Transcriptional Host Responses to Infection with Streptococcus suis in a Porcine Precision-Cut Lung Slice Model: Between-Strain Differences Suggest Association with Virulence Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Preparation of PCLS
2.3. Infection of PCLS
2.4. Isolation of Porcine and Bacterial RNA
2.5. Reverse Transcription and Pre-Amplification of cDNA
2.6. High-Throughput qPCR
2.7. qPCR Data Analysis
2.8. Immunofluorescence Staining
2.9. ELISA
2.10. Statistical Analysis and Data Visualization
3. Results
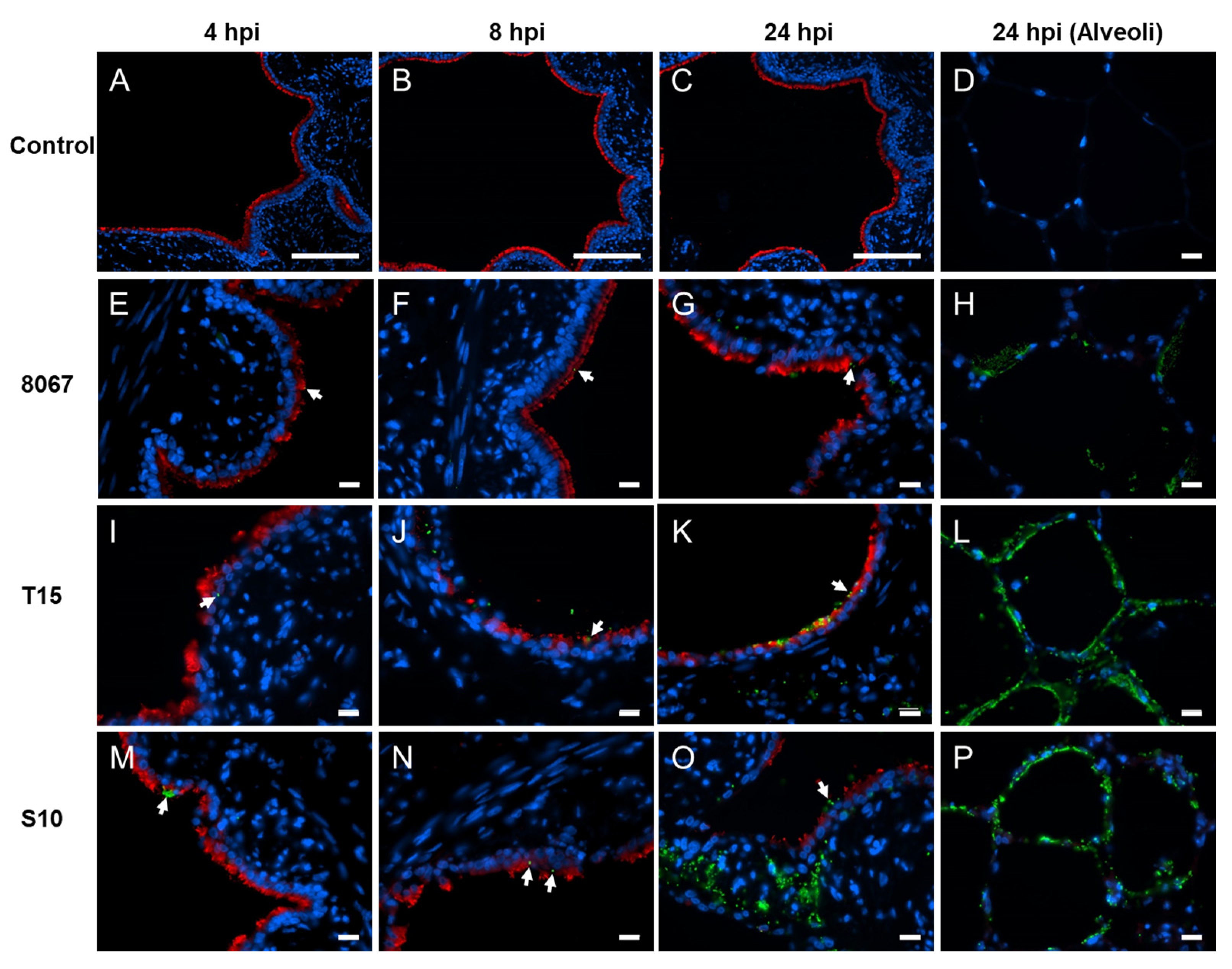
3.1. Growth, Adherence, and Colonization of S. suis in Porcine PCLS
3.2. Transcriptional Host Response to S. suis Infection in the PCLS Model
3.3. ELISA Quantification of Pro-Inflammatory Cytokines
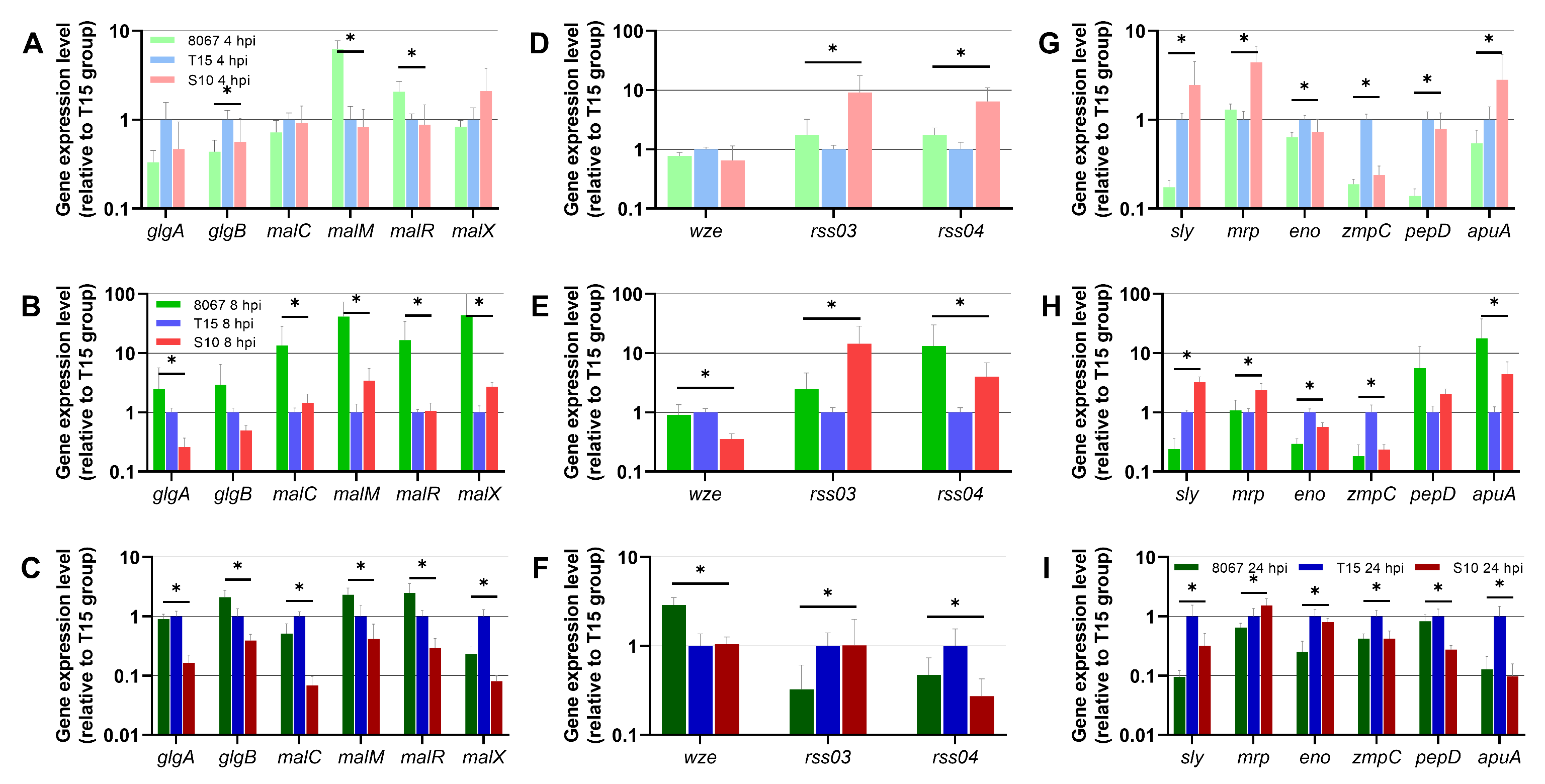
3.4. S. suis Gene Expression in the PCLS Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Votsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis—The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.; Grenier, D. Understanding the virulence of Streptococcus suis: A veterinary, medical, and economic challenge. Med. Et Mal. Infect. 2018, 48, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Hlebowicz, M.; Jakubowski, P.; Smiatacz, T. Streptococcus suis Meningitis: Epidemiology, Clinical Presentation and Treatment. Vector Borne Zoonotic Dis. 2019, 19, 557–562. [Google Scholar] [CrossRef] [PubMed]
- de Greeff, A.; Wisselink, H.J.; de Bree, F.M.; Schultsz, C.; Baums, C.G.; Thi, H.N.; Stockhofe-Zurwieden, N.; Smith, H.E. Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization. BMC Microbiol. 2011, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef] [PubMed]
- Baums, C.G.; Valentin-Weigand, P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim. Health Res. Rev. 2009, 10, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.M.; Baums, C.G.; Rehm, T.; Wisselink, H.J.; Goethe, R.; Valentin-Weigand, P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 2006, 115, 117–127. [Google Scholar] [CrossRef]
- Lecours, M.P.; Gottschalk, M.; Houde, M.; Lemire, P.; Fittipaldi, N.; Segura, M. Critical role for Streptococcus suis cell wall modifications and suilysin in resistance to complement-dependent killing by dendritic cells. J. Infect. Dis. 2011, 204, 919–929. [Google Scholar] [CrossRef]
