A SARS-CoV-2: Companion Animal Transmission and Variants Classification
Abstract
1. Introduction to the Human Coronavirus
2. Coronavirus Disease-2019 (COVID-19)
3. Morphology, Genome Organization, and Pathogenesis of SARS-CoV-2
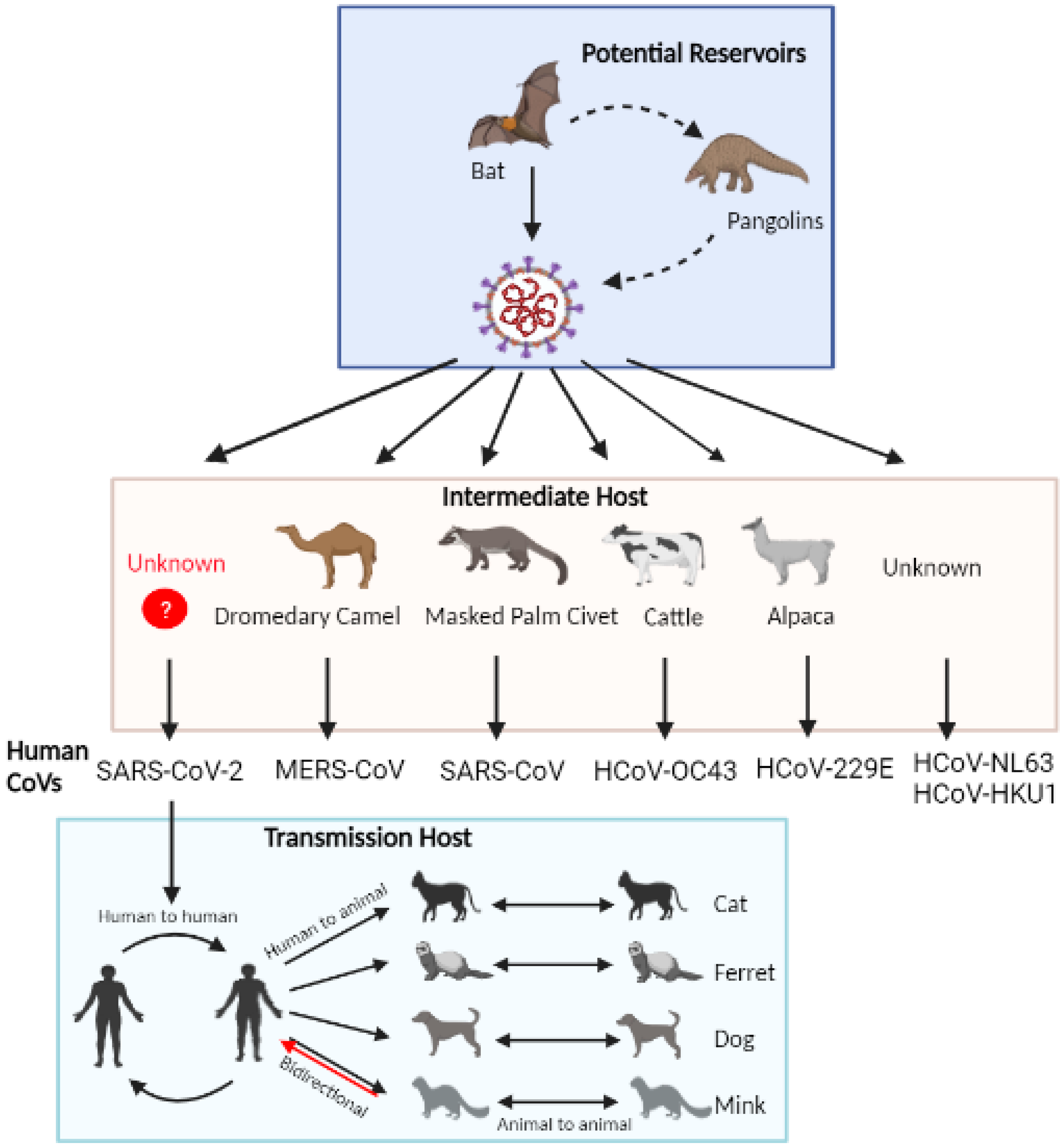
4. Potential Animal Reservoirs of SARS-CoV-2
4.1. Bats
4.2. Pangolins
5. Natural Infection in Companion and Farm Animals
5.1. Domestic Cats and Ferrets
5.2. Dogs
5.3. Minks
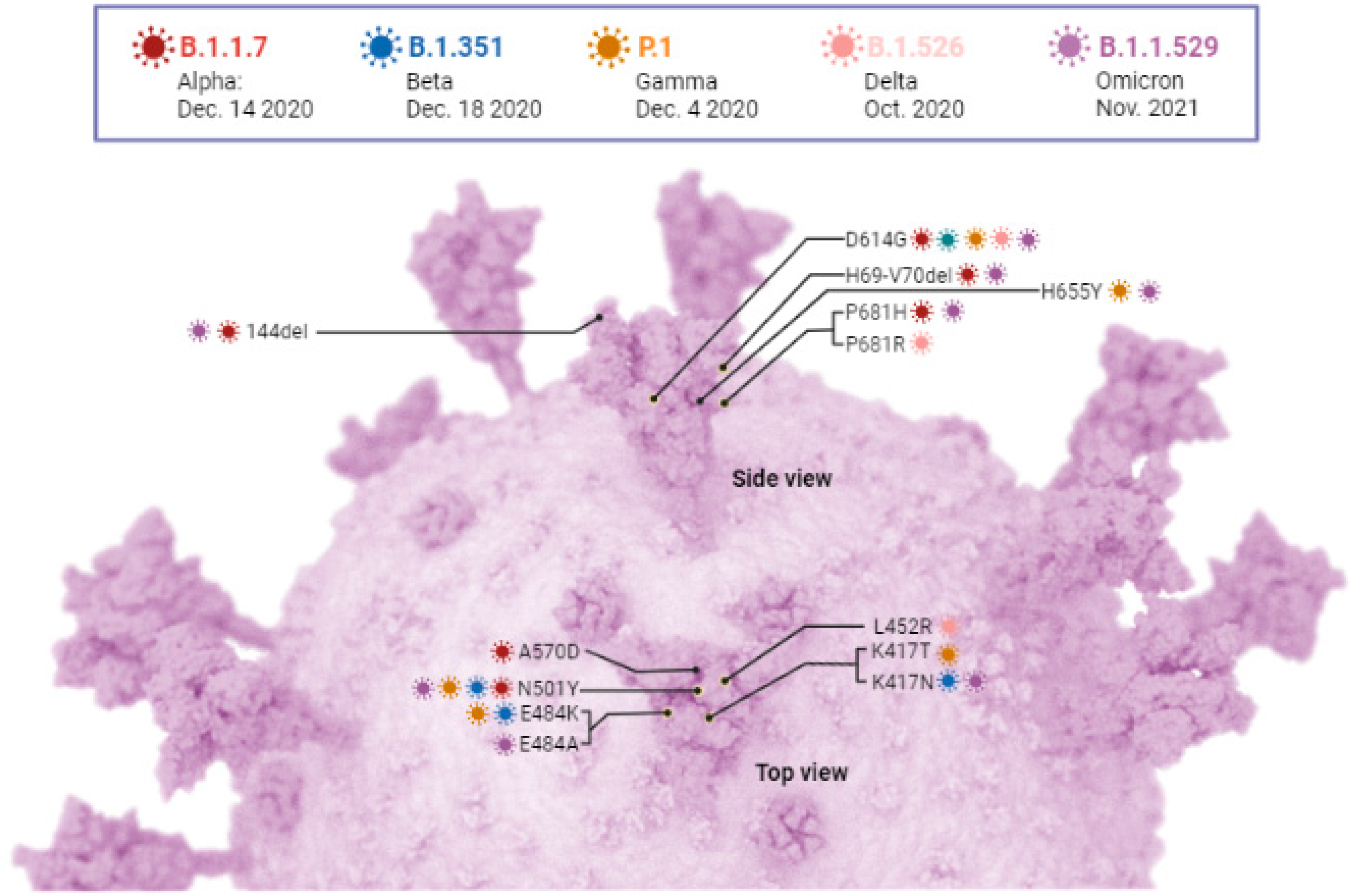
6. SARS-CoV-2 Variants
6.1. SAR-CoV-2 Variant Being Monitored and Variant of Concern
6.1.1. Alpha Variant (B.1.1.7)
6.1.2. Beta (B.1.351)
| Variants and Current Status | Lineages and Descendent Lineages | Country and Date of Earliest Identification/Documentation) | Key Mutations Reported | Phenotypes | References |
|---|---|---|---|---|---|
| Alpha VBM | B.1.1.7 and Q lineages | UK, September 2020 | 69/70 del **, Y144 del **, N501Y **, A570D, D614G **, P681H **, T716I, S982A, D1118H | Increased transmissibility (30–80%); increased risk of hospitalization (12–64%) and risk of death (30–70%); no adverse effects on vaccine efficacy | [85,86,87,90,92,95,96,97,98,101,104,115] |
| Beta VBM | B.1.351 and descendant lineages | South Africa, October 2020 | L18F **, D80A, D215G, 241–243 del, K417N **, E484K **, N501Y **, D614G **, A701V | Increased transmissibility; increased risk of death during hospitalization; immune escaping capacity | [101,107,109,110,111,112,113,114,119] |
| Gamma VBM | P.1 and descendent lineages | Brazil, May 2020 | L18F **, T20N, P26S, D138Y, R190S, K417T **, E484K **, N501Y **, D614G **, H655Y **, T1027I, V1176F | Increased transmissibility; less severe; immune escaping capacity | [86,90,120,121,122,123] |
| Delta and Delta Plus VBM | B.1.617.2 And descendent lineages | India, October 2020 | T19R, E156G, 157–158 del, K417N ++, L452R, T478K, D614G **, P681R, D950N | Increased transmissibility; less severe; immune escaping capacity | [86,89,115,124,125,126,127,128] |
| Omicron VOC | B.1.1.529 and descendent lineages (BA.2.75, BQ.1, BQ.1.1, BF.2.3.20, BF.7, BF.11, XBB, XBB.1.5, XBC, XAC BN.1, CH.1.1) | South Africa, November 2021 | D3G, P13L, Q19E, 31–33 del, A63T, A67V, 69–70 del **, T91, T95I, 142–144 del **, R203K, G204R, del211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N **, N440K, G446S, L452R ++, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y **, Y505H, T547K, D614G **, H655Y **, N679K, P681H **, N764K, D796Y, N856K, Q954H, N969K, L981F | Highly increased transmissibility, less severe disease development (>20%); increased impact on immunity and re-infection, potential reduction in neutralization | [43,86,129,130,131,132,133,134,135] |
6.1.3. Gamma (P.1)
6.1.4. Delta (B.1.617.2)
6.1.5. Omicron (B.1.1.529)
6.1.6. Mu (B.1.621)
6.1.7. Lambda (C.37)
6.1.8. Epsilon (B.1.427/B.1.429)
6.1.9. Kappa (B.1.617.1)
| SARS-CoV-2 Variants | Current Status Designation | Lineage | Earliest Identification/ Documentation (Country, Date) | Key Mutations | References |
|---|---|---|---|---|---|
| Mu | VBM | B.1.621 and B.1.621.1 | Colombia, January 2021 | T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N | [86,163,166,169] |
| Lambda | VBM | C.37 B.1.1.1.C37 | Peru, December 2020 | G75V, T76I, 246–253 delinsN, L452Q, F490S, D614G, T859N | [86,100,174,179] |
