef]
- Frasca, F.; Scordio, M.; Santinelli, L.; Gabriele, L.; Gandini, O.; Criniti, A.; Pierangeli, A.; Angeloni, A.; Mastroianni, C.M.; d’Ettorre, G.; et al. Anti-IFN-α/-ω neutralizing antibodies from COVID-19 patients correlate with downregulation of IFN response and laboratory biomarkers of disease severity. Eur. J. Immunol. 2022, 52, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Codd, A.S.; Hanna, S.J.; Compeer, E.B.; Richter, F.C.; Pring, E.J.; Gea-Mallorquí, E.; Borsa, M.; Moon, O.R.; Scourfield, D.O.; Oxford-Cardiff COVID-19 Literature Consortium; et al. Neutrophilia, lymphopenia and myeloid dysfunction: A living review of the quantitative changes to innate and adaptive immune cells which define COVID-19 pathology. Oxf. Open Immunol. 2021, 15, iqab016. [Google Scholar] [CrossRef]
- Schultze, J.L.; Aschenbrenner, A.C. COVID-19 and the human innate immune system. Cell 2021, 184, 1671–1692. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Dentone, C.; Vena, A.; Loconte, M.; Grillo, F.; Brunetti, I.; Barisione, E.; Tedone, E.; Mora, S.; Di Biagio, A.; Orsi, A.; et al. Bronchoalveolar lavage fluid characteristics and outcomes of invasively mechanically ventilated patients with COVID-19 pneumonia in Genoa, Italy. BMC Infect. Dis. 2021, 21, 353. [Google Scholar] [CrossRef] [PubMed]
- Saris, A.; Reijnders, T.D.Y.; Nossent, E.J.; Schuurman, A.R.; Verhoeff, J.; van Asten, S.; Bontkes, H.; Blok, S.; Duitman, J.; Bogaard, H.-J.; et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax 2021, 76, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Szabo, P.A.; Dogra, P.; Gray, J.I.; Wells, S.B.; Connors, T.J.; Weisberg, S.P.; Krupska, I.; Matsumoto, R.; Poon, M.M.; Idzikowski, E.; et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity 2021, 54, 797–814.e6. [Google Scholar] [CrossRef]
- Puk, O.; Nowacka, A.; Smulewicz, K.; Mocna, K.; Bursiewicz, W.; Kęsy, N.; Kwiecień, J.; Wiciński, M. Pulmonary artery targeted therapy in treatment of COVID-19 related ARDS. Literature review. Biomed. Pharmacother. 2022, 146, 112592. [Google Scholar] [CrossRef]
- Bos, L.D.J.; Sjoding, M.; Sinha, P.; Bhavani, S.V.; Lyons, P.G.; Bewley, A.F.; Botta, M.; Tsonas, A.M.; Serpa Neto, A.; PRoVENT-COVID Collaborative Group; et al. Longitudinal respiratory subphenotypes in patients with COVID-19-related acute respiratory distress syndrome: Results from three observational cohorts. Lancet Respir. Med. 2021, 9, 1377–1386. [Google Scholar] [CrossRef]
- Carsana, L.; Sonzogni, A.; Nasr, A.; Rossi, R.S.; Pellegrinelli, A.; Zerbi, P.; Rech, R.; Colombo, R.; Antinori, S.; Corbellino, M.; et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infect Dis. 2020, 20, 1135–1140. [Google Scholar] [CrossRef]
- Doglioni, C.; Ravaglia, C.; Chilosi, M.; Rossi, G.; Dubini, A.; Pedica, F.; Piciucchi, S.; Vizzuso, A.; Stella, F.; Maitan, S.; et al. Covid-19 Interstitial Pneumonia: Histological and Immunohistochemical Features on Cryobiopsies. Respiration 2021, 100, 488–498. [Google Scholar] [CrossRef]
- D’Ettorre, G.; Recchia, G.; Ridolfi, M.; Siccardi, G.; Pinacchio, C.; Innocenti, G.P.; Santinelli, L.; Frasca, F.; Bitossi, C.; Ceccarelli, G.; et al. Analysis of type I IFN response and T cell activation in severe COVID-19/HIV-1 coinfection: A case report. Medicine 2020, 99, e21803. [Google Scholar] [CrossRef]