- Li, G.; Wang, G.; Si, X.; Zhang, X.; Liu, W.; Li, L.; Wang, J. Inhibition of suilysin activity and inflammation by myricetin attenuates Streptococcus suis virulence. Life Sci. 2019, 223, 62–68. [Google Scholar] [CrossRef]
- Tenenbaum, T.; Asmat, T.; Seitz, M.; Schroten, H.; Schwerk, C. Biological activities of suilysin: Role in Streptococcus suis pathogenesis. Future Microbiol. 2016, 11, 941–954. [Google Scholar] [CrossRef] [PubMed]
- Tharavichitkul, P.; Wongsawan, K.; Takenami, N.; Pruksakorn, S.; Fongcom, A.; Gottschalk, M.; Khanthawa, B.; Supajatura, V.; Takai, S. Correlation between PFGE Groups and mrp/epf/sly Genotypes of Human Streptococcus suis Serotype 2 in Northern Thailand. J. Pathog. 2014, 2014, 350416. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.A.; Loeffen, P.L.; van den Berg, A.J.; Storm, P.K. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 1994, 62, 1742–1748. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Auger, J.P.; Segura, M.; Fittipaldi, N.; Takamatsu, D.; Okura, M.; Gottschalk, M. Role of the capsular polysaccharide as a virulence factor for Streptococcus suis serotype 14. Can. J. Vet. Res. 2015, 79, 141–146. [Google Scholar] [PubMed]
- Segura, M.; Gottschalk, M.; Olivier, M. Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect. Immun. 2004, 72, 5322–5330. [Google Scholar] [CrossRef]
- Smith, H.E.; Damman, M.; van der Velde, J.; Wagenaar, F.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Smits, M.A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: The capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 1999, 67, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Neila-Ibanez, C.; Brogaard, L.; Pailler-Garcia, L.; Martinez, J.; Segales, J.; Segura, M.; Heegaard, P.M.H.; Aragon, V. Piglet innate immune response to Streptococcus suis colonization is modulated by the virulence of the strain. Vet. Res. 2021, 52, 145. [Google Scholar] [CrossRef]
- Liu, M.; Tan, C.; Fang, L.; Xiao, S.; Chen, H. Microarray analyses of THP-1 cells infected with Streptococcus suis serotype 2. Vet. Microbiol. 2011, 150, 126–131. [Google Scholar] [CrossRef]