| Kappa | VBM | B.1.617.1 | India, October 2021 | T95I, G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H | [86,115,179,183,184] |
| Epsilon | VBM | B.1.427 and B.1.429 | USA, September 2020 | B.1.427: L452R, D614G B.1.429: S13I, W152C, L452R, D614G | [86,177,178,179,180] |
| Eta | VBM | B.1.525 | Nigeria, UK, December 2020 | Q52R, A67V, 69/70 del, 144 del, E484K, D614G, Q677H, F888L | [86,185,186] |
| Iota | VBM | B.1.526 | USA, December 2020 | L5F, T95I, D253G, S477N, E484K, D614G, A701V | [86,94,179] |
| Theta | VBM | P.3 | Philippines January 2021 | Del 141–143, E484K, N501Y, D614G, P681H | [86,187,188] |
| Zeta | VBM | P.2/B.1.1.28.2 | Brazil and Japan | L18F, T20N, P26S, A119S, F120F, F157L, M234I, E484K, D614G, S929I, V1176F, L3468V | [86,179,181,189] |
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- ICTV. Current ICTV Taxonomy Release. Available online: https://ictv.global/taxonomy (accessed on 15 May 2023).
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H. Bats Are Natural Reservoirs of SARS-like Coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar]
- Kesheh, M.M.; Hosseini, P.; Soltani, S.; Zandi, M. An overview on the seven pathogenic human coronaviruses. Rev. Med. Virol. 2022, 32, e2282. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- WHO. Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 15 May 2023).
- CDC about COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/your-health/about-covid-19.html (accessed on 15 May 2023).
- WHO. WHO Coronavirus (COVID-19) Dashboard; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- WHO. COVID-19-Landscape of Novel Coronavirus Candidate Vaccine Development Worldwide. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 16 May 2023).
- WHO. Coronavirus Disease (COVID-19): Vaccines Q&A. Available online: https://www.who.int/news-room/questions-and-answers/item/coronavirus-disease-(covid-19)-vaccines (accessed on 18 May 2023).
- Holder, J. Tracking Coronavirus Vaccinations around the World. The New York Times. The New York Times 2022. Available online: https://www.nytimes.com (accessed on 17 March 2023).
- Flores-Alanis, A.; Sandner-Miranda, L.; Delgado, G.; Cravioto, A.; Morales-Espinosa, R. The receptor binding domain of SARS-CoV-2 spike protein is the result of an ancestral recombination between the bat-CoV RaTG13 and the pangolin-CoV MP789. BMC Res. Notes 2020, 13, 398. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- te Velthuis, A.J.; van den Worm, S.H.; Snijder, E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012, 40, 1737–1747. [Google Scholar] [CrossRef]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal. Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Atri, D.; Siddiqi, H.K.; Lang, J.P.; Nauffal, V.; Morrow, D.A.; Bohula, E.A. COVID-19 for the Cardiologist: Basic Virology, Epidemiology, Cardiac Manifestations, and Potential Therapeutic Strategies. JACC. Basic Transl. Sci. 2020, 5, 518–536. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- USDA Confirmed Cases of SARS-CoV-2 in Animals in the United States. Available online: https://www.aphis.usda.gov/aphis/dashboards/tableau/sars-dashboard (accessed on 15 May 2023).
