Differences in Gut Microbiome Profile between Healthy Children and Children with Inflammatory Bowel Disease and/or Autoimmune Liver Disease: A Case-Control Study
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Population
2.2. Stool Sample and Data Collection
2.3. Microbial DNA Extraction and 16S rRNA Sequencing
2.4. Generation of Microbial Community Composition Profiles
2.5. Statistical Analysis
3. Results
3.1. Study Population
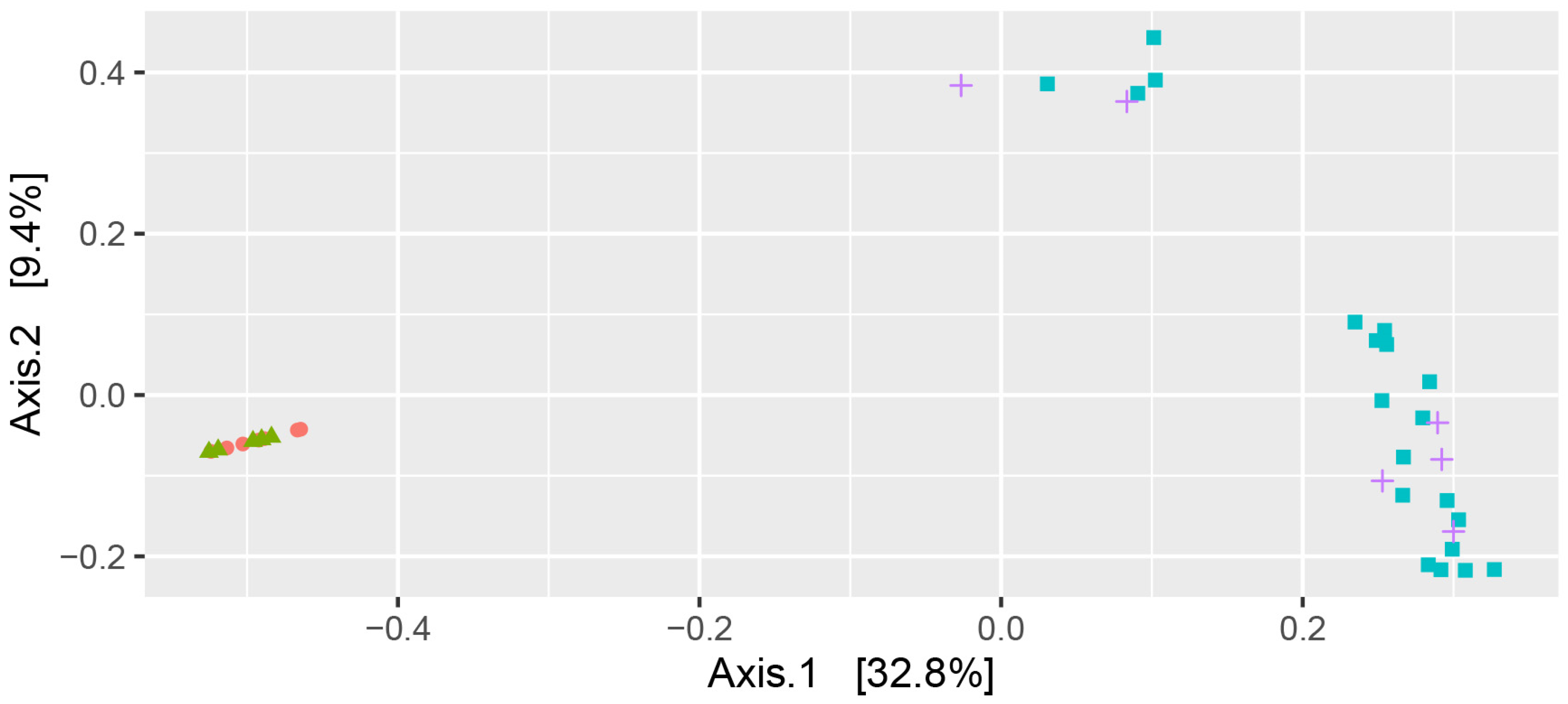
3.2. Microbiome Analysis—Alpha Diversity
3.3. Microbiome Analysis—Phyla Distribution
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mieli-Vergani, G.; Vergani, D.; Baumann, U.; Czubkowski, P.; Debray, D.; Dezsofi, A.; Fischler, B.; Gupte, G.; Hierro, L.; Indolfi, G.; et al. Diagnosis and Management of Pediatric Autoimmune Liver Disease: ESPGHAN Hepatology Committee Position Statement. J. Pediatr. Gastroenterol. Nutr. 2018, 66, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Deneau, M.R.; El-Matary, W.; Valentino, P.L.; Abdou, R.; Alqoaer, K.; Amin, M.; Amir, A.Z.; Auth, M.; Bazerbachi, F.; Broderick, A.; et al. The natural history of primary sclerosing cholangitis in 781 children: A multicenter, international collaboration. Hepatology 2017, 66, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, K.N.; LaRusso, N.F. Primary Sclerosing Cholangitis. N. Engl. J. Med. 2016, 375, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020, 202, 2147–2167. [Google Scholar] [CrossRef] [PubMed]
- Kummen, M.; Holm, K.; Anmarkrud, J.A.; Nygård, S.; Vesterhus, M.; Høivik, M.L.; Trøseid, M.; Marschall, H.U.; Schrumpf, E.; Moum, B.; et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2017, 66, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Sabino, J.; Vieira-Silva, S.; Machiels, K.; Joossens, M.; Falony, G.; Ballet, V.; Ferrante, M.; Van Assche, G.; Van der Merwe, S.; Vermeire, S.; et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016, 65, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Iwasawa, K.; Suda, W.; Tsunoda, T.; Oikawa-Kawamoto, M.; Umetsu, S.; Inui, A.; Fujisawa, T.; Morita, H.; Sogo, T.; Hattori, M. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut 2017, 66, 1344–1346. [Google Scholar] [CrossRef] [PubMed]