- Puliani, G.; Hasenmajer, V.; Sciarra, F.; Barbagallo, F.; Sbardella, E.; Pofi, R.; Gianfrilli, D.; Romagnoli, E.; Venneri, M.A.; Isidori, A.M. Impaired Immune Function in Patients with Chronic Postsurgical Hypoparathyroidism: Results of the EMPATHY Study. J. Clin. Endocrinol. Metab. 2021, 106, e2215–e2227. [Google Scholar] [CrossRef]
- Pinacchio, C.; Scheri, G.C.; Statzu, M.; Santinelli, L.; Ceccarelli, G.; Innocenti, G.P.; Vullo, V.; Antonelli, G.; Brenchley, J.M.; d’Ettorre, G.; et al. Type I/II Interferon in HIV-1-Infected Patients: Expression in Gut Mucosa and in Peripheral Blood Mononuclear Cells and Its Modification upon Probiotic Supplementation. J. Immunol. Res. 2018, 12, 1738676. [Google Scholar] [CrossRef]
- Santinelli, L.; Ceccarelli, G.; Borrazzo, C.; Innocenti, G.P.; Frasca, F.; Cavallari, E.N.; Celani, L.; Nonne, C.; Mastroianni, C.M.; d’Ettorre, G. Sex-related differences in markers of immune activation in virologically suppressed HIV-infected patients. Biol. Sex Differ. 2020, 11, 23. [Google Scholar] [CrossRef]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- Grifoni, E.; Valoriani, A.; Cei, F.; Vannucchi, V.; Moroni, F.; Pelagatti, L.; Tarquini, R.; Landini, G.; Masotti, L. The CALL Score for Predicting Outcomes in Patients with COVID-19. Clin. Infect. Dis. 2021, 72, 182–183. [Google Scholar] [CrossRef]
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and Macrophages in COVID-19. Front. Immunol. 2021, 21, 720109. [Google Scholar] [CrossRef]
- Krämer, B.; Knoll, R.; Bonaguro, L.; ToVinh, M.; Raabe, J.; Astaburuaga-García, R.; Schulte-Schrepping, J.; Melanie Kaiser, K.; Rieke, G.J.; Bischoff, J.; et al. Early IFN-alpha signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 2021, 54, 2650–2669.e14. [Google Scholar] [CrossRef]
- Niu, J.; Sareli, C.; Mayer, D.; Visbal, A.; Sareli, A. Lymphopenia as a Predictor for Adverse Clinical Outcomes in Hospitalized Patients with COVID-19: A Single Center Retrospective Study of 4485 Cases. J. Clin. Med. 2022, 11, 700. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, Z.; Prajapati, M.; Li, Y. Lymphopenia Caused by Virus Infections and the Mechanisms Beyond. Viruses 2021, 13, 1876. [Google Scholar] [CrossRef]
- Meidaninikjeh, S.; Sabouni, N.; Marzouni, H.Z.; Bengar, S.; Khalili, A.; Jafari, R. Monocytes and macrophages in COVID-19: Friends and foes. Life Sci. 2021, 269, 119010. [Google Scholar] [CrossRef]
- Sánchez-Cerrillo, I.; Landete, P.; Aldave, B.; Sánchez-Alonso, S.; Sánchez-Azofra, A.; Marcos-Jiménez, A.; Ávalos, E.; Alcaraz-Serna, A.; Santos, I.D.L.; Mateu-Albero, T.; et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J. Clin. Investig. 2020, 130, 6290–6300. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Karolinska COVID-19 Study Group; et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020, 21, eabd6832. [Google Scholar] [CrossRef]
- Li, M.; Guo, W.; Dong, Y.; Wang, X.; Dai, D.; Liu, X.; Wu, Y.; Li, M.; Zhang, W.; Zhou, H.; et al. Elevated Exhaustion Levels of NK and CD8+ T Cells as Indicators for Progression and Prognosis of COVID-19 Disease. Front. Immunol. 2020, 14, 580237. [Google Scholar] [CrossRef]
- Moon, C. Fighting COVID-19 exhausts T cells. Nat. Rev. Immunol. 2020, 20, 277. [Google Scholar] [CrossRef]