- El-Sayed, A.; Kamel, M. Coronaviruses in humans and animals: The role of bats in viral evolution. Environ. Sci. Pollut. Res. Int. 2021, 28, 19589–19600. [Google Scholar] [CrossRef] [PubMed]
- Leopardi, S.; Holmes, E.C.; Gastaldelli, M.; Tassoni, L.; Priori, P.; Scaravelli, D.; Zamperin, G.; De Benedictis, P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018, 58, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, B.; Yang, J.; Jin, Q. DBatVir: The database of bat-associated viruses. Database 2014, 2014, bau021. [Google Scholar] [CrossRef]
- Li, C.; Yang, Y.; Ren, L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect. Genet. Evol. 2020, 82, 104285. [Google Scholar] [CrossRef]
- Skirmuntt, E.C.; Escalera-Zamudio, M.; Teeling, E.C.; Smith, A.; Katzourakis, A. The Potential Role of Endogenous Viral Elements in the Evolution of Bats as Reservoirs for Zoonotic Viruses. Annu. Rev. Virol. 2020, 7, 103–119. [Google Scholar] [CrossRef]
- Worobey, M. Dissecting the early COVID-19 cases in Wuhan. Science 2021, 374, 1202–1204. [Google Scholar] [CrossRef]
- Touati, R.; Haddad-Boubaker, S.; Ferchichi, I.; Messaoudi, I.; Ouesleti, A.E.; Triki, H.; Lachiri, Z.; Kharrat, M. Comparative genomic signature representations of the emerging COVID-19 coronavirus and other coronaviruses: High identity and possible recombination between Bat and Pangolin coronaviruses. Genomics 2020, 112, 4189–4202. [Google Scholar] [CrossRef]
- Temmam, S.; Vongphayloth, K.; Baquero, E.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Chia, W.N.; Ampoot, W.; Lim, B.L. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Kitamura, T.; Suzuki, J.; Sato, R.; Aoi, T.; Fujii, M.; Matsugo, H.; Kamiki, H.; Ishida, H.; Takenaka-Uema, A. Detection and Characterization of Bat Sarbecovirus Phylogenetically Related to SARS-CoV-2, Japan. Emerg. Infect. Dis. J. 2020, 26, 3025. [Google Scholar] [CrossRef]
- Hul, V.; Delaune, D.; Karlsson, E.A.; Hassanin, A.; Tey, P.O.; Baidaliuk, A.; Gámbaro, F.; Tu, V.T.; Keatts, L.; Mazet, J. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Biorxiv Prepr. Serv. Biol. 2021, 12, 6563. [Google Scholar]
- Zou, J.; Kurhade, C.; Patel, S.; Kitchin, N.; Tompkins, K.; Cutler, M.; Cooper, D.; Yang, Q.; Cai, H.; Muik, A. Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N. Engl. J. Med. 2023, 388, 854–857. [Google Scholar] [CrossRef]
- Lytras, S.; Hughes, J.; Martin, D.; Swanepoel, P.; de Klerk, A.; Lourens, R.; Kosakovsky Pond, S.L.; Xia, W.; Jiang, X.; Robertson, D.L. Exploring the Natural Origins of SARS-CoV-2 in the Light of Recombination. Genome Biol. Evol. 2022, 14, evac018. [Google Scholar] [CrossRef]
- Malaiyan, J.; Arumugam, S.; Mohan, K.; Gomathi Radhakrishnan, G. An update on the origin of SARS-CoV-2: Despite closest identity, bat (RaTG13) and pangolin derived coronaviruses varied in the critical binding site and O-linked glycan residues. J. Med. Virol. 2021, 93, 499–505. [Google Scholar] [CrossRef]
- Niu, S.; Wang, J.; Bai, B.; Wu, L.; Zheng, A.; Chen, Q.; Du, P.; Han, P.; Zhang, Y.; Jia, Y. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J. 2021, 40, e107786. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-Q.; Lin, H.-F.; Li, J.; Chen, Y.; Luo, Y.; Zhang, W.; Hu, B.; Tian, F.-J.; Hu, Y.-J.; Liu, Y.-J. A SARS-CoV-2-Related Virus from Malayan Pangolin Causes Lung Infection without Severe Disease in Human ACE2-Transgenic Mice. J. Virol. 2023, 97, e01719–e01722. [Google Scholar] [CrossRef]
- Piplani, S.; Singh, P.K.; Winkler, D.A.; Petrovsky, N. In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin. Sci. Rep. 2021, 11, 13063. [Google Scholar] [CrossRef]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef]
- Frutos, R.; Serra-Cobo, J.; Chen, T.; Devaux, C.A. COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans. Infect. Genet. Evol. 2020, 84, 104493. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.; Smith, D.; Ghai, R.R.; Wallace, R.M.; Torchetti, M.K.; Loiacono, C.; Murrell, L.S.; Carpenter, A.; Moroff, S.; Rooney, J.A. First Reported Cases of SARS-CoV-2 Infection in Companion Animals—New York, March-April 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 710–713. [Google Scholar] [CrossRef]
- Hamer, S.A.; Ghai, R.R.; Zecca, I.B.; Auckland, L.D.; Roundy, C.M.; Davila, E.; Busselman, R.E.; Tang, W.; Pauvolid-Corrêa, A.; Killian, M.L. SARS-CoV-2 B.1.1.7 variant of concern detected in a pet dog and cat after exposure to a person with COVID-19, USA. Transbound. Emerg. Dis. 2022, 69, 1656–1658. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Garigliany, M.; Van Laere, A.S.; Clercx, C.; Giet, D.; Escriou, N.; Huon, C.; van der Werf, S.; Eloit, M.; Desmecht, D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. 2020, 26, 3069–3071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2021, 11, 100192. [Google Scholar] [CrossRef]
- Wendling, N.M.; Carpenter, A.; Liew, A.; Ghai, R.R.; Gallardo-Romero, N.; Stoddard, R.A.; Tao, Y.; Zhang, J.; Retchless, A.C.; Ahmad, A. Transmission of SARS-CoV-2 Delta variant (B.1.617.2) from a fully vaccinated human to a canine in Georgia, July 2021. Zoonoses Public Health 2022, 69, 587–592. [Google Scholar] [CrossRef]
- Jairak, W.; Chamsai, E.; Udom, K.; Charoenkul, K.; Chaiyawong, S.; Techakriengkrai, N.; Tangwangvivat, R.; Suwannakarn, K.; Amonsin, A. SARS-CoV-2 delta variant infection in domestic dogs and cats, Thailand. Sci. Rep. 2022, 12, 8403. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Arévalo, S.; Rivera, B.; Domínguez, L.; Sánchez-Vizcaíno, J.M. First Detection of SARS-CoV-2 B.1.1.7 Variant of Concern in an Asymptomatic Dog in Spain. Viruses 2021, 13, 1379. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, C.L.; Gao, Z.Y.; Qiao, H.X.; Wang, W.J.; Liu, X.Y.; Chuai, X. Ferrets: A powerful model of SARS-CoV-2. Zool. Res. 2023, 44, 323–330. [Google Scholar] [CrossRef] [PubMed]
