G-Quadruplexes in the Regulation of Viral Gene Expressions and Their Impacts on Controlling Infection
Abstract
:1. Introduction to G-Quadruplexes
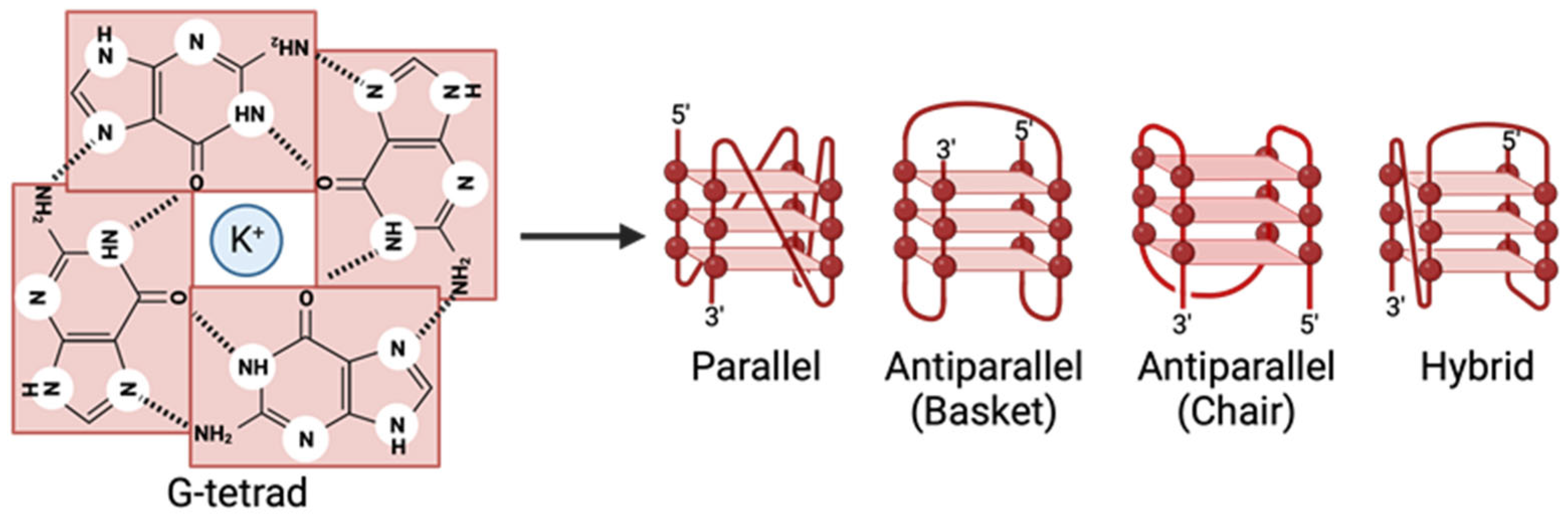
1.1. G4 Structure
1.2. Determining G4 Formation
1.3. G4s in Eukaryotes
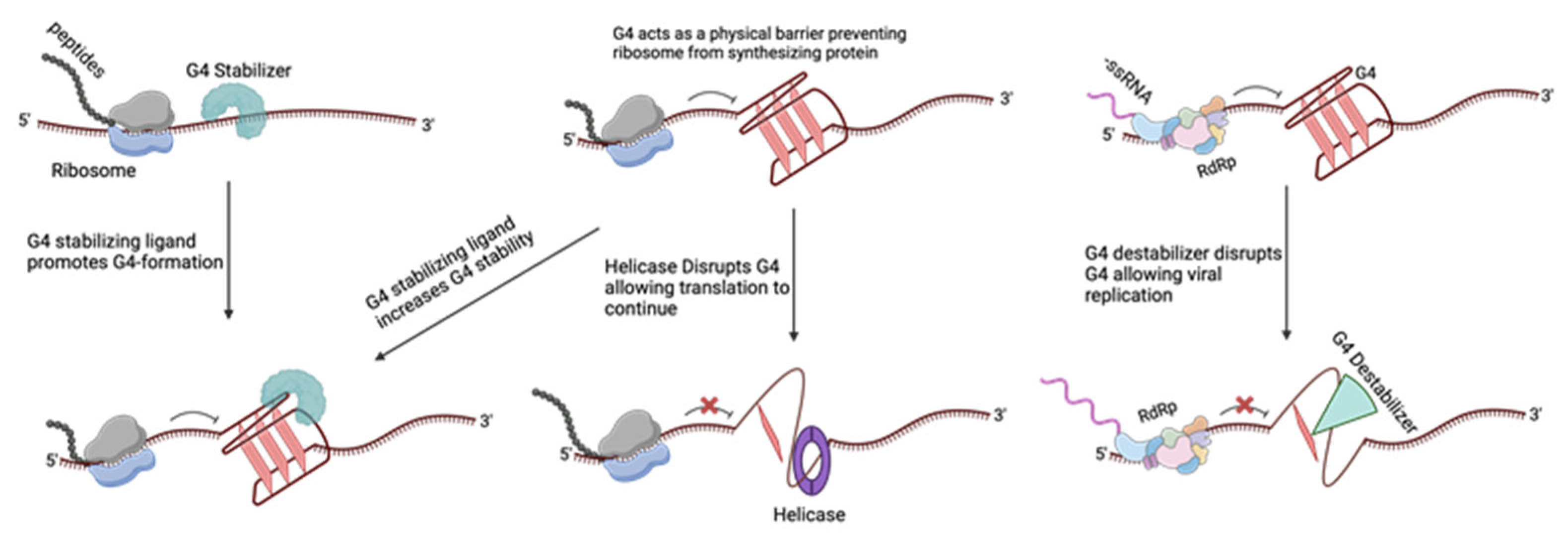
2. Commonly Studied G4 Ligands
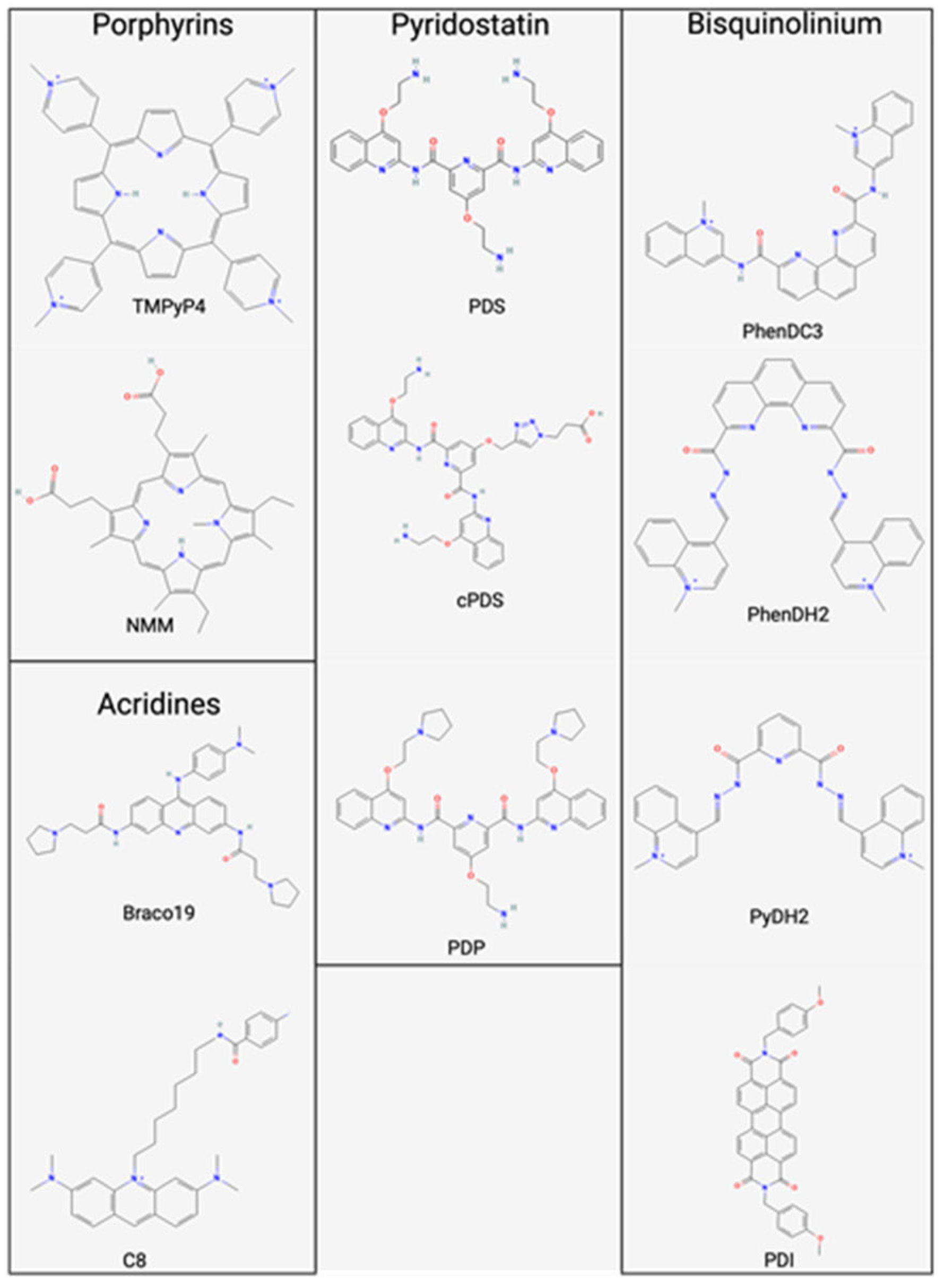
2.1. Porphyrins
2.2. Bisquinolinium Compounds
2.3. Napthalene Diimides
2.4. Acridines
2.5. Pyridostatin/Derivatives
3. Alternative Strategies and Ligands in Targeting G4s
3.1. Antisense Oligonucleotides
3.2. CRISPR–Cas9-Mediated G4 Stabilization
3.3. RNA Interference and Small Interfering RNA
3.4. Aptamers
4. DNA Viruses
4.1. Herpesviridae
4.2. Human Papillomavirus
4.3. Polyomavirus
4.4. Hepatitis B
4.5. Monkeypox Virus

5. RNA Viruses
5.1. HIV
5.2. Hepatitis C
5.3. Zika Virus and Other Flaviviruses
5.4. Influenza
5.5. Enterovirus
5.6. Coronaviruses
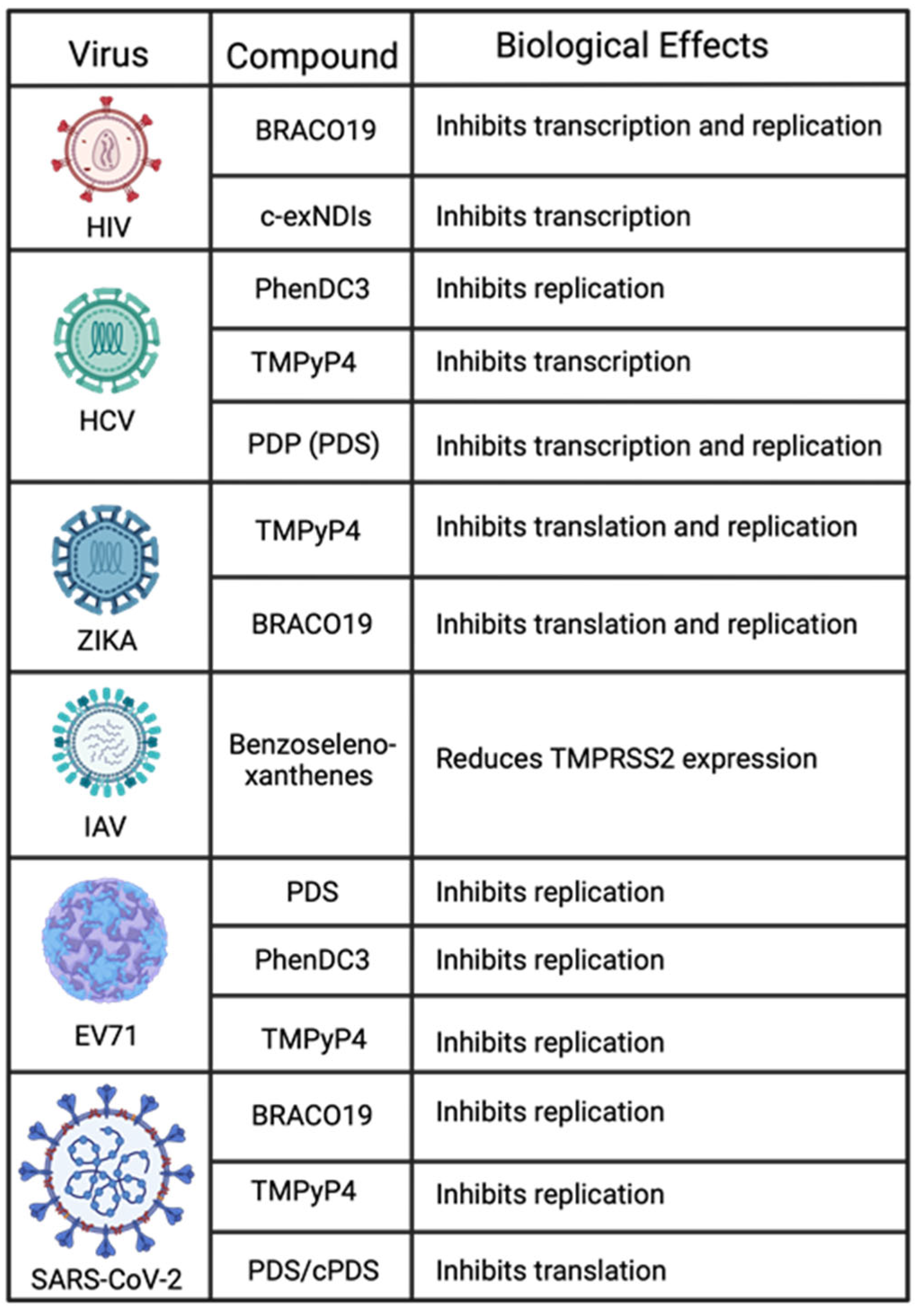
6. Discussion
7. Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Cui, Y.; Koirala, D.; Ghimire, C.; Kushwaha, S.; Yu, Z.; Yangyuoru, P.M.; Mao, H. Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 2013, 41, 3915–3923. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Howard, F.B.; Miles, H.T. Poly(inosinic acid) helices: Essential chelation of alkali metal ions in the axial channel. Biochemistry 1982, 21, 6736–6745. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, S.; Kettani, A.; Patel, D.J. A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 1998, 282, 637–652. [Google Scholar] [CrossRef]
- Plavec, J.; Tong, W.; Chattopadhyaya, J. How do the gauche and anomeric effects drive the pseudorotational equilibrium of the pentofuranose moiety of nucleosides? J. Am. Chem. Soc. 1993, 115, 9734–9746. [Google Scholar] [CrossRef]
- Zaccaria, F.; Paragi, G.; Fonseca Guerra, C. The role of alkali metal cations in the stabilization of guanine quadruplexes: Why K(+) is the best. Phys. Chem. Chem. Phys. 2016, 18, 20895–20904. [Google Scholar] [CrossRef]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef]
- Webba da Silva, M.; Trajkovski, M.; Sannohe, Y.; Ma’ani Hessari, N.; Sugiyama, H.; Plavec, J. Design of a G-Quadruplex Topology through Glycosidic Bond Angles. Angew. Chem. Int. Ed. 2009, 48, 9167–9170. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.M.; Neidle, S.; Parkinson, G.N. A structural analysis of G-quadruplex/ligand interactions. Biochimie 2011, 93, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Biver, T. Discriminating between Parallel, Anti-Parallel and Hybrid G-Quadruplexes: Mechanistic Details on Their Binding to Small Molecules. Molecules 2022, 27, 4165. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.T.; Varnai, P.; Bugaut, A.; Reszka, A.P.; Neidle, S.; Balasubramanian, S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009, 131, 13399–13409. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.W.; Alberti, P.; Guédin, A.; Lacroix, L.; Riou, J.F.; Royle, N.J.; Mergny, J.L.; Phan, A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 2009, 37, 6239–6248. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Li, M.H.; Hsu, S.T.; Chang, T.C. Structural basis of sodium-potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 2014, 42, 4723–4733. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Agarwala, P.; Maiti, S. Effect of Loops and G-Quartets on the Stability of RNA G-Quadruplexes. J. Phys. Chem. B 2013, 117, 6896–6905. [Google Scholar] [CrossRef]
- Da Silva, M.W. Geometric formalism for DNA quadruplex folding. Chemistry 2007, 13, 9738–9745. [Google Scholar] [CrossRef]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Dai, J.; Punchihewa, C.; Ambrus, A.; Chen, D.; Jones, R.A.; Yang, D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: A novel adenine triple formation. Nucleic Acids Res. 2007, 35, 2440–2450. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Noguchi, Y.; Sugiyama, H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006, 14, 5584–5591. [Google Scholar] [CrossRef] [PubMed]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Luu, K.N.; Patel, D.J. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006, 34, 5715–5719. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dai, J.; Veliath, E.; Jones, R.A.; Yang, D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: Insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010, 38, 1009–1021. [Google Scholar] [CrossRef]
- Hänsel, R.; Löhr, F.; Trantirek, L.; Dötsch, V. High-resolution insight into G-overhang architecture. J. Am. Chem. Soc. 2013, 135, 2816–2824. [Google Scholar] [CrossRef]
- Yang, D. G-Quadruplex DNA and RNA. Methods Mol. Biol. 2019, 2035, 1–24. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef] [PubMed]