- Graveline, R.; Segura, M.; Radzioch, D.; Gottschalk, M. TLR2-dependent recognition of Streptococcus suis is modulated by the presence of capsular polysaccharide which modifies macrophage responsiveness. Int. Immunol. 2007, 19, 375–389. [Google Scholar] [CrossRef]
- Zheng, H.; Luo, X.; Segura, M.; Sun, H.; Ye, C.; Gottschalk, M.; Xu, J. The role of toll-like receptors in the pathogenesis of Streptococcus suis. Vet. Microbiol. 2012, 156, 147–156. [Google Scholar] [CrossRef]
- Meijerink, M.; Ferrando, M.L.; Lammers, G.; Taverne, N.; Smith, H.E.; Wells, J.M. Immunomodulatory effects of Streptococcus suis capsule type on human dendritic cell responses, phagocytosis and intracellular survival. PLoS ONE 2012, 7, e35849. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Lachance, C.; Wang, Y.; Gagnon, C.A.; Savard, C.; Segura, M.; Grenier, D.; Gottschalk, M. Transcriptional approach to study porcine tracheal epithelial cells individually or dually infected with swine influenza virus and Streptococcus suis. BMC Vet. Res. 2014, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Fang, L.; Tan, C.; Long, T.; Chen, H.; Xiao, S. Understanding Streptococcus suis serotype 2 infection in pigs through a transcriptional approach. BMC Genom. 2011, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co Ltd.: London, UK, 1959. [Google Scholar]
- Meng, F.; Punyadarsaniya, D.; Uhlenbruck, S.; Hennig-Pauka, I.; Schwegmann-Wessels, C.; Ren, X.; Durrwald, R.; Herrler, G. Replication characteristics of swine influenza viruses in precision-cut lung slices reflect the virulence properties of the viruses. Vet. Res. 2013, 44, 110. [Google Scholar] [CrossRef] [PubMed]
- Weldearegay, Y.B.; Muller, S.; Hanske, J.; Schulze, A.; Kostka, A.; Ruger, N.; Hewicker-Trautwein, M.; Brehm, R.; Valentin-Weigand, P.; Kammerer, R.; et al. Host-Pathogen Interactions of Mycoplasma mycoides in Caprine and Bovine Precision-Cut Lung Slices (PCLS) Models. Pathogens 2019, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Reamon-Buettner, S.M.; Niehof, M.; Hirth, N.; Danov, O.; Obernolte, H.; Braun, A.; Warnecke, J.; Sewald, K.; Wronski, S. Transcriptomic Analysis Reveals Priming of The Host Antiviral Interferon Signaling Pathway by Bronchobini((R)) Resulting in Balanced Immune Response to Rhinovirus Infection in Mouse Lung Tissue Slices. Int. J. Mol. Sci. 2019, 20, 2242. [Google Scholar] [CrossRef] [PubMed]