- Mortezaee, K.; Majidpoor, J. CD8+ T Cells in SARS-CoV-2 Induced Disease and Cancer—Clinical Perspectives. Front. Immunol. 2022, 13, 864298. [Google Scholar] [CrossRef]
- Mohammed, R.N.; Tamjidifar, R.; Rahman, H.S.; Adili, A.; Ghoreishizadeh, S.; Saeedi, H.; Thangavelu, L.; Shomali, N.; Aslaminabad, R.; Marofi, F.; et al. A comprehensive review about immune responses and exhaustion during coronavirus disease (COVID-19). Cell Commun. Signal. 2022, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Kusnadi, A.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Meckiff, B.J.; Simon, H.; Pelosi, E.; Seumois, G.; Ay, F.; Vijayanand, P.; et al. Severely ill patients with COVID-19 display impaired exhaustion features in SARS-CoV-2–reactive CD8+ T cells. Sci. Immunol. 2021, 6, eabe4782. [Google Scholar] [CrossRef]
- Urra, J.; Cabrera, C.; Porras, L.; Ródenas, I. Selective CD8 cell reduction by SARS-CoV-2 is associated with a worse prognosis and systemic inflammation in COVID-19 patients. Clin. Immunol. 2020, 217, 108486. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Rumende, C.M.; Sharma, S.K.; Rengganis, I.; Amin, Z.; Loho, T.; Hermiyanti, E.; Harimurti, K.; Wibowo, H. Low BALF CD4 T cells count is associated with extubation failure and mortality in critically ill covid-19 pneumonia. Ann. Med. 2022, 54, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.H.M.; Wong, V.W.S.; Wong, C.K.; Chan, P.; Chu, C.M.; Hui, D.; Suen, M.W.M.; Sung, J.J.Y.; Chung, S.S.C.; Lam, C.W.K. Serum LD1 isoenzyme and blood lymphocyte subsets as prognostic indicators for severe acute respiratory syndrome. J. Intern. Med. 2004, 255, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Wauters, E.; Van Mol, P.; Garg, A.D.; Jansen, S.; Van Herck, Y.; Vanderbeke, L.; Bassez, A.; Boeckx, B.; Malengier-Devlies, B.; Timmerman, A.; et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 2021, 31, 272–290. [Google Scholar] [CrossRef]
- Peng, X.; Ouyang, J.; Isnard, S.; Lin, J.; Fombuena, B.; Zhu, B.; Routy, J.-P. Sharing CD4+ T Cell Loss: When COVID-19 and HIV Collide on Immune System. Front. Immunol. 2020, 11, 596631. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef]
- Liu, Y.; Pang, Y.; Hu, Z.; Wu, M.; Wang, C.; Feng, Z.; Mao, C.; Tan, Y.; Liu, Y.; Chen, L.; et al. Thymosin Alpha 1 Reduces the Mortality of Severe Coronavirus Disease 2019 by Restoration of Lymphocytopenia and Reversion of Exhausted T Cells. Clin. Infect. Dis. 2020, 71, 2150–2157. [Google Scholar] [CrossRef]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef]
- Laing, A.G.; Lorenc, A.; del Barrio, I.D.M.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef]
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses prior to patient recovery: A case report of non-severe COVID-19. Nat. Med. 2020, 26, 453–455. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Z.; Liu, H.; Luo, Y.; Kang, K.; Li, X.; Yang, W.; Fei, D.; Wang, C.; Yu, K. Decreased T Cell Levels in Critically Ill Coronavirus Patients: Single-Center, Prospective and Observational Study. J. Inflamm. Res. 2021, 14, 1331–1340. [Google Scholar] [CrossRef]
- Ganji, A.; Farahani, I.; Khansarinejad, B.; Ghazavi, A.; Mosayebi, G. Increased expression of CD8 marker on T-cells in COVID-19 patients. Blood Cells Mol. Dis. 2020, 83, 102437. [Google Scholar] [CrossRef]