- James, J.; Byrne, A.M.P.; Goharriz, H.; Golding, M.; Cuesta, J.M.A.; Mollett, B.C.; Shipley, R.; McElhinney, L.M.; Fooks, A.R.; Brookes, S.M. Infectious droplet exposure is an inefficient route for SARS-CoV-2 infection in the ferret model. J. Gen. Virol. 2022, 103, 001799. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Belser, J.A.; Kieran, T.J.; Brock, N.; Pulit-Penaloza, J.A.; Pappas, C.; Basu Thakur, P.; Jones, J.; Wentworth, D.E.; Zhou, B. Enhanced fitness of SARS-CoV-2 B.1.617.2 Delta variant in ferrets. Virology 2023, 582, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Sila, T.; Sunghan, J.; Laochareonsuk, W.; Surasombatpattana, S.; Kongkamol, C.; Ingviya, T.; Siripaitoon, P.; Kositpantawong, N.; Kanchanasuwan, S.; Hortiwakul, T. Suspected Cat-to-Human Transmission of SARS-CoV-2, Thailand, July–September 2021. Emerg. Infect. Dis. J. 2022, 28, 1485. [Google Scholar] [CrossRef]
- Alfano, F.; Fusco, G.; Mari, V.; Occhiogrosso, L.; Miletti, G.; Brunetti, R.; Galiero, G.; Desario, C.; Cirilli, M.; Decaro, N. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound. Emerg. Dis. 2020, 67, 1991–1999. [Google Scholar] [CrossRef]
- Decaro, N.; Cordonnier, N.; Demeter, Z.; Egberink, H.; Elia, G.; Grellet, A.; Le Poder, S.; Mari, V.; Martella, V.; Ntafis, V. European surveillance for pantropic canine coronavirus. J. Clin. Microbiol. 2013, 51, 83–88. [Google Scholar] [CrossRef]
- Xia, X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020, 37, 2699–2705. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef]
- USDA Confirmation of COVID-19 in Pet Dog in New York. Available online: https://content.govdelivery.com/accounts/USDAAPHIS/bulletins/28ead4f (accessed on 1 September 2021).
- Liew, A.Y.; Carpenter, A.; Moore, T.A.; Wallace, R.M.; Hamer, S.A.; Hamer, G.L.; Fischer, R.S.B.; Zecca, I.B.; Davila, E.; Auckland, L.D. Clinical and epidemiologic features of SARS-CoV-2 in dogs and cats compiled through national surveillance in the United States. J. Am. Vet. Med. Assoc. 2023, 261, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Doerksen, T.; Lu, A.; Noll, L.; Almes, K.; Bai, J.; Upchurch, D.; Palinski, R. Near-Complete Genome of SARS-CoV-2 Delta (AY.3) Variant Identified in a Dog in Kansas, USA. Viruses 2021, 13, 2104. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Mediannikov, O.; Maurin, M.; Devaux, C.; Colson, P.; Levasseur, A.; Fournier, P.E.; Raoult, D. Mink, SARS-CoV-2, and the Human-Animal Interface. Front. Microbiol. 2021, 12, 663815. [Google Scholar] [CrossRef] [PubMed]
- Sparrer, M.N.; Hodges, N.F.; Sherman, T.; VandeWoude, S.; Bosco-Lauth, A.M.; Mayo, C.E. Role of Spillover and Spillback in SARS-CoV-2 Transmission and the Importance of One Health in Understanding the Dynamics of the COVID-19 Pandemic. J. Clin. Microbiol 2023, e0161022. [Google Scholar] [CrossRef]
- Larsen, C.S.; Paludan, S.R. Corona’s new coat: SARS-CoV-2 in Danish minks and implications for travel medicine. Travel Med. Infect. Dis. 2020, 38, 101922. [Google Scholar] [CrossRef]
- Boklund, A.; Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Lohse, L.; Strandbygaard, B.; Jørgensen, C.S.; Olesen, A.S.; Hjerpe, F.B.; Petersen, H.H. SARS-CoV-2 in Danish Mink Farms: Course of the Epidemic and a Descriptive Analysis of the Outbreaks in 2020. Anim. Open Access J. MDPI 2021, 11, 164. [Google Scholar] [CrossRef]
- Lu, L.; Sikkema, R.S.; Velkers, F.C.; Nieuwenhuijse, D.F.; Fischer, E.A.J.; Meijer, P.A.; Bouwmeester-Vincken, N.; Rietveld, A.; Wegdam-Blans, M.C.A.; Tolsma, P. Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat. Commun. 2021, 12, 6802. [Google Scholar] [CrossRef]
- Clayton, E.; Ackerley, J.; Aelmans, M.; Ali, N.; Ashcroft, Z.; Ashton, C.; Barker, R.; Budryte, V.; Burrows, C.; Cai, S. Structural Bases of Zoonotic and Zooanthroponotic Transmission of SARS-CoV-2. Viruses 2022, 14, 418. [Google Scholar] [CrossRef]
- Xiang, B.; Yang, L.; Ye, Z.; Ren, T.; Ye, Y. Vaccination of susceptible animals against SARS-CoV-2. J. Infect. 2022, 84, e48–e49. [Google Scholar] [CrossRef]
- van Aart, A.E.; Velkers, F.C.; Fischer, E.A.J.; Broens, E.M.; Egberink, H.; Zhao, S.; Engelsma, M.; Hakze-van der Honing, R.W.; Harders, F.; de Rooij, M.M.T. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound. Emerg. Dis. 2022, 69, 3001–3007. [Google Scholar] [CrossRef]
- Amman, B.R.; Cossaboom, C.M.; Wendling, N.M.; Harvey, R.R.; Rettler, H.; Taylor, D.; Kainulainen, M.H.; Ahmad, A.; Bunkley, P.; Godino, C. GPS Tracking of Free-Roaming Cats (Felis catus) on SARS-CoV-2-Infected Mink Farms in Utah. Viruses 2022, 14, 2131. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H. Viral infections at the animal–human interface—Learning lessons from the SARS-CoV-2 pandemic. Microb. Biotechnol. 2023, 1–15. [Google Scholar] [CrossRef]
- Chan, J.F.W.; Siu, G.K.H.; Yuan, S.; Ip, J.D.; Cai, J.P.; Chu, A.W.H.; Chan, W.M.; Abdullah, S.M.U.; Luo, C.; Chan, B.P.C.; et al. Probable Animal-to-Human Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Delta Variant AY.127 Causing a Pet Shop-Related Coronavirus Disease 2019 (COVID-19) Outbreak in Hong Kong. Clin. Infect. Dis. 2022, 75, e76–e81. [Google Scholar] [CrossRef]
- Yen, H.L.; Sit, T.H.C.; Brackman, C.J.; Chuk, S.S.Y.; Gu, H.; Tam, K.W.S.; Law, P.Y.T.; Leung, G.M.; Peiris, M.; Poon, L.L.M. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: A case study. Lancet 2022, 399, 1070–1078. [Google Scholar] [CrossRef]
- Wei, C.; Shan, K.J.; Wang, W.; Zhang, S.; Huan, Q.; Qian, W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genom. 2021, 48, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Tang, H.; McDanal, C.; Wagh, K.; Fischer, W.; Theiler, J.; Yoon, H.; Li, D.; Haynes, B.F.; Sanders, K.O. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 2021, 29, 529–539.e3. [Google Scholar] [CrossRef]
- CDC. SARS-CoV-2 Variant Classifications and Definitions; CDC: Atlanta, GA, USA, 2023. [Google Scholar]
- O’Toole, Á.; Hill, V.; Pybus, O.G.; Watts, A.; Bogoch, I.I.; Khan, K.; Messina, J.P.; Tegally, H.; Lessells, R.R.; Giandhari, J.; et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch. Wellcome Open Res. 2021, 6, 121. [Google Scholar] [CrossRef]
- Willis, O. How Much more Contagious is the New Strain of COVID B117 Detected in the UK? Australian Broadcasting Corporation: Sydney, NSW, Australia, 2021. [Google Scholar]
- CDC. What You Need to Know about Variants; CDC: Atlanta, GA, USA, 2021. [Google Scholar]
- Altmann, D.M.; Boyton, R.J.; Beale, R. Immunity to SARS-CoV-2 variants of concern. Science 2021, 371, 1103–1104. [Google Scholar] [CrossRef]