- Henjakovic, M.; Martin, C.; Hoymann, H.G.; Sewald, K.; Ressmeyer, A.R.; Dassow, C.; Pohlmann, G.; Krug, N.; Uhlig, S.; Braun, A. Ex vivo lung function measurements in precision-cut lung slices (PCLS) from chemical allergen-sensitized mice represent a suitable alternative to in vivo studies. Toxicol. Sci. 2008, 106, 444–453. [Google Scholar] [CrossRef]
- Henjakovic, M.; Sewald, K.; Switalla, S.; Kaiser, D.; Muller, M.; Veres, T.Z.; Martin, C.; Uhlig, S.; Krug, N.; Braun, A. Ex vivo testing of immune responses in precision-cut lung slices. Toxicol. Appl. Pharmacol. 2008, 231, 68–76. [Google Scholar] [CrossRef]
- Lyons-Cohen, M.R.; Thomas, S.Y.; Cook, D.N.; Nakano, H. Precision-cut Mouse Lung Slices to Visualize Live Pulmonary Dendritic Cells. J. Vis. Exp. 2017, 122, e55465. [Google Scholar] [CrossRef]
- Temann, A.; Golovina, T.; Neuhaus, V.; Thompson, C.; Chichester, J.A.; Braun, A.; Yusibov, V. Evaluation of inflammatory and immune responses in long-term cultured human precision-cut lung slices. Hum. Vaccines Immunother. 2017, 13, 351–358. [Google Scholar] [CrossRef]
- Brogaard, L.; Klitgaard, K.; Heegaard, P.M.; Hansen, M.S.; Jensen, T.K.; Skovgaard, K. Concurrent host-pathogen gene expression in the lungs of pigs challenged with Actinobacillus pleuropneumoniae. BMC Genom. 2015, 16, 417. [Google Scholar] [CrossRef] [PubMed]
- Vecht, U.; Arends, J.P.; van der Molen, E.J.; van Leengoed, L.A. Differences in virulence between two strains of Streptococcus suis type II after experimentally induced infection of newborn germ-free pigs. Am. J. Vet. Res. 1989, 50, 1037–1043. [Google Scholar] [PubMed]
- Paddenberg, R.; Mermer, P.; Goldenberg, A.; Kummer, W. Videomorphometric analysis of hypoxic pulmonary vasoconstriction of intra-pulmonary arteries using murine precision cut lung slices. J. Vis. Exp. 2014, 83, e50970. [Google Scholar] [CrossRef]
- Punyadarsaniya, D.; Liang, C.H.; Winter, C.; Petersen, H.; Rautenschlein, S.; Hennig-Pauka, I.; Schwegmann-Wessels, C.; Wu, C.Y.; Wong, C.H.; Herrler, G. Infection of differentiated porcine airway epithelial cells by influenza virus: Differential susceptibility to infection by porcine and avian viruses. PLoS ONE 2011, 6, e28429. [Google Scholar] [CrossRef] [PubMed]