- Martadinata, H.; Phan, A.T. Structure of human telomeric RNA (TERRA): Stacking of two G-quadruplex blocks in K(+) solution. Biochemistry 2013, 52, 2176–2183. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Haider, S.M.; Neidle, S.; Parkinson, G.N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010, 38, 5569–5580. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, D. Sequence, Stability, and Structure of G-Quadruplexes and Their Interactions with Drugs. Curr. Protoc. Nucleic Acid. Chem. 2012, 50, 17.5.1–17.5.17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.H.; Fujimoto, T.; Saxena, S.; Yu, H.Q.; Miyoshi, D.; Sugimoto, N. Monomorphic RNA G-quadruplex and polymorphic DNA G-quadruplex structures responding to cellular environmental factors. Biochemistry 2010, 49, 4554–4563. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef]
- Nazia, P.; Amen, S.; Seunghee Cho and Kyeong Kyu, K. Computational Approaches to Predict the Non-canonical DNAs. Curr. Bioinform. 2019, 14, 470–479. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovská, I.; Renciuk, D.; Vorlícková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. UV Melting of G-Quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009, 17, 17.1.1–17.1.15. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Hurley, L.H. Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods Mol. Biol. 2010, 608, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Patel, D.J. DNA architecture: From G to Z. Curr. Opin. Struct. Biol. 2006, 16, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.K.; Shayo, Y.; Vankayalapati, H.; Hurley, L.H.; Schaefer, J. Structure of a quinobenzoxazine-G-quadruplex complex by REDOR NMR. Biochemistry 2004, 43, 11953–11958. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.M.; Parkinson, G.N.; Neidle, S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003, 326, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Collie, G.W. X-Ray Crystallographic Studies of G-Quadruplex Structures. Methods Mol. Biol. 2019, 2035, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Parkinson, G.N. Crystallographic studies of quadruplex nucleic acids. Methods 2007, 43, 252–263. [Google Scholar] [CrossRef]
- Millevoi, S.; Moine, H.; Vagner, S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA 2012, 3, 495–507. [Google Scholar] [CrossRef]
- Yu, C.-H.; Teulade-Fichou, M.-P.; Olsthoorn, R.C.L. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res. 2014, 42, 1887–1892. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Metifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Sun, X.; Wang, L.; Li, Q.; Guan, A.; Shen, G.; Tang, Y. Selective recognition of c-myc promoter G-quadruplex and down-regulation of oncogene c-myc transcription in human cancer cells by 3,8a-disubstituted indolizinone. RSC Adv. 2017, 7, 51965–51969. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef]
- Membrino, A.; Cogoi, S.; Pedersen, E.B.; Xodo, L.E. G4-DNA Formation in the HRAS Promoter and Rational Design of Decoy Oligonucleotides for Cancer Therapy. PLoS ONE 2011, 6, e24421. [Google Scholar] [CrossRef]
- Nagesh, N.; Sharma, V.K.; Ganesh Kumar, A.; Lewis, E.A. Effect of Ionic Strength on Porphyrin Drugs Interaction with Quadruplex DNA Formed by the Promoter Region of C-myc and Bcl2 Oncogenes. J. Nucleic Acids 2010, 2010, 146418. [Google Scholar] [CrossRef]
- Ou, T.M.; Lu, Y.J.; Tan, J.H.; Huang, Z.S.; Wong, K.Y.; Gu, L.Q. G-quadruplexes: Targets in anticancer drug design. Chem. Med. Chem. 2008, 3, 690–713. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Doria, F.; Butovskaya, E.; Frasson, I.; Botti, S.; Scalabrin, M.; Lago, S.; Grande, V.; Nadai, M.; Freccero, M.; et al. Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 2015, 58, 9639–9652. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. Annu. Rep. Med. Chem. 2020, 54, 101–131. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Zanin, I.; Terreri, M.; Richter, S.N. G-Quadruplex Targeting in the Fight against Viruses: An Update. Int. J. Mol. Sci. 2021, 22, 10984. [Google Scholar] [CrossRef] [PubMed]
- Abiri, A.; Lavigne, M.; Rezaei, M.; Nikzad, S.; Zare, P.; Mergny, J.L.; Rahimi, H.R. Unlocking G-Quadruplexes as Antiviral Targets. Pharmacol. Rev. 2021, 73, 897–923. [Google Scholar] [CrossRef]
- Kingsbury, C.J.; Senge, M.O. The shape of porphyrins. Coord. Chem. Rev. 2021, 431, 213760. [Google Scholar] [CrossRef]
- Cavallari, M.; Garbesi, A.; Di Felice, R. Porphyrin Intercalation in G4-DNA Quadruplexes by Molecular Dynamics Simulations. J. Phys. Chem. B 2009, 113, 13152–13160. [Google Scholar] [CrossRef]
- Fujiwara, N.; Mazzola, M.; Cai, E.; Wang, M.; Cave, J.W. TMPyP4, a Stabilizer of Nucleic Acid Secondary Structure, Is a Novel Acetylcholinesterase Inhibitor. PLoS ONE 2015, 10, e0139167. [Google Scholar] [CrossRef]
- Le, V.H.; Nagesh, N.; Lewis, E.A. Bcl-2 promoter sequence G-quadruplex interactions with three planar and non-planar cationic porphyrins: TMPyP4, TMPyP3, and TMPyP2. PLoS ONE 2013, 8, e72462. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.; Trent, J.O.; Chowdhry, B.Z.; Jenkins, T.C. Intercalative G-Tetraplex Stabilization of Telomeric DNA by a Cationic Porphyrin1. J. Am. Chem. Soc. 1999, 121, 1768–1779. [Google Scholar] [CrossRef]
- Arora, A.; Maiti, S. Effect of Loop Orientation on Quadruplex−TMPyP4 Interaction. J. Phys. Chem. B 2008, 112, 8151–8159. [Google Scholar] [CrossRef] [PubMed]
- Grand, C.L.; Han, H.; Muñoz, R.M.; Weitman, S.; Von Hoff, D.D.; Hurley, L.H.; Bearss, D.J. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002, 1, 565–573. [Google Scholar]
