Bioreactor Production Process of Spodoptera frugiperda multiple nucleopolyhedrovirus Biopesticide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line, Virus, and Medium
2.2. Evaluation of Culture Media Supplementation for Sf9 Cells Growth
2.3. Effect of Multiplicity of Infection (MOI) on SfMNPV In Vitro Production
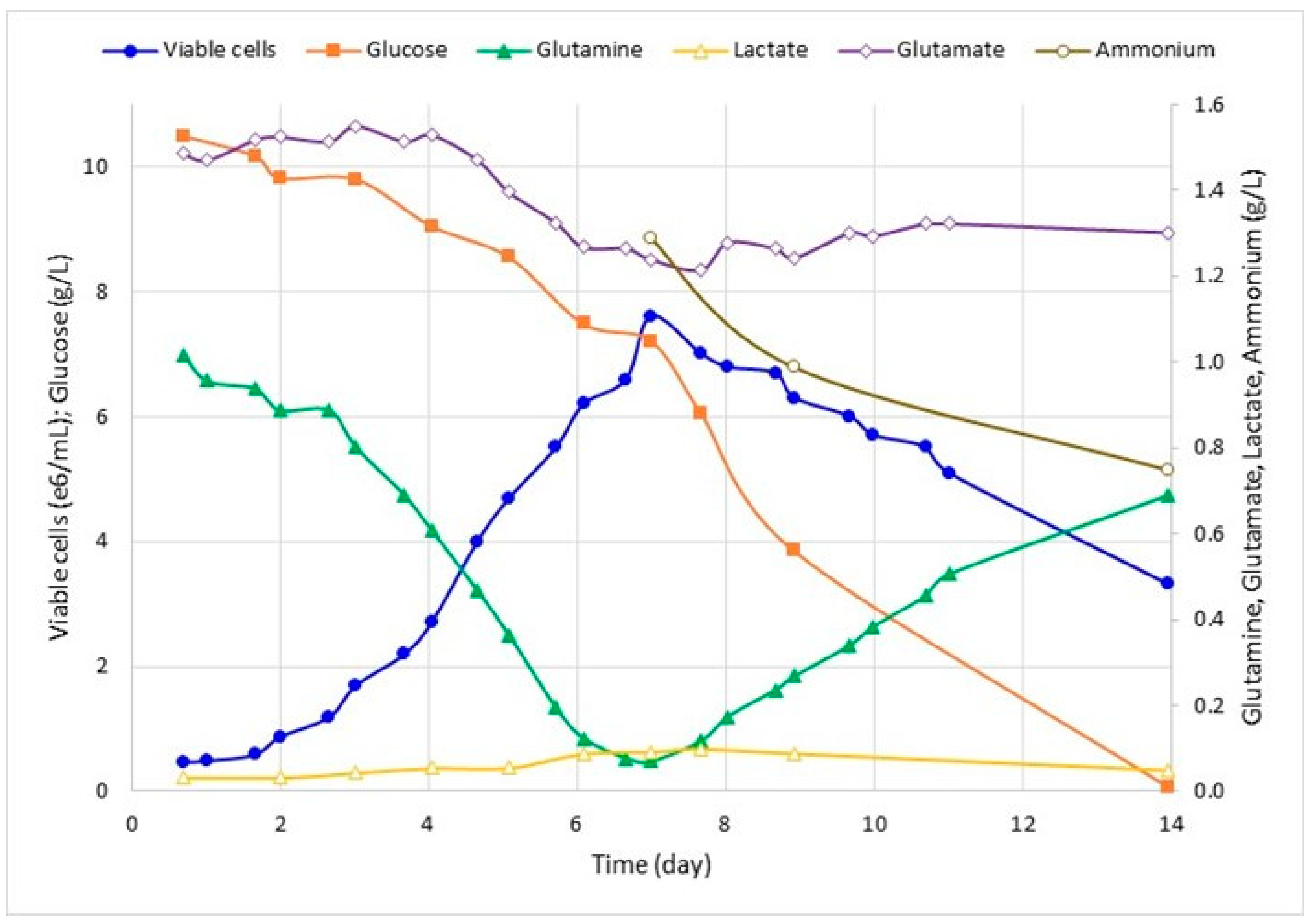
2.4. Analysis of Growth, Nutrients and Metabolites
2.5. Morphological Analysis of the Infection
2.6. PCR Analysis
2.7. Quantitative Real-Time PCR (qPCR)
3. Results
3.1. Evaluation of Culture Media Supplementation for Sf9 Cells Growth
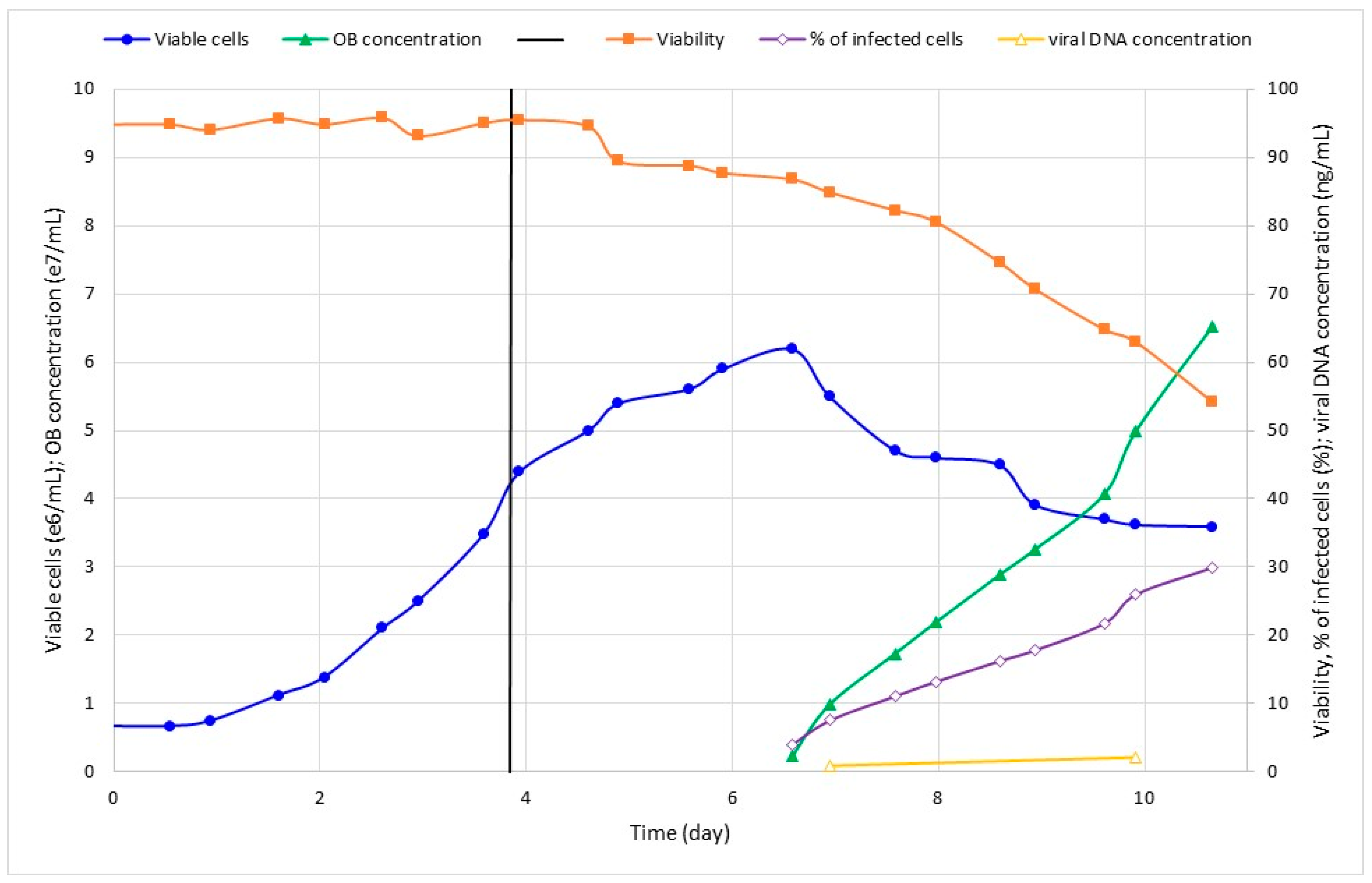
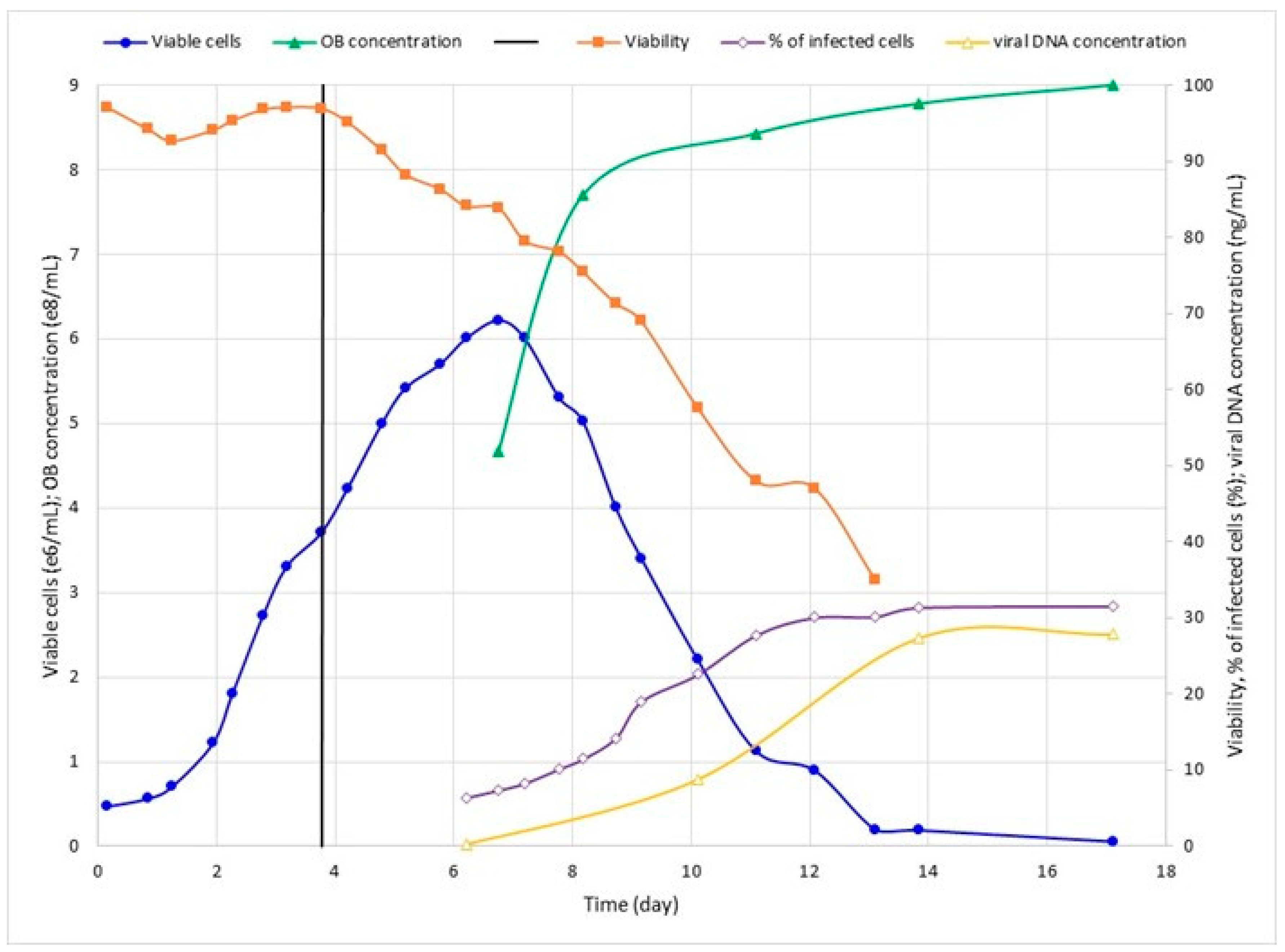