- Davies, N.G.; Jarvis, C.I.; van Zandvoort, K.; Clifford, S.; Sun, F.Y.; Funk, S.; Medley, G.; Jafari, Y.; Meakin, S.R.; Lowe, R.; et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Baric, R.S. Emergence of a Highly Fit SARS-CoV-2 Variant. N. Engl. J. Med. 2020, 383, 2684–2686. [Google Scholar] [CrossRef] [PubMed]
- Petrone, M.E.; Rothman, J.E.; Breban, M.I.; Ott, I.M.; Russell, A.; Lasek-Nesselquist, E.; Badr, H.; Kelly, K.; Omerza, G.; Renzette, N.; et al. Combining genomic and epidemiological data to compare the transmissibility of SARS-CoV-2 variants Alpha and Iota. Commun. Biol. 2022, 5, 439. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. = Eur. Commun. Dis. Bull. 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Kemp, S.A.; Meng, B.; Ferriera, I.A.; Datir, R.; Harvey, W.T.; Papa, G.; Lytras, S.; Collier, D.A.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70. Biorxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, J.; Hasnain, S.E.; Sundar, D. Possible Link between Higher Transmissibility of Alpha, Kappa and Delta Variants of SARS-CoV-2 and Increased Structural Stability of Its Spike Protein and hACE2 Affinity. Int. J. Mol. Sci. 2021, 22, 9131. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef]
- Collier, D.A.; De Marco, A.; Ferreira, I.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef]
- Carreño, J.M.; Alshammary, H.; Singh, G.; Raskin, A.; Amanat, F.; Amoako, A.; Gonzalez-Reiche, A.S.; van de Guchte, A.; PARIS study group; Srivastava, K.; et al. Evidence for retained spike-binding and neutralizing activity against emerging SARS-CoV-2 variants in serum of COVID-19 mRNA vaccine recipients. EBioMedicine 2021, 73, 103626. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ (Clin. Res. Ed.) 2021, 372, n296. [Google Scholar] [CrossRef]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 2021, 29, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Supasa, P.; Zhou, D.; Dejnirattisai, W.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Nutalai, R.; Tuekprakhon, A.; et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, 184, 2201–2211.e7. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T.; Kläser, K.; Canas, L.S.; Molteni, E.; Modat, M.; et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: An ecological study. Lancet Public Health 2021, 6, e335–e345. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; France, A.M.; Aoki, Y.; Batra, D.; Biggerstaff, M.; Dugan, V.; Galloway, S.; Hall, A.J.; Johansson, M.A.; Kondor, R.J.; et al. Genomic Surveillance for SARS-CoV-2 Variants Circulating in the United States, December 2020–May 2021. MMWR Morb Mortal Wkly. Rep. 2021, 2021, 846–850. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Moyo-Gwete, T.; Madzivhandila, M.; Makhado, Z.; Ayres, F.; Mhlanga, D.; Oosthuysen, B.; Lambson, B.E.; Kgagudi, P.; Tegally, H.; Iranzadeh, A.; et al. SARS-CoV-2 501Y.V2 (B.1.351) elicits cross-reactive neutralizing antibodies. Biorxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Alshammary, H.; Amoako, A.A.; Awawda, M.H.; Beach, K.F.; Bermúdez-González, M.C.; Chernet, R.L.; et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2021, 2, e283–e284. [Google Scholar] [CrossRef]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.-M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488.e4. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef]
- Hu, J.; Peng, P.; Wang, K.; Fang, L.; Luo, F.-Y.; Jin, A.-S.; Liu, B.-Z.; Tang, N.; Huang, A.-L. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021, 18, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Bruel, T.; Grzelak, L.; Guivel-Benhassine, F.; Staropoli, I.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 27, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, J.; Wang, P.; Zhang, Z. The mechanisms of immune response and evasion by the main SARS-CoV-2 variants. iScience 2022, 25, 105044. [Google Scholar] [CrossRef]
- Hall, V.J.; Foulkes, S.; Saei, A.; Andrews, N.; Oguti, B.; Charlett, A.; Wellington, E.; Stowe, J.; Gillson, N.; Atti, A.; et al. COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): A prospective, multicentre, cohort study. Lancet 2021, 397, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Burgess, J.; Naleway, A.; Tyner, H.; Yoon, S.; Meece, J.; Olsho, L.; Caban-Martinez, A.; Fowlkes, A.; Lutrick, K.; et al. Interim Estimates of Vaccine Effectiveness of BNT162b2 and mRNA-1273 COVID-19 Vaccines in Preventing SARS-CoV-2 Infection among Health Care Personnel, First Responders, and Other Essential and Frontline Workers—Eight, U.S. Locations, December 2020–March 2021. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 495. [Google Scholar]
- Amit, S.; Regev-Yochay, G.; Afek, A.; Kreiss, Y.; Leshem, E. Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. Lancet 2021, 397, 875–877. [Google Scholar] [CrossRef]
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernán, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 mRNA COVID-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021, 384, 1412–1423. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Singh, J.; Samal, J.; Kumar, V.; Sharma, J.; Agrawal, U.; Ehtesham, N.Z.; Sundar, D.; Rahman, S.A.; Hira, S.; Hasnain, S.E. Structure-Function Analyses of New SARS-CoV-2 Variants, B.1.1.7, B.1.351 and B.1.1.28.1: Clinical, Diagnostic, Therapeutic and Public Health Implications. Viruses 2021, 13, 439. [Google Scholar] [CrossRef]
- Naveca, F.G.; Nascimento, V.; de Souza, V.C.; Corado, A.D.L.; Nascimento, F.; Silva, G.; Costa, Á.; Duarte, D.; Pessoa, K.; Mejía, M.; et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021, 27, 1230–1238. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021, 184, 2939–2954.e9. [Google Scholar] [CrossRef]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M.; The COVID-19 Genomics UK (COG-UK) Consortium; Peacock, T.P.; et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021, 17, e1010022. [Google Scholar] [CrossRef]
- Jeong, M. Delta Plus Variant of SARS-CoV-2: How Does it Compare with the Delta Variant? Medical News Today 2021. Today 2021. Delta Plus Variant of SARS-CoV-2: What Do We Know So Far? Available online: https://www.medicalnewstoday.com (accessed on 15 May 2023).