- Izbicka, E.; Wheelhouse, R.T.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of Cationic Porphyrins as G-Quadruplex Interactive Agents in Human Tumor Cells1. Cancer Res. 1999, 59, 639–644. [Google Scholar]
- Han, F.X.; Wheelhouse, R.T.; Hurley, L.H. Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on Telomerase Inhibition. J. Am. Chem. Soc. 1999, 121, 3561–3570. [Google Scholar] [CrossRef]
- Nicoludis, J.M.; Miller, S.T.; Jeffrey, P.D.; Barrett, S.P.; Rablen, P.R.; Lawton, T.J.; Yatsunyk, L.A. Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA. J. Am. Chem. Soc. 2012, 134, 20446–20456. [Google Scholar] [CrossRef]
- Nicoludis, J.M.; Barrett, S.P.; Mergny, J.-L.; Yatsunyk, L.A. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012, 40, 5432–5447. [Google Scholar] [CrossRef]
- Sabharwal, N.C.; Savikhin, V.; Turek-Herman, J.R.; Nicoludis, J.M.; Szalai, V.A.; Yatsunyk, L.A. N-methylmesoporphyrin IX fluorescence as a reporter of strand orientation in guanine quadruplexes. FEBS J. 2014, 281, 1726–1737. [Google Scholar] [CrossRef]
- Zhang, Q.S.; Manche, L.; Xu, R.M.; Krainer, A.R. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA 2006, 12, 1116–1128. [Google Scholar] [CrossRef]
- Chung, W.J.; Heddi, B.; Hamon, F.; Teulade-Fichou, M.-P.; Phan, A.T. Solution Structure of a G-quadruplex Bound to the Bisquinolinium Compound Phen-DC3. Angew. Chem. Int. Ed. 2014, 53, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; DeLemos, E.; Mergny, J.-L.; Teulade-Fichou, M.-P.; Monchaud, D. Highly Efficient G-Quadruplex Recognition by Bisquinolinium Compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Kim, Y.E.; Bansal, V.; Ghosh, A.; Hur, J.; Subramani, V.K.; Pradhan, S.; Lee, M.K.; Kim, K.K.; Ahn, J.H. Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS Pathog. 2018, 14, e1007334. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Boulé, J.B.; Lopes, J.; Mingo, K.; Largy, E.; Teulade-Fichou, M.P.; Nicolas, A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010, 38, 4337–4348. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Cristofari, G.; Reichenbach, P.; De Lemos, E.; Monchaud, D.; Teulade-Fichou, M.P.; Shin-Ya, K.; Lacroix, L.; Lingner, J.; Mergny, J.L. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. USA 2007, 104, 17347–17352. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.G.; Hymøller, K.M.; Thorsager, M.E.; Hansen, N.Y.; Erlandsen, J.U.; Tesauro, C.; Simonsen, A.K.W.; Andersen, A.B.; Vandsø Petersen, K.; Holm, L.L.; et al. Topoisomerase 1 inhibits MYC promoter activity by inducing G-quadruplex formation. Nucleic Acids Res. 2022, 50, 6332–6342. [Google Scholar] [CrossRef] [PubMed]
- Al Kobaisi, M.; Bhosale, S.V.; Latham, K.; Raynor, A.M.; Bhosale, S.V. Functional Naphthalene Diimides: Synthesis, Properties, and Applications. Chem. Rev. 2016, 116, 11685–11796. [Google Scholar] [CrossRef] [PubMed]
- Pirota, V.; Nadai, M.; Doria, F.; Richter, S.N. Naphthalene Diimides as Multimodal G-Quadruplex-Selective Ligands. Molecules 2019, 24, 426. [Google Scholar] [CrossRef]
- Read, M.; Harrison, R.J.; Romagnoli, B.; Tanious, F.A.; Gowan, S.H.; Reszka, A.P.; Wilson, W.D.; Kelland, L.R.; Neidle, S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 4844–4849. [Google Scholar] [CrossRef]
- White, E.W.; Tanious, F.; Ismail, M.A.; Reszka, A.P.; Neidle, S.; Boykin, D.W.; Wilson, W.D. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys. Chem. 2007, 126, 140–153. [Google Scholar] [CrossRef]
- Harrison, R.J.; Cuesta, J.; Chessari, G.; Read, M.A.; Basra, S.K.; Reszka, A.P.; Morrell, J.; Gowan, S.M.; Incles, C.M.; Tanious, F.A.; et al. Trisubstituted acridine derivatives as potent and selective telomerase inhibitors. J. Med. Chem. 2003, 46, 4463–4476. [Google Scholar] [CrossRef] [PubMed]
- Hänsel, R.; Löhr, F.; Foldynová-Trantírková, S.; Bamberg, E.; Trantírek, L.; Dötsch, V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011, 39, 5768–5775. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Correia, J.J.; Wang, L.; Trent, J.O.; Chaires, J.B. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005, 33, 4649–4659. [Google Scholar] [CrossRef] [PubMed]
- Machireddy, B.; Kalra, G.; Jonnalagadda, S.; Ramanujachary, K.; Wu, C. Probing the Binding Pathway of BRACO19 to a Parallel-Stranded Human Telomeric G-Quadruplex Using Molecular Dynamics Binding Simulation with AMBER DNA OL15 and Ligand GAFF2 Force Fields. J. Chem. Inf. Model. 2017, 57, 2846–2864. [Google Scholar] [CrossRef] [PubMed]
- Gowan, S.M.; Harrison, J.R.; Patterson, L.; Valenti, M.; Read, M.A.; Neidle, S.; Kelland, L.R. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol Pharmacol. 2002, 61, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Incles, C.M.; Schultes, C.M.; Kempski, H.; Koehler, H.; Kelland, L.R.; Neidle, S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol. Cancer Ther. 2004, 3, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Incles, C.M.; Schultes, C.M.; Kelland, L.R.; Neidle, S. Acquired cellular resistance to flavopiridol in a human colon carcinoma cell line involves up-regulation of the telomerase catalytic subunit and telomere elongation. Sensitivity of resistant cells to combination treatment with a telomerase inhibitor. Mol. Pharmacol. 2003, 64, 1101–1108. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Wang, X.N.; Cheng, S.Q.; Su, X.X.; Ou, T.M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid—Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef]