- Little, R.; Wine, E.; Kamath, B.M.; Griffiths, A.M.; Ricciuto, A. Gut microbiome in primary sclerosing cholangitis: A review. World J. Gastroenterol. 2020, 26, 2768–2780. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Koletzko, S.; Turner, D.; Escher, J.C.; Cucchiara, S.; de Ridder, L.; Kolho, K.L.; Veres, G.; Russell, R.K.; Paerregaard, A.; et al. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Engevik, M.A.; Spinler, J.K.; Versalovic, J. Healthy Human Gastrointestinal Microbiome: Composition and Function After a Decade of Exploration. Dig. Dis. Sci. 2020, 65, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ran, Y.; Zhang, H.; Wang, B.; Zhou, L. The Microbiome in Autoimmune Liver Diseases: Metagenomic and Metabolomic Changes. Front. Physiol. 2021, 12, 715852. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, Y.; Yan, L.; Sun, C.; Miao, Q.; Wang, Q.; Xiao, X.; Lian, M.; Li, B.; Chen, Y.; et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020, 69, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Jiang, Y.; Rao, B.; Li, A.; Ding, S.; Yan, H.; Zhou, H.; Liu, Z.; Shi, Q.; Cui, G.; et al. Fecal Microbiomes Distinguish Patients With Autoimmune Hepatitis From Healthy Individuals. Front. Cell. Infect. Microbiol. 2020, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Cortez, R.V.; Moreira, L.N.; Padilha, M.; Bibas, M.D.; Toma, R.K.; Porta, G.; Taddei, C.R. Gut Microbiome of Children and Adolescents With Primary Sclerosing Cholangitis in Association With Ulcerative Colitis. Front. Immunol. 2020, 11, 598152. [Google Scholar] [CrossRef] [PubMed]
- Su, J.W.; Ma, J.J.; Zhang, H.J. Use of antibiotics in patients with Crohn’s disease: A systematic review and meta-analysis. J. Dig. Dis. 2015, 16, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Gionchetti, P.; Calabrese, C.; Lauri, A.; Rizzello, F. The therapeutic potential of antibiotics and probiotics in the treatment of pouchitis. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Tabibian, J.H.; Weeding, E.; Jorgensen, R.A.; Petz, J.L.; Keach, J.C.; Talwalkar, J.A.; Lindor, K.D. Randomised clinical trial: Vancomycin or metronidazole in patients with primary sclerosing cholangitis—A pilot study. Aliment. Pharmacol. Ther. 2013, 37, 604–612. [Google Scholar] [CrossRef] [PubMed]


| AILD | AILD-IBD | IBD | HC | |
|---|---|---|---|---|
| Number | 7 | 6 | 22 | 5 |
| M/F | 1M/6F | 5M/1F | 4M/18F | 3M/2F |
| Age in years: Mean (SD) [Range] | 13.3 (3.1) [7.0–16.2] | 9.1 (4.2) [11.1–18.0] | 11.7 (3.1) [5.4–16.6] | 8.1 (5.0) [5.2–16.9] |