- England, P.H. Investigation of SARS-CoV-2 Variants of Concern: Technical Briefings. Available online: https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201#full-publication-update-history (accessed on 15 May 2023).
- Singh, J.; Rahman, S.A.; Ehtesham, N.Z.; Hira, S.; Hasnain, S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021, 27, 1131–1133. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.S.; Marwal, R.; Vs, R.; Ponnusamy, K.; Jolly, B.; Bhoyar, R.C.; Sardana, V.; Naushin, S.; Rophina, M.; Mellan, T.A.; et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science 2021, 374, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Akbar Samad, A.B.; Vaqar, S. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; StatPearls Publishing, Copyright © 2023; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- European Centre for Disease Prevention and Control. Implications of the Emergence and Spread of the SARS-CoV-2 B.1.1. 529 Variant of Concern (Omicron), for the EU/EEA. Available online: https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-emergence-sars-cov-2-variant-b.1.1.529 (accessed on 15 May 2023).
- ECDC. Implications for the EU/EEA of the Spread of the SARSCoV-2 Omicron XBB.1.5 Sub-Lineage for the EU/EEA–13 January 2023; ECDC: Stockholm, Sweden, 2023. [Google Scholar]
- Shah, M.; Woo, H.G. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front. Immunol. 2021, 12, 830527. [Google Scholar] [CrossRef] [PubMed]
- Venkatakrishnan, A.J.; Anand, P.; Lenehan, P.J.; Suratekar, R.; Raghunathan, B.; Niesen, M.J.M.; Soundararajan, V. Omicron variant of SARS-CoV-2 harbors a unique insertion mutation of putative viral or human genomic origin. OSF Prepr. 2021, 1–16. [Google Scholar] [CrossRef]
- Chavda, V.P.; Balar, P.; Vaghela, D.; Solanki, H.K.; Vaishnav, A.; Hala, V.; Vora, L. Omicron Variant of SARS-CoV-2: An Indian Perspective of Vaccination and Management. Vaccines 2023, 11, 160. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Wi, Y.M.; Park, H.; Lee, J.E.; Kim, S.H.; Lee, K.S. Current and Emerging Knowledge in COVID-19. Radiology 2023, 306, e222462. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393. [Google Scholar] [CrossRef]
- CDC US. COVID-19 Cases Caused by Variants. Available online: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html (accessed on 9 September 2022).
- Koyama, T.; Platt, D.; Parida, L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef]
- Rees-Spear, C.; Muir, L.; Griffith, S.A.; Heaney, J.; Aldon, Y.; Snitselaar, J.L.; Thomas, P.; Graham, C.; Seow, J.; Lee, N.; et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021, 34, 108890. [Google Scholar] [CrossRef]
- Shao, W.; Chen, X.; Zheng, C.; Liu, H.; Wang, G.; Zhang, B.; Li, Z.; Zhang, W. Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern in real-world: A literature review and meta-analysis. Emerg. Microbes Infect. 2022, 11, 2383–2392. [Google Scholar] [CrossRef] [PubMed]
- CDC. Monitoring Variant Proportions; CDC: Atlanta, GA, USA, 2021. [Google Scholar]
- Sharma, H.N.; Latimore, C.O.D.; Matthews, Q.L. Biology and Pathogenesis of SARS-CoV-2: Understandings for Therapeutic Developments against COVID-19. Pathogens 2021, 10, 1218. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Gao, Q.; Gao, F.; Wang, Q.; He, Q.; Wu, X.; Mao, Q.; Xu, M.; Liang, Z. Impact of the Delta variant on vaccine efficacy and response strategies. Expert Rev. Vaccines 2021, 20, 1201–1209. [Google Scholar] [CrossRef]
- Mathew, J.C. Bharat Biotech’s Covaxin 78% Effective; Works against Most COVID-19 Variants. BusinessToday, 21 April 2021. [Google Scholar]
- Wang, C.; Liu, B.; Zhang, S.; Huang, N.; Zhao, T.; Lu, Q.-B.; Cui, F. Differences in incidence and fatality of COVID-19 by SARS-CoV-2 Omicron variant versus Delta variant in relation to vaccine coverage: A world-wide review. J. Med. Virol. 2023, 95, e28118. [Google Scholar] [CrossRef] [PubMed]
- Hyams, C.; Challen, R.; Marlow, R.; Nguyen, J.; Begier, E.; Southern, J.; King, J.; Morley, A.; Kinney, J.; Clout, M.; et al. Severity of Omicron (B.1.1.529) and Delta (B.1.617.2) SARS-CoV-2 infection among hospitalised adults: A prospective cohort study in Bristol, United Kingdom. Lancet Reg. Health. Eur. 2023, 25, 100556. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet. Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Allen, H.; Vusirikala, A.; Flannagan, J.; Twohig, K.A.; Zaidi, A.; Chudasama, D.; Lamagni, T.; Groves, N.; Turner, C.; Rawlinson, C.; et al. Household transmission of COVID-19 cases associated with SARS-CoV-2 delta variant (B.1.617.2): National case-control study. Lancet Reg. Health—Eur. 2022, 12, 100252. [Google Scholar] [CrossRef]
- Antonelli, M.; Pujol, J.C.; Spector, T.D.; Ourselin, S.; Steves, C.J. Risk of long COVID associated with delta versus omicron variants of SARS-CoV-2. Lancet 2022, 399, 2263–2264. [Google Scholar] [CrossRef]
- Reuters. UK Study Finds Vaccines Offer High Protection against Hospitalisation from Delta Variant; Reuters: London, UK, 2021. [Google Scholar]
- Ren, W.; Ju, X.; Gong, M.; Lan, J.; Yu, Y.; Long, Q.; Kenney, D.J.; O’Connell, A.K.; Zhang, Y.; Zhong, J.; et al. Characterization of SARS-CoV-2 Variants, B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618 by Cell Entry and Immune Evasion. mBio 2022, 13, e0009922. [Google Scholar] [CrossRef]
- Goto, A.; Miyakawa, K.; Nakayama, I.; Yagome, S.; Xu, J.; Kaneko, M.; Ohtake, N.; Kato, H.; Ryo, A. Prediction models for neutralization activity against emerging SARS-CoV-2 variants: A cross-sectional study. Front. Microbiol. 2023, 14, 1126527. [Google Scholar] [CrossRef] [PubMed]
- Brussow, H. COVID-19: Omicron—The latest, the least virulent, but probably not the last variant of concern of SARS-CoV-2. Microb. Biotechnol. 2022, 15, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.D.; Brunkard, J.M.; Boehmer, T.K.; Peterson, E.; Adjei, S.; Binder, A.M.; Cobb, S.; Graff, P.; Hidalgo, P.; Panaggio, M.J.; et al. Trends in Disease Severity and Health Care Utilization During the Early Omicron Variant Period Compared with Previous SARS-CoV-2 High Transmission Periods—United States, December 2020–January 2022. MMWR. Morb. Mortal. Wkly. Rep. 2022, 71, 146–152. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.; Fang, Y.; Liu, J.; Ye, Q.; Ding, L. SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal. Transduct. Target. Ther. 2022, 7, 76. [Google Scholar] [CrossRef]