- Moruno-Manchon, J.F.; Koellhoffer, E.C.; Gopakumar, J.; Hambarde, S.; Kim, N.; McCullough, L.D.; Tsvetkov, A.S. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging 2017, 9, 1957–1970. [Google Scholar] [CrossRef]
- De Magis, A.; Manzo, S.G.; Russo, M.; Marinello, J.; Morigi, R.; Sordet, O.; Capranico, G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Müller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.F.; Balasubramanian, S. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, J.; Tacconi, E.M.C.; Folio, C.; Badie, S.; Porru, M.; Klare, K.; Tumiati, M.; Markkanen, E.; Halder, S.; Ryan, A.; et al. Targeting BRCA1 and BRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol. Cell. 2016, 61, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.S.; Balasubramanian, S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 2016, 13, 841–844. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers--the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010, 38, D750–D753. [Google Scholar] [CrossRef] [PubMed]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. bioRxiv 2018, 14, 344127. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Alandijany, T.; Roberts, A.P.E.; Conn, K.L.; Loney, C.; McFarlane, S.; Orr, A.; Boutell, C. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 2018, 14, e1006769. [Google Scholar] [CrossRef]
- Shipley, M.M.; Renner, D.W.; Ott, M.; Bloom, D.C.; Koelle, D.M.; Johnston, C.; Szpara, M.L. Genome-Wide Surveillance of Genital Herpes Simplex Virus Type 1 From Multiple Anatomic Sites Over Time. J. Infect. Dis. 2018, 218, 595–605. [Google Scholar] [CrossRef]
- Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M.A.; Palù, G.; Flamand, L.; Calistri, A.; Richter, S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral. Res. 2015, 118, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Artusi, S.; Perrone, R.; Lago, S.; Raffa, P.; Di Iorio, E.; Palù, G.; Richter, S.N. Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res. 2016, 44, 10343–10353. [Google Scholar] [CrossRef]
- Bagga, S.; Bouchard, M.J. Cell cycle regulation during viral infection. Methods Mol. Biol. 2014, 1170, 165–227. [Google Scholar] [CrossRef] [PubMed]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Domíniguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. Targeting G-quadruplexes to achieve antiviral activity. Bioorg. Med. Chem. Lett. 2023, 79, 129085. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, S.G.; Beaudoin, J.D.; Bisaillon, M.; Perreault, J.P. Small antisense oligonucleotides against G-quadruplexes: Specific mRNA translational switches. Nucleic Acids Res. 2015, 43, 595–606. [Google Scholar] [CrossRef]
- Duellman, S.J.; Thompson, K.L.; Coon, J.J.; Burgess, R.R. Phosphorylation sites of Epstein-Barr virus EBNA1 regulate its function. J. Gen. Virol. 2009, 90, 2251–2259. [Google Scholar] [CrossRef]
- Kennedy, G.; Sugden, B. EBNA-1, a bifunctional transcriptional activator. Mol. Cell Biol. 2003, 23, 6901–6908. [Google Scholar] [CrossRef]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus–encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef]
- Cadoni, E.; De Paepe, L.; Manicardi, A.; Madder, A. Beyond small molecules: Targeting G-quadruplex structures with oligonucleotides and their analogues. Nucleic Acids Res. 2021, 49, 6638–6659. [Google Scholar] [CrossRef]
- Kupryushkin, M.S.; Filatov, A.V.; Mironova, N.L.; Patutina, O.A.; Chernikov, I.V.; Chernolovskaya, E.L.; Zenkova, M.A.; Pyshnyi, D.V.; Stetsenko, D.A.; Altman, S.; et al. Antisense oligonucleotide gapmers containing phosphoryl guanidine groups reverse MDR1-mediated multiple drug resistance of tumor cells. Mol. Ther. Nucleic Acids 2022, 27, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Balci, H.; Globyte, V.; Joo, C. Targeting G-quadruplex Forming Sequences with Cas9. ACS Chem. Biol. 2021, 16, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.E.; Mustafa, G.; Basu, S.; Balci, H. Encounters between Cas9/dCas9 and G-Quadruplexes: Implications for Transcription Regulation and Cas9-Mediated DNA Cleavage. ACS Synth. Biol. 2021, 10, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cui, S.; Qi, Q.; Lei, H.; Zhang, Y.; Shen, W.; Fu, F.; Tian, T.; Zhou, X. G-quadruplex-guided RNA engineering to modulate CRISPR-based genomic regulation. Nucleic Acids Res. 2022, 50, 11387–11400. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; Zamore, P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet Dev. 2002, 12, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Chery, J. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc. J. 2016, 4, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, L.; Karelsky, S.; Andino, R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 2002, 418, 430–434. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1191–1203.e12. [Google Scholar] [CrossRef]