- Gelarden, I.; Nguyen, J.; Gao, J.; Chen, Q.; Morales-Nebreda, L.; Wunderink, R.; Li, L.; Chmiel, J.S.; Hrisinko, M.; Marszalek, L.; et al. Comprehensive evaluation of bronchoalveolar lavage from patients with severe COVID-19 and correlation with clinical outcomes. Hum. Pathol. 2021, 113, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Poloni, T.E.; Moretti, M.; Medici, V.; Turturici, E.; Belli, G.; Cavriani, E.; Visonà, S.D.; Rossi, M.; Fantini, V.; Ferrari, R.R.; et al. COVID-19 Pathology in the Lung, Kidney, Heart and Brain: The Different Roles of T-Cells, Macrophages, and Microthrombosis. Cells 2022, 11, 3124. [Google Scholar] [CrossRef] [PubMed]
| Item a | Survivors (n = 6) | Non-Survivors (n = 12) | p Value b |
|---|---|---|---|
| Gender Male/Female [number (%)] | [4 (66.7)]/[2(33.3)] | [7 (62.5)]/[5(37.5)] | 0.86/0.86 |
| Age (years) | 71 (±16) | 72 (±7) | |
| White blood cells (cells/mm3) | 8735 (6875–10,115) | 9200 (4545–14,410) | 0.925 |
| Neutrophils (cells/mm3) | 7260 (5717–8525) | 7775 (3702–12,930) | 1.00 |
| Lymphocytes (cells/mm3) | 805 (535–1007) | 600 (417–750) | 0.223 |
| Monocytes (cells/mm3) | 235 (222–435) | 290 (267–577) | 0.301 |
| C-reactive protein (mg/L) (cells/mm3) | 68,100 (37,925–104,125) | 90,765 (49,365–182,720) | 0.453 |
| D-dimer (mg/dL) | 4473 (4404–4473) | 1949 (1576–4109) | 0.034 |
| Albumin (mg/dL) | 34 (31–34) | 30 (29–32) | 0.115 |
| Lactate dehydrogenase (U/L) | 423 (305–452) | 500 (350–533) | 0.134 |
| Platelets (cells/mm3) | 220,500 (175,250–226,750) | 264,500 (201,500–332,000) | 0.160 |
| Length of hospitalization (days) | 101 (66–130) | 33 (12–49) | 0.002 |
| CHARLSON index | 4 (2–5) | 4 (2–5) | 0.658 |
| CURB-65 | 2 (1–2) | 2 (2–2) | 0.653 |
| EXP CURB-65 | 4 (3.5–4.5) | 4 (4–4) | 1.00 |
| PSI | 110 (79–112) | 96 (87–100) | 0.446 |
| CALL score | 10 (9–12) | 11 (10–13) | 0.224 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santinelli, L.; Lazzaro, A.; Sciarra, F.; Maddaloni, L.; Frasca, F.; Fracella, M.; Moretti, S.; Borsetti, A.; Bugani, G.; Alessandri, F.; et al. Cellular Immune Profiling of Lung and Blood Compartments in Patients with SARS-CoV-2 Infection. Pathogens 2023, 12, 442. https://doi.org/10.3390/pathogens12030442
Santinelli L, Lazzaro A, Sciarra F, Maddaloni L, Frasca F, Fracella M, Moretti S, Borsetti A, Bugani G, Alessandri F, et al. Cellular Immune Profiling of Lung and Blood Compartments in Patients with SARS-CoV-2 Infection. Pathogens. 2023; 12(3):442. https://doi.org/10.3390/pathogens12030442
Chicago/Turabian StyleSantinelli, Letizia, Alessandro Lazzaro, Francesca Sciarra, Luca Maddaloni, Federica Frasca, Matteo Fracella, Sonia Moretti, Alessandra Borsetti, Ginevra Bugani, Francesco Alessandri, and et al. 2023. "Cellular Immune Profiling of Lung and Blood Compartments in Patients with SARS-CoV-2 Infection" Pathogens 12, no. 3: 442. https://doi.org/10.3390/pathogens12030442
APA StyleSantinelli, L., Lazzaro, A., Sciarra, F., Maddaloni, L., Frasca, F., Fracella, M., Moretti, S., Borsetti, A., Bugani, G., Alessandri, F., Zullino, V., Ruberto, F., Pugliese, F., Sorrentino, L., Gianfrilli, D., Isidori, A., Venneri, M. A., Mastroianni, C. M., Ceccarelli, G., & d’Ettorre, G. (2023). Cellular Immune Profiling of Lung and Blood Compartments in Patients with SARS-CoV-2 Infection. Pathogens, 12(3), 442. https://doi.org/10.3390/pathogens12030442