3.2. Effect of Viral Inoculum on SfMNPV Production
3.3. Quantitative PCR Standardization
3.4. Effect of SfMNPV Infection on Cell Metabolic Activities
3.5. Sequencing Analysis of SfMNPV Produced in Bioreactor
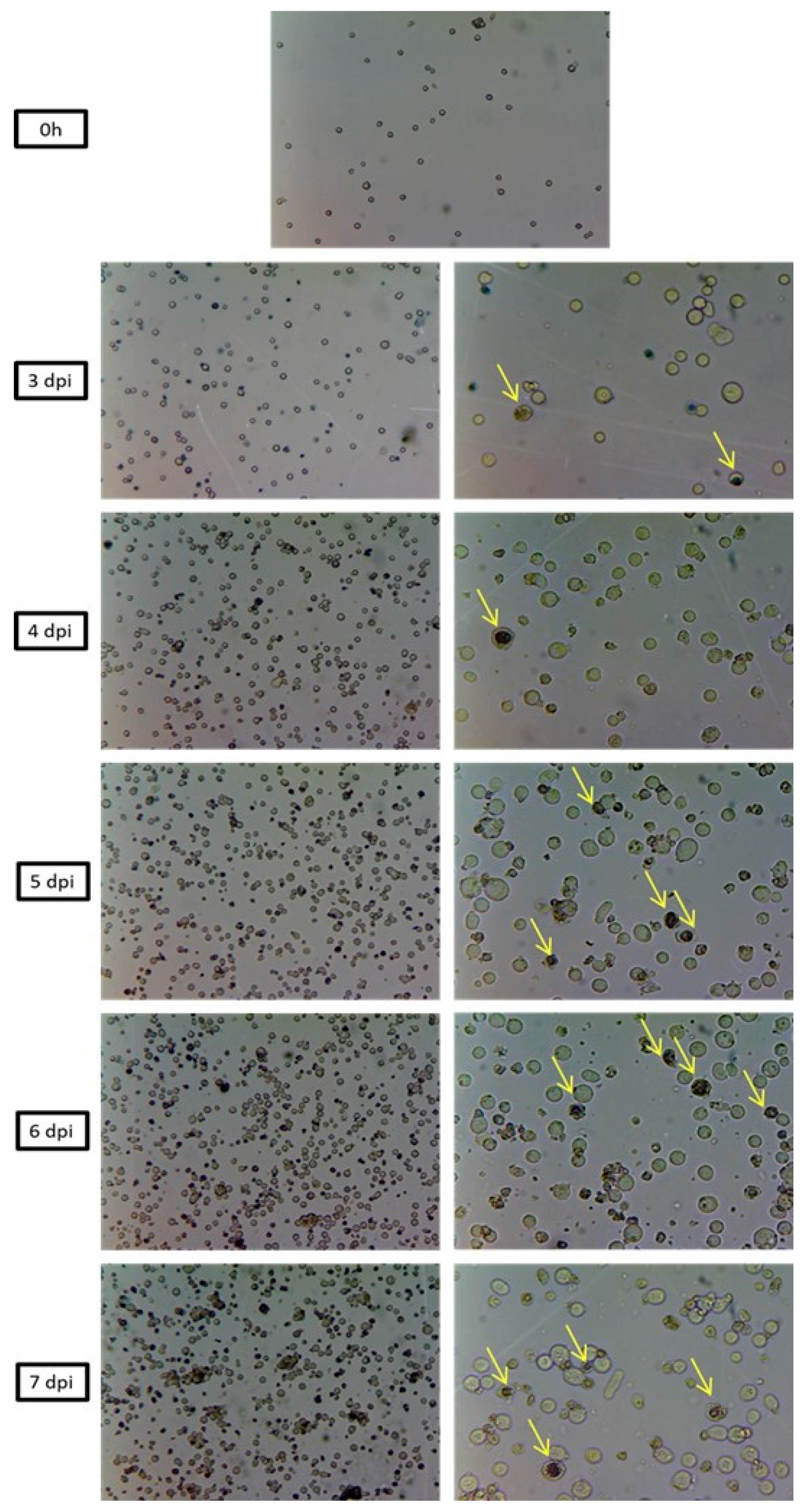
3.6. Morphological Analysis of SfMNPV Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farias, J.R.; Andow, D.A.; Horikoshi, R.J.; Sorgatto, R.J.; Fresia, P.; Dos Santos, A.C.; Omoto, C. Field evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot. 2014, 64, 150–158. [Google Scholar] [CrossRef]
- Cruz, I.; Figueiredo, M.; Oliveira, A.C.; Vasconcelos, C.A. Damage of Spodoptera frugiperda (Smith) in different maize genotypes cultivated in soil under three levels of aluminum saturation. Int. J. Pest. Manag. 