- Heale, B.S.E.; Soifer, H.S.; Bowers, C.; Rossi, J.J. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res. 2005, 33, e30. [Google Scholar] [CrossRef]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Dumas, L.; Herviou, P.; Dassi, E.; Cammas, A.; Millevoi, S. G-Quadruplexes in RNA Biology: Recent Advances and Future Directions. Trends Biochem. Sci. 2021, 46, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Rader, C. Chemically programmed antibodies. Trends Biotechnol. 2014, 32, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt B, 1429–1447. [Google Scholar] [CrossRef]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-Forming Aptamers-Characteristics, Applications, and Perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.I.V.; Almeida, S.P.; Lourenço, L.M.O.; Pereira, P.M.R.; Fernandes, R.; Faustino, M.A.F.; Tomé, J.P.C.; Carvalho, J.; Cruz, C.; Neves, M.G.P.M.S. Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures. Molecules 2019, 24, 733. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt B, 1414–1428. [Google Scholar] [CrossRef]
- Qiu, W.; Zhou, F.; Zhang, Q.; Sun, X.; Shi, X.; Liang, Y.; Wang, X.; Yue, L. Overexpression of nucleolin and different expression sites both related to the prognosis of gastric cancer. Apmis 2013, 121, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Otake, Y.; Soundararajan, S.; Sengupta, T.K.; Kio, E.A.; Smith, J.C.; Pineda-Roman, M.; Stuart, R.K.; Spicer, E.K.; Fernandes, D.J. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood 2007, 109, 3069–3075. [Google Scholar] [CrossRef] [PubMed]
- Oleinick, N.L.; Morris, R.L.; Belichenko, I. The role of apoptosis in response to photodynamic therapy: What, where, why, and how. Photochem. Photobiol. Sci. 2002, 1, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Shieh, Y.A.; Yang, S.J.; Wei, M.F.; Shieh, M.J. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano 2010, 4, 1433–1442. [Google Scholar] [CrossRef]
- Carvalho, J.; Lopes-Nunes, J.; Lopes, A.C.; Campello, M.P.C.; Paulo, A.; Queiroz, J.A.; Cruz, C. Aptamer-guided acridine derivatives for cervical cancer. Org. Biomol. Chem. 2019, 17, 2992–3002. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Nunes, J.; Carvalho, J.; Figueiredo, J.; Ramos, C.I.V.; Lourenço, L.M.O.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Mergny, J.L.; Queiroz, J.A.; Salgado, G.F.; et al. Phthalocyanines for G-quadruplex aptamers binding. Bioorg. Chem. 2020, 100, 103920. [Google Scholar] [CrossRef] [PubMed]
- Do, N.Q.; Chung, W.J.; Truong, T.H.A.; Heddi, B.; Phan, A.T. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017, 45, 7487–7493. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Cho, Y.L.; Chae, J.R.; Moon, S.H.; Cho, W.G.; Choi, Y.J.; Lee, S.J.; Kang, W.J. Gemcitabine-Incorporated G-Quadruplex Aptamer for Targeted Drug Delivery into Pancreas Cancer. Mol. Ther. Nucleic Acids 2018, 12, 543–553. [Google Scholar] [CrossRef]
- García-Recio, E.M.; Pinto-Díez, C.; Pérez-Morgado, M.I.; García-Hernández, M.; Fernández, G.; Martín, M.E.; González, V.M. Characterization of MNK1b DNA Aptamers That Inhibit Proliferation in MDA-MB231 Breast Cancer Cells. Mol. Ther. Nucleic Acids 2016, 5, e275. [Google Scholar] [CrossRef]
- Esposito, V.; Russo, A.; Vellecco, V.; Bucci, M.; Russo, G.; Mayol, L.; Virgilio, A.; Galeone, A. Thrombin binding aptamer analogues containing inversion of polarity sites endowed with antiproliferative and anti-motility properties against Calu-6 cells. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2645–2650. [Google Scholar] [CrossRef]
- Louten, J. Virus Replication. In Essential Human Virology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 49–70. [Google Scholar]
- Speck, S.H.; Ganem, D. Viral latency and its regulation: Lessons from the gamma-herpesviruses. Cell Host Microbe 2010, 8, 100–115. [Google Scholar] [CrossRef]
- Cohen, J.I. Herpesvirus latency. J. Clin. Investig. 2020, 130, 3361–3369. [Google Scholar] [CrossRef]
- Sevvana, M.; Klose, T.; Rossmann, M.G. Principles of Virus Structure. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 257–277. [Google Scholar]
- Biswas, B.; Kandpal, M.; Jauhari, U.K.; Vivekanandan, P. Genome-wide analysis of G-quadruplexes in herpesvirus genomes. BMC Genom. 2016, 17, 949. [Google Scholar] [CrossRef]
- Norberg, P. Divergence and genotyping of human α-herpesviruses: An overview. Infect. Genet. Evol. 2010, 10, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Estep, K.N.; Butler, T.J.; Ding, J.; Brosh, R.M. G4-Interacting DNA Helicases and Polymerases: Potential Therapeutic Targets. Curr. Med. Chem. 2019, 26, 2881–2897. [Google Scholar] [CrossRef] [PubMed]
- Frasson, I.; Soldà, P.; Nadai, M.; Lago, S.; Richter, S.N. Parallel G-quadruplexes recruit the HSV-1 transcription factor ICP4 to promote viral transcription in herpes virus-infected human cells. Commun. Biol. 2021, 4, 510. [Google Scholar] [CrossRef] [PubMed]
- Frasson, I.; Soldà, P.; Nadai, M.; Tassinari, M.; Scalabrin, M.; Gokhale, V.; Hurley, L.H.; Richter, S.N. Quindoline-derivatives display potent G-quadruplex-mediated antiviral activity against herpes simplex virus 1. Antivir. Res. 2022, 208, 105432. [Google Scholar] [CrossRef]