- Niehof, M.; Hildebrandt, T.; Danov, O.; Arndt, K.; Koschmann, J.; Dahlmann, F.; Hansen, T.; Sewald, K. RNA isolation from precision-cut lung slices (PCLS) from different species. BMC Res. Notes 2017, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Dresen, M.; Schenk, J.; Berhanu Weldearegay, Y.; Votsch, D.; Baumgartner, W.; Valentin-Weigand, P.; Nerlich, A. Streptococcus suis Induces Expression of Cyclooxygenase-2 in Porcine Lung Tissue. Microorganisms 2021, 9, 366. [Google Scholar] [CrossRef]
- Votsch, D.; Willenborg, M.; Baumgartner, W.; Rohde, M.; Valentin-Weigand, P. Bordetella bronchiseptica promotes adherence, colonization, and cytotoxicity of Streptococcus suis in a porcine precision-cut lung slice model. Virulence 2021, 12, 84–95. [Google Scholar] [CrossRef]
- Viana, F.; O’Kane, C.M.; Schroeder, G.N. Precision-cut lung slices: A powerful ex vivo model to investigate respiratory infectious diseases. Mol. Microbiol. 2022, 117, 578–588. [Google Scholar] [CrossRef]
- Haapasalo, K.; Meri, S. Regulation of the Complement System by Pentraxins. Front. Immunol. 2019, 10, 1750. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Mu, Y.; Zhang, Y.; Dong, W.; Zhu, Y.; Ma, J.; Song, W.; Pan, Z.; Lu, C.; Yao, H. Antibacterial effect of porcine PTX3 against Streptococcus suis type 2 infection. Microb. Pathog. 2015, 89, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Beninati, C.; Zummo, S.; Biondo, C. Protective role of IL-18 in host defenses against group B Streptococcus. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2657–2663. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zeng, Y.; Gao, P.; Lu, X.; Xia, K.; Zhou, L.; Zhang, C.; Yi, C.; Zhang, A. IL-18 Signaling Is Essential for Causing Streptococcal Toxic Shock-like Syndrome (STSLS). Life 2022, 12, 1324. [Google Scholar] [CrossRef] [PubMed]
- Lecours, M.P.; Segura, M.; Fittipaldi, N.; Rivest, S.; Gottschalk, M. Immune receptors involved in Streptococcus suis recognition by dendritic cells. PLoS ONE 2012, 7, e44746. [Google Scholar] [CrossRef] [PubMed]