1999, 45, 293–296. [Google Scholar] [CrossRef]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamo, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E smith) (Lepidoptera, Noctuidae), a new alien invasive pest in west and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Jiang, C.; Guo, X.; Chen, D.; You, C.; Zhang, Y.; Wang, M.; Li, Q. Potential distribution of Spodoptera frugiperda (J.E. Smith) in China and the major factors influencing distribution. Glob. Ecol. Conserv. 2020, 21, e00865. [Google Scholar] [CrossRef]
- Miranda, G.R.B.; Raetano, C.G.; Silva, E.; Daam, M.A.; Cerejeira, M.J. Environmental fate of neonicotinoids and classification of their potential risks to hypogean, epygean, and surface water ecosystems in Brazil. Hum. Ecol. Risk Assess. 2011, 17, 981–995. [Google Scholar] [CrossRef]
- Muraro, D.S.; Salmeron, E.; Cruz, J.V.S.; Amaral, F.S.A.; Guidolin, A.S.; Nascimento, A.R.B.; Malaquias, J.B.; Bernardi, O.; Omoto, C. Evidence of field-evolved resistance in Spodoptera frugiperda (Lepidoptera: Noctuidae) to emamectin benzoate in Brazil. Crop Prot. 2022, 162, 106071. [Google Scholar] [CrossRef]
- Barrera, G.; Simón, O.; Villamizar, L.; Williams, T.; Caballero, P. Spodoptera frugiperda multiple nucleopolyhedrovirus as a potential biological insecticide: Genetic and phenotypic comparison of field isolates from Colombia. Biol. Control 2011, 58, 113–120. [Google Scholar] [CrossRef]
- Cuartas-Otalora, P.E.; Gomez-Valderrama, J.A.; Ramos, A.E.; Barrera-Cubillos, G.P.; Villamizar-Rivero, L.F. Bio-insecticidal potential of nucleopolyhedrovirus and granulovirus mixtures to control the fall armyworm Spodoptera frugiperda (J.E. Smith, 1797) (Lepidoptera: Noctuidae). Viruses 2019, 11, 684. [Google Scholar] [CrossRef] [Green Version]
- Valicente, F.H.; Tuelher, E.S. Controle Biológico da Lagarta do Cartucho Spodoptera frugiperda, com baculovírus. Circular Técnica 114 da Embrapa Milho & Sorgo. Available online: https://ainfo.cnptia.embrapa.br/digital/bitstream/CNPMS-2010/22431/1/Circ-114.pdf (accessed on 12 June 2023).