| CD/UC/IBDU/VEO-IBD | NA | 2/0/4/0 | 11/2/7/2 | NA |
| Antibiotics | NA | 67% | 14% | NA |
| Steriods | 100% | 60% | 32% | NA |
| EEN | NA | 40% | 23% | NA |
| ASA | NA | 60% | 77% | NA |
| MTX | 0% | 40% | 5% | NA |
| Tacrolimus | 14% | 20% | 9% | NA |
| Azathioprine | 18% | 60% | 59% | NA |
| Anti-TNF | 0% | 20% | 5% | NA |
| Tukey’s Multiple Comparisons Test | Significant | Summary | Adjusted p Value |
|---|---|---|---|
| AILD vs. IBD-AILD | Yes | **** | <0.0001 |
| AILD vs. IBD | Yes | **** | <0.0001 |
| AILD vs. HC | No | ns | 0.9919 |
| IBD-AILD vs. IBD | No | ns | 0.9922 |
| IBD-AILD vs. HC | Yes | **** | <0.0001 |
| IBD vs. HC | Yes | **** | <0.0001 |
| Tukey’s Multiple Comparisons Test | Below Threshold? | Summary | Adjusted p Value |
|---|---|---|---|
| AILD vs. IBD-AILD | Yes | **** | <0.0001 |
| AILD vs. IBD | Yes | **** | <0.0001 |
| AILD vs. HC | No | ns | 0.9997 |
| IBD-AILD vs. IBD | No | ns | 0.9811 |
| IBD-AILD vs. HC | Yes | **** | <0.0001 |
| IBD vs. HC | Yes | **** | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez, R.N.; Leach, S.T.; Bowcock, N.; Coker, E.; Shapiro, A.J.; Day, A.S.; Lemberg, D.A. Differences in Gut Microbiome Profile between Healthy Children and Children with Inflammatory Bowel Disease and/or Autoimmune Liver Disease: A Case-Control Study. Pathogens 2023, 12, 585. https://doi.org/10.3390/pathogens12040585
Lopez RN, Leach ST, Bowcock N, Coker E, Shapiro AJ, Day AS, Lemberg DA. Differences in Gut Microbiome Profile between Healthy Children and Children with Inflammatory Bowel Disease and/or Autoimmune Liver Disease: A Case-Control Study. Pathogens. 2023; 12(4):585. https://doi.org/10.3390/pathogens12040585
Chicago/Turabian StyleLopez, Robert N., Steven T. Leach, Nerissa Bowcock, Elise Coker, Amanda J. Shapiro, Andrew S. Day, and Daniel A. Lemberg. 2023. "Differences in Gut Microbiome Profile between Healthy Children and Children with Inflammatory Bowel Disease and/or Autoimmune Liver Disease: A Case-Control Study" Pathogens 12, no. 4: 585. https://doi.org/10.3390/pathogens12040585
APA StyleLopez, R. N., Leach, S. T., Bowcock, N., Coker, E., Shapiro, A. J., Day, A. S., & Lemberg, D. A. (2023). Differences in Gut Microbiome Profile between Healthy Children and Children with Inflammatory Bowel Disease and/or Autoimmune Liver Disease: A Case-Control Study. Pathogens, 12(4), 585. https://doi.org/10.3390/pathogens12040585