- Lavagna, A.; Auger, J.P.; Girardin, S.E.; Gisch, N.; Segura, M.; Gottschalk, M. Recognition of Lipoproteins by Toll-like Receptor 2 and DNA by the AIM2 Inflammasome Is Responsible for Production of Interleukin-1beta by Virulent Suilysin-negative Streptococcus suis Serotype 2. Pathogens 2020, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Auger, J.P.; Boa, A.C.; Segura, M.; Gottschalk, M. Antigen I/II Participates in the Interactions of Streptococcus suis Serotype 9 With Phagocytes and the Development of Systemic Disease. Front. Cell Infect. Microbiol. 2019, 9, 124. [Google Scholar] [CrossRef]
- Auger, J.P.; Benoit-Biancamano, M.O.; Bedard, C.; Segura, M.; Gottschalk, M. Differential role of MyD88 signaling in Streptococcus suis serotype 2-induced systemic and central nervous system diseases. Int. Immunol. 2019, 31, 697–714. [Google Scholar] [CrossRef]
- Lavagna, A.; Auger, J.P.; Dumesnil, A.; Roy, D.; Girardin, S.E.; Gisch, N.; Segura, M.; Gottschalk, M. Interleukin-1 signaling induced by Streptococcus suis serotype 2 is strain-dependent and contributes to bacterial clearance and inflammation during systemic disease in a mouse model of infection. Vet. Res. 2019, 50, 52. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weldearegay, Y.B.; Brogaard, L.; Nerlich, A.; Schaaf, D.; Heegaard, P.M.H.; Valentin-Weigand, P. Transcriptional Host Responses to Infection with Streptococcus suis in a Porcine Precision-Cut Lung Slice Model: Between-Strain Differences Suggest Association with Virulence Potential. Pathogens 2024, 13, 4. https://doi.org/10.3390/pathogens13010004
Weldearegay YB, Brogaard L, Nerlich A, Schaaf D, Heegaard PMH, Valentin-Weigand P. Transcriptional Host Responses to Infection with Streptococcus suis in a Porcine Precision-Cut Lung Slice Model: Between-Strain Differences Suggest Association with Virulence Potential. Pathogens. 2024; 13(1):4. https://doi.org/10.3390/pathogens13010004
Chicago/Turabian StyleWeldearegay, Yenehiwot Berhanu, Louise Brogaard, Andreas Nerlich, Désirée Schaaf, Peter M. H. Heegaard, and Peter Valentin-Weigand. 2024. "Transcriptional Host Responses to Infection with Streptococcus suis in a Porcine Precision-Cut Lung Slice Model: Between-Strain Differences Suggest Association with Virulence Potential" Pathogens 13, no. 1: 4. https://doi.org/10.3390/pathogens13010004