- Rockett, R.J.; Draper, J.; Gall, M.; Sim, E.M.; Arnott, A.; Agius, J.E.; Johnson-Mackinnon, J.; Fong, W.; Martinez, E.; Drew, A.P.; et al. Co-infection with SARS-CoV-2 Omicron and Delta variants revealed by genomic surveillance. Nat. Commun. 2022, 13, 2745. [Google Scholar] [CrossRef]
- Jørgensen, S.B.; Nygård, K.; Kacelnik, O.; Telle, K. Secondary Attack Rates for Omicron and Delta Variants of SARS-CoV-2 in Norwegian Households. Jama 2022, 327, 1610–1611. [Google Scholar] [CrossRef]
- Lauring, A.S.; Tenforde, M.W.; Chappell, J.D.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Clinical severity of, and effectiveness of mRNA vaccines against, covid-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: Prospective observational study. BMJ (Clin. Res. Ed.) 2022, 376, e069761. [Google Scholar] [CrossRef]
- Regev-Yochay, G.; Gonen, T.; Gilboa, M.; Mandelboim, M.; Indenbaum, V.; Amit, S.; Meltzer, L.; Asraf, K.; Cohen, C.; Fluss, R.; et al. Efficacy of a Fourth Dose of Covid-19 mRNA Vaccine against Omicron. N. Engl. J. Med. 2022, 386, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Pajon, R.; Doria-Rose, N.A.; Shen, X.; Schmidt, S.D.; O’Dell, S.; McDanal, C.; Feng, W.; Tong, J.; Eaton, A.; Maglinao, M.; et al. SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. N. Engl. J. Med. 2022, 386, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.A.; Morden, E.; Rosseau, P.; Arendse, J.; Bam, J.L.; Boloko, L.; Cloete, K.; Cohen, C.; Chetty, N.; Dane, P.; et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection during resurgence driven by Omicron lineages BA.4 and BA.5 compared with previous waves in the Western Cape Province, South Africa. Int. J. Infect. Dis. 2023, 127, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Altarawneh, H.N.; Chemaitelly, H.; Hasan, M.R.; Ayoub, H.H.; Qassim, S.; AlMukdad, S.; Coyle, P.; Yassine, H.M.; Al-Khatib, H.A.; Benslimane, F.M.; et al. Protection against the Omicron Variant from Previous SARS-CoV-2 Infection. N. Engl. J. Med. 2022, 386, 1288–1290. [Google Scholar] [CrossRef]
- Laiton-Donato, K.; Franco-Muñoz, C.; Álvarez-Díaz, D.A.; Ruiz-Moreno, H.A.; Usme-Ciro, J.A.; Prada, D.A.; Reales-González, J.; Corchuelo, S.; Herrera-Sepúlveda, M.T.; Naizaque, J.; et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect. Genet. Evol. 2021, 95, 105038. [Google Scholar] [CrossRef]
- WHO. COVID-19 Weekly Epidemiological Update; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Uriu, K.; Kimura, I.; Shirakawa, K.; Takaori-Kondo, A.; Nakada, T.-A.; Kaneda, A.; Nakagawa, S.; Sato, K. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. N. Engl. J. Med. 2021, 385, 2397–2399. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Winkler, M.S.; Lier, M.; Schulz, S.; Jäck, H.M.; Cossmann, A.; Stankov, M.V.; et al. SARS-CoV-2 variants C.1.2 and B.1.621 (Mu) partially evade neutralization by antibodies elicited upon infection or vaccination. Cell Rep. 2022, 39, 110754. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Kuroda, M.; Armbrust, T.; Theiler, J.; Balaram, A.; Moreno, G.K.; Accola, M.A.; Iwatsuki-Horimoto, K.; Valdez, R.; Stoneman, E.; et al. Characterization of the SARS-CoV-2 B.1.621 (Mu) variant. Sci. Transl. Med. 2022, 14, eabm4908. [Google Scholar] [CrossRef] [PubMed]
- Edara, V.-V.; Pinsky, B.A.; Suthar, M.S.; Lai, L.; Davis-Gardner, M.E.; Floyd, K.; Flowers, M.W.; Wrammert, J.; Hussaini, L.; Ciric, C.R.; et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N. Engl. J. Med. 2021, 385, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Outbreak.info Mu Variant Report. Available online: https://outbreak.info/situation-reports/mu?loc=COL&loc=GBR&selected=Worldwide&overlay=false (accessed on 18 May 2023).
- Romero, P.E.; Dávila-Barclay, A.; Salvatierra, G.; González, L.; Cuicapuza, D.; Solís, L.; Marcos-Carbajal, P.; Huancachoque, J.; Maturrano, L.; Tsukayama, P. The Emergence of Sars-CoV-2 Variant Lambda (C.37) in South America. Microbiol. Spectr. 2021, 9, e00789–e00821. [Google Scholar] [CrossRef]
- Takuya, T.; Hao, Z.; Belinda, M.D.; Marie, I.S.; Mark, J.M.; Nathaniel, R.L. SARS-CoV-2 Lambda Variant Remains Susceptible to Neutralization by mRNA Vaccine-elicited Antibodies and Convalescent Serum. Biorxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Kimura, I.; Kosugi, Y.; Wu, J.; Zahradnik, J.; Yamasoba, D.; Butlertanaka, E.P.; Tanaka, Y.L.; Uriu, K.; Liu, Y.; Morizako, N.; et al. The SARS-CoV-2 Lambda variant exhibits enhanced infectivity and immune resistance. Cell Rep. 2022, 38, 110218. [Google Scholar] [CrossRef]
- García-Mendoza, M.; Merino-Sarmiento, N.; De Lucio-Burga, G.; Fernández-Navarro, M.G.; Pampa-Espinoza, L.; Solis-Sánchez, G.; Huaringa-Núñez, M.; Palomino-Rodríguez, M.; Ríos-Monteza, P.; Solari, L. IgG antibody response by ELISA using Wuhan and Lambda variant antigens in BBIBP-CORV vaccinated health care workers. Rev. Peru. Med. Exp. Salud Publica 2022, 39, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Mónica, L.A.; Luis, A.-P.; Andrés, B.; Aldo, G.; Fabio, P.; Claudia, P.C.; Fernando, V.-E.; Ricardo, S.-R. Infectivity and immune escape of the new SARS-CoV-2 variant of interest Lambda. Medrxiv Prepr. Serv. Health Sci. 2021. [Google Scholar] [CrossRef]
- Tada, T.; Zhou, H.; Samanovic, M.I.; Dcosta, B.M.; Cornelius, A.; Mulligan, M.J.; Landau, N.R. Comparison of Neutralizing Antibody Titers Elicited by mRNA and Adenoviral Vector Vaccine against SARS-CoV-2 Variants. Biorxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Zhang, W.; Davis, B.D.; Chen, S.S.; Sincuir Martinez, J.M.; Plummer, J.T.; Vail, E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. Jama 2021, 325, 1324–1326. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. Medrxiv Prepr. Serv. Health Sci. 2021. [Google Scholar] [CrossRef]