- Faria, M.; Souza, D.A.; Sanches, M.M.; Schmidt, F.G.V.; Oliveira, C.M.; Benito, N.P.; Lopes, R.B. Evaluation of key parameters for developing a Metarhizium rileyi -based biopesticide against Spodoptera frugiperda (Lepidoptera: Noctuidae) in maize: Laboratory, greenhouse, and field trials. Pest. Manag. Sci. 2022, 78, 1146–1154. [Google Scholar] [CrossRef]
- Valicente, F.H.; Tuelher, E.S.; Pena, R.C.; Andreazza, R.; Guimarães, M.R.F. Cannibalism and virus production in Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) larvae fed with two leaf substrates inoculated with Baculovirus spodoptera. Neotrop. Entomol. 2013, 42, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Grzywacz, D. Basic and applied research: Baculovirus. In Microbial Control of Insect and Mite Pests; Lacey, L.A., Ed.; Academic Press: Cambridge, UK, 2016; pp. 27–46. [Google Scholar]
- Reid, S.; Malmanche, H.; Chan, L.; Popham, H.; van Oers, M.M. Production of entomopathogenic viruses. In Mass Production of Beneficial Organisms, 2nd ed.; Morales-Ramos, J.A., Rojas, M.G., Shapiro-Ilan, D.I., Eds.; Academic Press: Cambridge, UK, 2023; pp. 375–406. [Google Scholar] [CrossRef]
- de Almeida, A.F.; de Macedo, G.R.; Chan, L.C.L.; Pedrini, M.R.d.S. Kinetic analysis of in vitro production of wild-type Spodoptera frugiperda nucleopolyhedrovirus. Braz. Arch. Biol. Technol. 2010, 53, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Sihler, W.; Souza, M.L.; Valicente, F.H.; Falcao, R.; Sanches, M.M. In vitro infectivity of Spodoptera frugiperda multiple nucleopolyhedrovirus to different insect cell lines. Pesq. Agropec. Bras. 2018, 53, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Pedrini, M.R.S.; Wolff, J.L.C.; Reid, S. Fast accumulation of few polyhedra mutants during passage of a Spodoptera frugiperda multicapsid nucleopolyhedrovirus (Baculoviridae) in Sf9 cell cultures. Ann. Appl. Biol. 2004, 145, 107–112. [Google Scholar]
- Sanches, M.M.; Guimarães, G.C.; Sihler, W.; Souza, M.L. Successful co-infection of two different baculovirus species in the same cell line reveals a potential strategy for large in vitro production. Braz. J. Microbiol. 2021, 52, 1835–1843. [Google Scholar] [CrossRef]
- Barreto, M.R.; Guimaraes, C.T.; Teixeira, F.F.; Paiva, E.; Valicente, F.H. Effect of Baculovirus spodoptera isolates in Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) larvae and their characterization by RAPD. Neotrop. Entomol. 2005, 34, 67–75. [Google Scholar] [CrossRef] [Green Version]
- O’Reilly, D.R.; Miller, L.K.; Luckow, V.A. Baculovirus Expression Vectors: A Laboratory Manual; W.H. Freeman and Co.: Salt Lake City, UT, USA, 1992; pp. 1–347. [Google Scholar]
- Reed, L.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938, 27, 493–4978. [Google Scholar]
- Bozzola, J.J.; Russel, L.D. Specimen Preparation for Transmission Electron Microscopy. In Electron Microscopy Principles and Techniques for Biologists, 2nd ed.; Bozzola, J.J., Russel, L.D., Eds.; Jones and Bartlett Publishers: Sudbury, MA, USA, 1992; pp. 14–37. [Google Scholar]
- Wolff, J.L.C.; Valicente, F.H.; Martins, R.; Oliveira, J.V.C.; Zanotto, P.M.A. Analysis of the genome of Spodoptera frugiperda nucleopolyhedrovirus (SfMNPV-19) and of the high genomic heterogeneity in group II nucleopolyhedroviruses. J. Gen. Virol. 2008, 89, 1202–1211. [Google Scholar] [CrossRef]