- Artusi, S.; Ruggiero, E.; Nadai, M.; Tosoni, B.; Perrone, R.; Ferino, A.; Zanin, I.; Xodo, L.; Flamand, L.; Richter, S.N. Antiviral Activity of the G-Quadruplex Ligand TMPyP4 against Herpes Simplex Virus-1. Viruses 2021, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Frasson, I.; Nadai, M.; Richter, S.N. Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses. Molecules 2019, 24, 2375. [Google Scholar] [CrossRef]
- Chung, W.C.; Ravichandran, S.; Park, D.; Lee, G.M.; Kim, Y.E.; Choi, Y.; Song, M.J.; Kim, K.K.; Ahn, J.H. G-quadruplexes formed by Varicella-Zoster virus reiteration sequences suppress expression of glycoprotein C and regulate viral cell-to-cell spread. PLoS Pathog. 2023, 19, e1011095. [Google Scholar] [CrossRef]
- Nogalski, M.T.; Collins-McMillen, D.; Yurochko, A.D. Overview of human cytomegalovirus pathogenesis. Methods Mol. Biol. 2014, 1119, 15–28. [Google Scholar] [CrossRef]
- De Bolle, L.; Naesens, L.; De Clercq, E. Update on Human Herpesvirus 6 Biology, Clinical Features, and Therapy. Clin. Microbiol. Rev. 2005, 18, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Agut, H.; Bonnafous, P.; Gautheret-Dejean, A. Human Herpesviruses 6A, 6B, and 7. Microbiol. Spectr. 2016, 4, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E. Kaposi’s Sarcoma-Associated Herpesvirus microRNAs. Front. Microbiol. 2012, 3, 165. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Daibata, M.; Taguchi, T.; Nemoto, Y.; Taguchi, H.; Miyoshi, I. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 1999, 94, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Luppi, M.; Barozzi, P.; Marasca, R.; Torelli, G. Integration of human herpesvirus-6 (HHV-6) genome in chromosome 17 in two lymphoma patients. Leukemia 1994, 8 (Suppl. S1), S41–S45. [Google Scholar] [PubMed]
- Gilbert-Girard, S.; Gravel, A.; Artusi, S.; Richter, S.N.; Wallaschek, N.; Kaufer, B.B.; Flamand, L. Stabilization of Telomere G-Quadruplexes Interferes with Human Herpesvirus 6A Chromosomal Integration. J. Virol. 2017, 91, e00402-17. [Google Scholar] [CrossRef]
- Weed, D.J.; Damania, B. Pathogenesis of Human Gammaherpesviruses: Recent Advances. Curr. Clin. Microbiol. Rep. 2019, 6, 166–174. [Google Scholar] [CrossRef]
- Lista, M.J.; Martins, R.P.; Billant, O.; Contesse, M.A.; Findakly, S.; Pochard, P.; Daskalogianni, C.; Beauvineau, C.; Guetta, C.; Jamin, C.; et al. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8, 16043. [Google Scholar] [CrossRef]
- Reznichenko, O.; Quillévéré, A.; Martins, R.P.; Loaëc, N.; Kang, H.; Lista, M.J.; Beauvineau, C.; González-García, J.; Guillot, R.; Voisset, C.; et al. Novel cationic bis(acylhydrazones) as modulators of Epstein–Barr virus immune evasion acting through disruption of interaction between nucleolin and G-quadruplexes of EBNA1 mRNA. Eur. J. Med. Chem. 2019, 178, 13–29. [Google Scholar] [CrossRef]
- Zheng, A.J.; Thermou, A.; Guixens Gallardo, P.; Malbert-Colas, L.; Daskalogianni, C.; Vaudiau, N.; Brohagen, P.; Granzhan, A.; Blondel, M.; Teulade-Fichou, M.P.; et al. The different activities of RNA G-quadruplex structures are controlled by flanking sequences. Life Sci. Alliance 2022, 5, e202101232. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Choudhary, D.; Patra, A.; Bhavesh, N.S.; Vivekanandan, P. Analysis of G-quadruplexes upstream of herpesvirus miRNAs: Evidence of G-quadruplex mediated regulation of KSHV miR-K12–1-9,11 cluster and HCMV miR-US33. BMC Mol. Cell Biol. 2020, 21, 67. [Google Scholar] [CrossRef] [PubMed]
- Dabral, P.; Babu, J.; Zareie, A.; Verma, S.C. LANA and hnRNP A1 Regulate the Translation of LANA mRNA through G-Quadruplexes. J. Virol. 2020, 94, e01508-19. [Google Scholar] [CrossRef] [PubMed]
- Kwun Hyun, J.; da Silva Suzane, R.; Shah Ishita, M.; Blake, N.; Moore Patrick, S.; Chang, Y. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen 1 Mimics Epstein-Barr Virus EBNA1 Immune Evasion through Central Repeat Domain Effects on Protein Processing. J. Virol. 2007, 81, 8225–8235. [Google Scholar] [CrossRef] [PubMed]
- Kwun, H.J.; da Silva, S.R.; Qin, H.; Ferris, R.L.; Tan, R.; Chang, Y.; Moore, P.S. The central repeat domain 1 of Kaposi’s sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology 2011, 412, 357–365. [Google Scholar] [CrossRef]
- Ballestas, M.E.; Kaye, K.M. The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Future Microbiol. 2011, 6, 1399–1413. [Google Scholar] [CrossRef]
- Lee, H.R.; Lee, S.; Chaudhary, P.M.; Gill, P.; Jung, J.U. Immune evasion by Kaposi’s sarcoma-associated herpesvirus. Future Microbiol. 2010, 5, 1349–1365. [Google Scholar] [CrossRef]
- Krüger, A.C.; Raarup, M.K.; Nielsen, M.M.; Kristensen, M.; Besenbacher, F.; Kjems, J.; Birkedal, V. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur. Biophys. J. 2010, 39, 1343–1350. [Google Scholar] [CrossRef]
- De Leo, A.; Deng, Z.; Vladimirova, O.; Chen, H.S.; Dheekollu, J.; Calderon, A.; Myers, K.A.; Hayden, J.; Keeney, F.; Kaufer, B.B.; et al. LANA oligomeric architecture is essential for KSHV nuclear body formation and viral genome maintenance during latency. PLoS Pathog. 2019, 15, e1007489. [Google Scholar] [CrossRef]