- Martin Webb, L.; Matzinger, S.; Grano, C.; Kawasaki, B.; Stringer, G.; Bankers, L.; Herlihy, R. Identification of and Surveillance for the SARS-CoV-2 Variants, B.1.427 and B.1.429—Colorado, January–March 2021. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 717–718. [Google Scholar] [CrossRef]
- Monajjemi, M.; Sayiner, H.S.; Kandemirli, F.; Mollaamin, F. An overview on lambda, epsilon, kappa, iota and zeta variants of covid-19 and its probability to merge with delta & delta plus, why it is a concern. Biointerface Res. Appl. Chem. 2022, 12, 6895–6914. [Google Scholar]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383. [Google Scholar] [CrossRef]
- McCallum, M.; Bassi, J.; Marco, A.; Chen, A.; Walls, A.C.; Iulio, J.D.; Tortorici, M.A.; Navarro, M.J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. Science 2021, 373, 648–654. [Google Scholar] [CrossRef]
- Yadav, P.D.; Sapkal, G.N.; Abraham, P.; Deshpande, G.; Nyayanit, D.A.; Patil, D.Y.; Gupta, N.; Sahay, R.R.; Shete, A.M.; Kumar, S.; et al. Neutralization Potential of Covishield Vaccinated Individuals Sera against, B.1.617.1. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2022, 74, 558–559. [Google Scholar] [CrossRef]
- McCallum, M.; Walls, A.C.; Sprouse, K.R.; Bowen, J.E.; Rosen, L.E.; Dang, H.V.; De Marco, A.; Franko, N.; Tilles, S.W.; Logue, J.; et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 2021, 374, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Egon, A.O.; Lacy, M.S.; Olubusuyi, M.A.; Adeola, A.F.; Ewean, C.O.; Johnson, A.A.; Taylor, J.D.; Janet, Z.; Pavan, P.B.; Michelle, K.A.; et al. Coincident rapid expansion of two SARS-CoV-2 lineages with enhanced infectivity in Nigeria. Medrxiv Prepr. Serv. Health Sci. 2021. [Google Scholar] [CrossRef]
- Pereira, F.; Tosta, S.; Lima, M.M.; Reboredo de Oliveira da Silva, L.; Nardy, V.B.; Gómez, M.K.A.; Lima, J.G.; Fonseca, V.; de Oliveira, T.; Lourenço, J.; et al. Genomic surveillance activities unveil the introduction of the SARS-CoV-2 B.1.525 variant of interest in Brazil: Case report. J. Med. Virol. 2021, 93, 5523–5526. [Google Scholar] [CrossRef]
- Rose, R.; Nolan, D.J.; LaFleur, T.M.; Lamers, S.L. Outbreak of P.3 (Theta) SARS-CoV-2 emerging variant of concern among service workers in Louisiana. J. Infect. Public Health 2022, 15, 7–9. [Google Scholar] [CrossRef]
- Haw, N.J.L.; Cañal, E.M.R.; Zuasula, J., Jr.; Loreche, M.J.; Bernadas, J. Epidemiological Characteristics of the SARS-CoV-2 Theta Variant (P.3) in the Central Visayas Region, Philippines, 30 October 2020–16 February 2021. West. Pac. Surveill. Response J. WPSAR 2022, 13, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Mohandas, S.; Sarkale, P.; Nyayanit, D.; Shete, A.; Sahay, R.; Potdar, V.; Baradkar, S.; Gupta, N.; Sapkal, G.; et al. Isolation of SARS-CoV-2 B.1.1.28.2 (P2) variant and pathogenicity comparison with D614G variant in hamster model. J. Infect. Public Health 2022, 15, 164–171. [Google Scholar] [CrossRef] [PubMed]


| Gene/Proteins | Function | Gene/Proteins | Function | |
|---|---|---|---|---|
| ORF-1a and ORF-1b | Encodes replicase polyprotein 1a (pp1a) and polyprotein 1b (pp1b), pp1a (nsp 1–11), and pp1b (nsp 11–16) cleaved into nsps 1–16 (Figure 1B) | Nsp-14 | 3′-5′ exoribonuclease; RNA cap formation; N-7-guanine methyltransferase | References for SARS-CoV-2 genes or proteins [17,18,19,20,21,22] |
| Nsp-1 | Restricts host innate immune response; Host mRNA degradation, translation inhibition | Nsp-15 | Endoribonuclease; chymotrypsin-like proteinase; evasion of immune response | |
| Nsp-2 | Binds to prohibitin (PHB)-1 and PHB-2; modulates host survival signaling pathway | Nsp-16 | 2′O-ribose methyltransferase; RNA cap formation; restricts host innate response | |
| Nsp-3 | Interacts with N-protein for double-membrane vesicle (DMV) formation; Papain-like proteinase; cleaves viral polyprotein and assists in immune escape | Spike protein | Forms spike complexes on the virion surface; attaches to host receptor ACE2 for virus entry and subsequent infection | |
| Nsp-4, 6 | Viral replication-transcription transmembrane scaffold protein; assembles DMV | ORF-3a and 3d | Forms ion channels for viral exit; induces apoptosis and pathogenesis | |
| Nsp-5 | Mpro/3C-like proteinase; cleave viral polyprotein at C-terminus; Inhibition of interferon signaling | Envelope protein | Forms the viral envelop; interferes with host immune responses (apoptosis) | |
| Nsp-7 | Complexes with nsp-8 and nsp-12; Cofactor for RNA-dependent RNA polymerase | Membrane protein | Most abundant; bind to the N for virus morphogenesis and assembly; transmembrane transport of nutrients and bud release | |
| Nsp-8 | Makes heterodimer with nsp-7 and nsp-12; primase | ORF-6 | Membrane-associated protein in the ER and Golgi compartments | |
| Nsp-9 | Complexes with Nsp-8; binding of single-stranded RNA; protein phosphatase | ORF-7a and 7b | Transmembrane protein; involved in viral escape | |
| Nsp-10 | Cofactor for nsp-14 and nsp-16 to cap viral RNA | ORF-8 | Role in host immune response | |
| Nsp-12 | RNA-dependent RNA polymerase/replicase; nucleotidyltransferase | Nucleocapsid protein | Warps the viral RNA; facilitates M proteins during virus assembly; Restricts antiviral response | |
| Nsp-13 | Helicase; RNA 5′ triphosphatase | ORF-10 | Role in antiviral immune response |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandit, R.; Matthews, Q.L. A SARS-CoV-2: Companion Animal Transmission and Variants Classification. Pathogens 2023, 12, 775. https://doi.org/10.3390/pathogens12060775
Pandit R, Matthews QL. A SARS-CoV-2: Companion Animal Transmission and Variants Classification. Pathogens. 2023; 12(6):775. https://doi.org/10.3390/pathogens12060775
Chicago/Turabian StylePandit, Rachana, and Qiana L. Matthews. 2023. "A SARS-CoV-2: Companion Animal Transmission and Variants Classification" Pathogens 12, no. 6: 775. https://doi.org/10.3390/pathogens12060775
APA StylePandit, R., & Matthews, Q. L. (2023). A SARS-CoV-2: Companion Animal Transmission and Variants Classification. Pathogens, 12(6), 775. https://doi.org/10.3390/pathogens12060775