- Sambrook, J.; Russel, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor: New York, NY, USA, 2001. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Bernal, V.; Carinhas, N.; Yokomizo, A.Y.; Carrondo, M.J.T.; Alves, P.M. Cell density effect in the baculovirus-insect cells system: A quantitative analysis of energetic metabolism. Biotechnol. Bioeng. 2009, 104, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.C.L.; Young, P.R.; Bletchy, C.; Reid, S. Production of the baculovirus-expressed dengue virus glycoprotein NS1 can be improved dramatically with optimised regimes for fed-batch cultures and the addition of the insect moulting hormone, 20-Hydroxyecdysone. J. Virol. Methods 2002, 105, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Ikonomou, L.; Schneider, Y.J.; Agathos, S.N. Insect cell culture for industrial production of recombinant proteins. Appl. Microbiol. Biotechnol. 2003, 62, 1–20. [Google Scholar] [CrossRef]
- Mena, J.A.; Kamen, A.A. Insect cell technology is a versatile and robust vaccine manufacturing platform. Expert Rev. Vaccines 2011, 10, 1063–1081. [Google Scholar] [CrossRef] [PubMed]
- Drugmand, J.C.; Schneider, Y.-J.; Agathos, S.N. Insect cells as factories for biomanufacturing. Biotechnol. Adv. 2012, 30, 1140–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Kanhaisingh, A.; Greenfield, P.F.; Reid, S.; Monsour, C.J.; Teakle, R. In vitro production of wild type Heliothis baculoviruses for use as biopesticides. Australas. Biotechnol. 1995, 5, 82–86. [Google Scholar]
- Rodas, V.M.; Marques, F.H.; Honda, M.T.; Soares, D.M.; Jorge, S.A.; Antoniazzi, M.M.; Medugno, C.; Castro, M.E.; Ribeiro, B.M.; Souza, M.L.; et al. Cell Culture Derived AgMNPV Bioinsecticide: Biological Constraints and Bioprocess Issues. Cytotechnology 2005, 48, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micheloud, G.A.; Gioria, V.V.; Eberhardt, I.; Visnovsky, G.; Claus, J.D. Production of the Anticarsia gemmatalis multiple nucleopolyhedrovirus in serum-free suspension cultures of the saUFL-AG-286 cell line in stirred reactor and airlift reactor. J. Virol. Methods 2011, 178, 106–116. [Google Scholar] [CrossRef]
- Castro, M.E.B.; Ribeiro, Z.M.A.; Souza, M.L. Infectivity of Anticarsia gemmatalis nucleopolyhedrovirus to different insect cell lines: Morphology, viral production, and protein synthesis. Biol. Control 2006, 36, 299–304. [Google Scholar] [CrossRef]
- Lua, L.H.; Reid, S. Effect of time of harvest of budded virus on the selection of baculovirus FP mutants in cell culture. Biotechnol. Prog. 2003, 19, 238–242. [Google Scholar] [CrossRef]
- Doverskog, M.; Bertram, E.; Ljunggren, J.; Ohman, L.; Sennerstam, R.; Haggstrom, L. Cell cycle progression in serum-free cultures of Sf-9 insect cells: Modulation by conditioned medium factors and implications for proliferation and productivity. Biotechnol. Prog. 2000, 16, 837–846. [Google Scholar] [CrossRef]
- Jardin, B.A.; Montes, J.; Lanthier, S.; Tran, R.; Elias, C. High cell density fed batch and perfusion processes for stable non-viral expression of secreted alkaline phosphatase (SEAP) using insect cells: Comparison to a batch Sf-9-BEV system. Biotechnol. Bioeng. 2007, 97, 332–345. [Google Scholar] [CrossRef]