- Zareie, A.R.; Verma, S.C. Nucleolin Regulates the Expression of Kaposi’s Sarcoma-Associated Herpesvirus’ Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA. Viruses 2023, 15, 2438. [Google Scholar] [CrossRef]
- Kumar, S.; Ramamurthy, C.; Choudhary, D.; Sekar, A.; Patra, A.; Bhavesh, N.S.; Vivekanandan, P. Contrasting roles for G-quadruplexes in regulating human Bcl-2 and virus homologues KSHV KS-Bcl-2 and EBV BHRF1. Sci. Rep. 2022, 12, 5019. [Google Scholar] [CrossRef]
- Madireddy, A.; Purushothaman, P.; Loosbroock, C.P.; Robertson, E.S.; Schildkraut, C.L.; Verma, S.C. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 2016, 44, 3675–3694. [Google Scholar] [CrossRef]
- Tlučková, K.; Marušič, M.; Tóthová, P.; Bauer, L.; Šket, P.; Plavec, J.; Viglasky, V. Human Papillomavirus G-Quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef] [PubMed]
- Marušič, M.; Hošnjak, L.; Krafčikova, P.; Poljak, M.; Viglasky, V.; Plavec, J. The effect of single nucleotide polymorphisms in G-rich regions of high-risk human papillomaviruses on structural diversity of DNA. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt B, 1229–1236. [Google Scholar] [CrossRef]
- Carvalho, J.; Lopes-Nunes, J.; Campello, M.P.C.; Paulo, A.; Milici, J.; Meyers, C.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Human Papillomavirus G-Rich Regions as Potential Antiviral Drug Targets. Nucleic Acid Ther. 2021, 31, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Shah, K.V. Polyomaviruses and human diseases. Adv. Exp. Med. Biol. 2006, 577, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Abend, J.R.; Johnson, S.F.; Imperiale, M.J. The role of polyomaviruses in human disease. Virology 2009, 384, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Saribas, A.S.; Coric, P.; Bouaziz, S.; Safak, M. Expression of novel proteins by polyomaviruses and recent advances in the structural and functional features of agnoprotein of JC virus, BK virus, and simian virus 40. J. Cell Physiol. 2019, 234, 8295–8315. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.F.; You, J. Regulation of Polyomavirus Transcription by Viral and Cellular Factors. Viruses 2020, 12, 1072. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.K.; Bhavesh, N.S.; Hosur, R.V. NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG. Biochem. Biophys. Res. Commun. 2000, 270, 967–971. [Google Scholar] [CrossRef]
- Topalis, D.; Andrei, G.; Snoeck, R. The large tumor antigen: A “Swiss Army knife” protein possessing the functions required for the polyomavirus life cycle. Antivir. Res. 2013, 97, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Plyler, J.; Jasheway, K.; Tuesuwan, B.; Karr, J.; Brennan, J.S.; Kerwin, S.M.; David, W.M. Real-time investigation of SV40 large T-antigen helicase activity using surface plasmon resonance. Cell Biochem. Biophys. 2009, 53, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Tuesuwan, B.; Kern, J.T.; Thomas, P.W.; Rodriguez, M.; Li, J.; David, W.M.; Kerwin, S.M. Simian Virus 40 Large T-Antigen G-Quadruplex DNA Helicase Inhibition by G-Quadruplex DNA-Interactive Agents. Biochemistry 2008, 47, 1896–1909. [Google Scholar] [CrossRef] [PubMed]
- Sunbul, M. Hepatitis B virus genotypes: Global distribution and clinical importance. World J. Gastroenterol. 2014, 20, 5427–5434. [Google Scholar] [CrossRef] [PubMed]
- Biswas, B.; Kandpal, M.; Vivekanandan, P. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Res. 2017, 45, 11268–11280. [Google Scholar] [CrossRef] [PubMed]
- Meier-Stephenson, V.; Badmalia, M.D.; Mrozowich, T.; Lau, K.C.K.; Schultz, S.K.; Gemmill, D.L.; Osiowy, C.; van Marle, G.; Coffin, C.S.; Patel, T.R. Identification and characterization of a G-quadruplex structure in the pre-core promoter region of hepatitis B virus covalently closed circular DNA. J. Biol. Chem. 2021, 296, 100589. [Google Scholar] [CrossRef] [PubMed]
- Molnár, O.R.; Végh, A.; Somkuti, J.; Smeller, L. Characterization of a G-quadruplex from hepatitis B virus and its stabilization by binding TMPyP4, BRACO19 and PhenDC3. Sci. Rep. 2021, 11, 23243. [Google Scholar] [CrossRef] [PubMed]
- Wenham, C.; Eccleston-Turner, M. Monkeypox as a PHEIC: Implications for global health governance. Lancet 2022, 400, 2169–2171. [Google Scholar] [CrossRef]
- Lv, L.; Zhang, L. G-quadruplexes in the monkeypox virus are potential antiviral targets. J. Med. Virol. 2023, 95, e28299. [Google Scholar] [CrossRef]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.C.; Sarin, P.S.; Gelmann, E.P.; Robert-Guroff, M.; Richardson, E.; Kalyanaraman, V.S.; Mann, D.; Sidhu, G.D.; Stahl, R.E.; Zolla-Pazner, S.; et al. Isolation of Human T-Cell Leukemia Virus in Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Coffin, J.; Haase, A.; Levy, J.A.; Montagnier, L.; Oroszlan, S.; Teich, N.; Temin, H.; Toyoshima, K.; Varmus, H.; Vogt, P. What to call the AIDS virus? Nature 1986, 321, 10. [Google Scholar] [CrossRef] [PubMed]
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Rev. Investig. Clín. Organo Hosp. Enfermedades Nutr. 1983, 56, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Prevention CfDCa. HIV Surveillance Report, 2018 (Preliminary). Volume 30. Available online: http://www.cdc.gov/hiv/library/reports/hiv-surveillance.html (accessed on 27 February 2023).