- Gioria, V.V.; Jäger, V.; Claus, J.D. Growth, metabolism and baculovirus production in suspension cultures of an Anticarsia gemmatalis cell line. Cytotechnology 2006, 52, 113–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, F.; Bernal, V.; Alves, P.M. The Role of Host Cell Physiology in the Productivity of the Baculovirus-Insect Cell System: Fluxome Analysis of Trichoplusia ni and Spodoptera frugiperda Cell Lines. Biotechnol. Bioeng. 2016, 114, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Rezende, S.H.; Castro, M.E.; Souza, M.L. Accumulation of few-polyhedra mutants upon serial passage of Anticarsia gemmatalis multiple nucleopolyhedrovirus in cell culture. J. Invertebr. Pathol. 2009, 100, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Radford, K.M.; Cavegn, C.; Bertrand, M.; Bernard, A.R.; Reid, S.; Greenfield, P.F. The indirect effects of multiplicity of infection on baculovirus expressed proteins in insect cells: Secreted and non-secreted products. Cytotechnology 1997, 24, 73–81. [Google Scholar] [CrossRef]


| Gene | Primer Name | Primer Sequence (5′-3′) |
|---|---|---|
| polh | polhFw_Sf_2013 polhRv_Sf_2013 | (AATGTATACTCGTTACAGCTATAACCCA) (GTGGTATGGTTTATTAGTACGCGGG) |
| protf | fproteinFw2_Sf_2013 fproteinRv2_Sf_2013 | (GCCGAACGTAAGTTGTTGTT) (CATACACAGATCCATTAACATTTACA) |
| fp25k | 25kfpFw_Sf_2013 25kfpRv_Sf_2013 | (CATAAACTAACATGACGACTGCCACTG) (CGTTTATCGCGTTGCGCACTCATC) |
| pp34 | pp34Fw_Sf_2013 pp34Rv_Sf_2013 | (GTTACAATATAATGTCGTTGATTAC) (CTTGGATAATCCTTTGATTG) |
| p10 | p10Fw_Sf_2013 p10Rv_Sf_2013 | (CGCATTCGATTAGACGGACC) (GGCCACGATACAGAATTACGC) |
| Run | MOI | Maximum Viable Cell Concentration (×106 cell/mL) | Maximum SfMNPV Concentrations (×106 OBs/mL) (×107) | Percentage of Infected Cells (%) | OBs/ Infected Cell | Viral DNA (ng/mL) |
|---|---|---|---|---|---|---|
| 3 | 0.1 | 6.2 | 6.5 | 30.0 | 30.4 | 2.03 |
| 4 | 1.0 | 6.2 | 90.0 | 32.0 | 339 | 8.72 |
| Cell Metabolic Parameter | Cell Consumption/Production Rate (nmol/106 cel/h) | |
|---|---|---|
| MOI 0.1 (Run 3) | MOI 1.0 (Run 4) | |
| Glucose | 25.8 | 115.5 |
| Lactate | 0.9 | 1.7 |
| Glutamine | 9.8 | 12.3 |
| Glutamate | 2.7 | 1.5 |
| Ammonium | 0.9 | 13.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klafke, K.; Sanches, M.M.; Sihler, W.; Souza, M.L.d.; Tonso, A. Bioreactor Production Process of Spodoptera frugiperda multiple nucleopolyhedrovirus Biopesticide. Pathogens 2023, 12, 1001. https://doi.org/10.3390/pathogens12081001
Klafke K, Sanches MM, Sihler W, Souza MLd, Tonso A. Bioreactor Production Process of Spodoptera frugiperda multiple nucleopolyhedrovirus Biopesticide. Pathogens. 2023; 12(8):1001. https://doi.org/10.3390/pathogens12081001
Chicago/Turabian StyleKlafke, Karina, Marcio Martinello Sanches, William Sihler, Marlinda Lobo de Souza, and Aldo Tonso. 2023. "Bioreactor Production Process of Spodoptera frugiperda multiple nucleopolyhedrovirus Biopesticide" Pathogens 12, no. 8: 1001. https://doi.org/10.3390/pathogens12081001
APA StyleKlafke, K., Sanches, M. M., Sihler, W., Souza, M. L. d., & Tonso, A. (2023). Bioreactor Production Process of Spodoptera frugiperda multiple nucleopolyhedrovirus Biopesticide. Pathogens, 12(8), 1001. https://doi.org/10.3390/pathogens12081001