- Gao, F.; Bailes, E.; Robertson, D.L.; Chen, Y.; Rodenburg, C.M.; Michael, S.F.; Cummins, L.B.; Arthur, L.O.; Peeters, M.; Shaw, G.M.; et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 1999, 397, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Muldoon, M.; Theiler, J.; Gao, F.; Gupta, R.; Lapedes, A.; Hahn, B.H.; Wolinsky, S.; Bhattacharya, T. Timing the Ancestor of the HIV-1 Pandemic Strains. Science 2000, 288, 1789. [Google Scholar] [CrossRef]
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 15035. [Google Scholar] [CrossRef]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palù, G.; Richter, S.N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef]
- Perrone, R.; Nadai, M.; Poe, J.A.; Frasson, I.; Palumbo, M.; Palù, G.; Smithgall, T.E.; Richter, S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: Implications for antiviral activity. PLoS ONE 2013, 8, e73121. [Google Scholar] [CrossRef]
- Quiñones-Mateu, M.E.; Mas, A.; Lain de Lera, T.; Soriano, V.; Alcamí, J.; Lederman, M.M.; Domingo, E. LTR and tat variability of HIV-1 isolates from patients with divergent rates of disease progression. Virus Res. 1998, 57, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Nonnemacher, M.R.; Irish, B.P.; Liu, Y.; Mauger, D.; Wigdahl, B. Specific sequence configurations of HIV-1 LTR G/C box array result in altered recruitment of Sp isoforms and correlate with disease progression. J. Neuroimmunol. 2004, 157, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Heaphy, S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 3393–3397. [Google Scholar] [CrossRef] [PubMed]
- Piekna-Przybylska, D.; Sharma, G.; Bambara, R.A. Mechanism of HIV-1 RNA dimerization in the central region of the genome and significance for viral evolution. J. Biol. Chem. 2013, 288, 24140–24150. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Gao, L.; Balakrishnan, M.; Bambara, R.A. A recombination hot spot in HIV-1 contains guanosine runs that can form a G-quartet structure and promote strand transfer in vitro. J. Biol. Chem. 2009, 284, 33883–33893. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.D.; Warmerdam, M.T.; Gaston, I.; Greene, W.C.; Feinberg, M.B. The human immunodeficiency virus-1 nef gene product: A positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 1994, 179, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Manzourolajdad, A.; Gonzalez, M.; Spouge, J.L. Changes in the Plasticity of HIV-1 Nef RNA during the Evolution of the North American Epidemic. PLoS ONE 2016, 11, e0163688. [Google Scholar] [CrossRef]
- Khullar, V.; Firpi, R.J. Hepatitis C cirrhosis: New perspectives for diagnosis and treatment. World J. Hepatol. 2015, 7, 1843–1855. [Google Scholar] [CrossRef]
- Vassilaki, N.; Friebe, P.; Meuleman, P.; Kallis, S.; Kaul, A.; Paranhos-Baccalà, G.; Leroux-Roels, G.; Mavromara, P.; Bartenschlager, R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 2008, 82, 11503–11515. [Google Scholar] [CrossRef]
- Moradpour, D.; Penin, F.; Rice, C.M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007, 5, 453–463. [Google Scholar] [CrossRef]
- Jaubert, C.; Bedrat, A.; Bartolucci, L.; Di Primo, C.; Ventura, M.; Mergny, J.L.; Amrane, S.; Andreola, M.L. RNA synthesis is modulated by G-quadruplex formation in Hepatitis C virus negative RNA strand. Sci. Rep. 2018, 8, 8120. [Google Scholar] [CrossRef] [PubMed]
- Pirakitikulr, N.; Kohlway, A.; Lindenbach, B.D.; Pyle, A.M. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol. Cell. 2016, 62, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.R.; Min, Y.Q.; Wang, J.Q.; Liu, C.X.; Fu, B.S.; Wu, F.; Wu, L.Y.; Qiao, Z.X.; Song, Y.Y.; Xu, G.H.; et al. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti-hepatitis C target. Sci. Adv. 2016, 2, e1501535. [Google Scholar] [CrossRef] [PubMed]
- Bian, W.X.; Xie, Y.; Wang, X.N.; Xu, G.H.; Fu, B.S.; Li, S.; Long, G.; Zhou, X.; Zhang, X.L. Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication. Nucleic Acids Res. 2019, 47, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Belachew, B.; Gao, J.; Byrd, A.K.; Raney, K.D. Hepatitis C virus nonstructural protein NS3 unfolds viral G-quadruplex RNA structures. J. Biol. Chem. 2022, 298, 102486. [Google Scholar] [CrossRef] [PubMed]
- Noorbakhsh, F.; Abdolmohammadi, K.; Fatahi, Y.; Dalili, H.; Rasoolinejad, M.; Rezaei, F.; Salehi-Vaziri, M.; Shafiei-Jandaghi, N.Z.; Gooshki, E.S.; Zaim, M.; et al. Zika Virus Infection, Basic and Clinical Aspects: A Review Article. Iran J. Public Health 2019, 48, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Alenko, A.; Burrows, C.J. Zika Virus Genomic RNA Possesses Conserved G-Quadruplexes Characteristic of the Flaviviridae Family. ACS Infect. Dis. 2016, 2, 674–681. [Google Scholar] [CrossRef]
- Majee, P.; Pattnaik, A.; Sahoo, B.R.; Shankar, U.; Pattnaik, A.K.; Kumar, A.; Nayak, D. Inhibition of Zika virus replication by G-quadruplex-binding ligands. Mol. Ther. Nucleic Acids 2021, 23, 691–701. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef]
- Szabo, R.; Bugge, T.H. Type II transmembrane serine proteases in development and disease. Int. J. Biochem. Cell Biol. 2008, 40, 1297–1316. [Google Scholar] [CrossRef] [PubMed]
- Böttcher-Friebertshäuser, E.; Klenk, H.D.; Garten, W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013, 69, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.W.; Mao, H.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie 2017, 142, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.W.; Qian, M.Q.; Yu, K.; Narva, S.; Yu, F.; Wu, Y.L.; Zhang, W. Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci. Rep. 2020, 10, 7635. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewska, M.; Szabat, M.; Zielińska, K.; Kierzek, R. Identification and Structural Aspects of G-Quadruplex-Forming Sequences from the Influenza A Virus Genome. Int. J. Mol. Sci. 2021, 22, 6031. [Google Scholar] [CrossRef] [PubMed]
- Lugo, D.; Krogstad, P. Enteroviruses in the early 21st century: New manifestations and challenges. Curr. Opin. Pediatr. 2016, 28, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Zhang, L. Characterization of G-Quadruplexes in Enterovirus A71 Genome and Their Interaction with G-Quadruplex Ligands. Microbiol. Spectr. 2022, 10, e0046022. [Google Scholar] [CrossRef] [PubMed]
- Bezzi, G.; Piga, E.J.; Binolfi, A.; Armas, P. CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. Int. J. Mol. Sci. 2021, 22, 2614. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603. [Google Scholar] [CrossRef]
- Zhang, R.; Xiao, K.; Gu, Y.; Liu, H.; Sun, X. Whole Genome Identification of Potential G-Quadruplexes and Analysis of the G-Quadruplex Binding Domain for SARS-CoV-2. Front. Genet. 2020, 11, 587829. [Google Scholar] [CrossRef]
- Ji, D.; Juhas, M.; Tsang, C.M.; Kwok, C.K.; Li, Y.; Zhang, Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinform. 2021, 22, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhang, L. G-Quadruplexes Are Present in Human Coronaviruses Including SARS-CoV-2. Front. Microbiol. 2020, 11, 567317. [Google Scholar] [CrossRef] [PubMed]
- Kusov, Y.; Tan, J.; Alvarez, E.; Enjuanes, L.; Hilgenfeld, R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication–transcription complex. Virology 2015, 484, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Ma-Lauer, Y.; Han, Y.; Thoms, M.; Buschauer, R.; Jores, J.; Thiel, V.; Beckmann, R.; Deng, W.; Leonhardt, H.; et al. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. Embo. J. 2021, 40, e102277. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, M.; Helynck, O.; Rigolet, P.; Boudria-Souilah, R.; Nowakowski, M.; Baron, B.; Brülé, S.; Hoos, S.; Raynal, B.; Guittat, L.; et al. SARS-CoV-2 Nsp3 unique domain SUD interacts with guanine quadruplexes and G4-ligands inhibit this interaction. Nucleic Acids Res. 2021, 49, 7695–7712. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Du, W.; Sang, X.; Tong, Q.; Wang, Y.; Chen, G.; Yuan, Y.; Jiang, L.; Cheng, W.; Liu, D.; et al. RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection. Nat. Commun. 2022, 13, 1444. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19–Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Qin, G.; Zhao, C.; Liu, Y.; Zhang, C.; Yang, G.; Yang, J.; Wang, Z.; Wang, C.; Tu, C.; Guo, Z.; et al. RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models. Cell Discov. 2022, 8, 86. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef]
- Oliva, R.; Mukherjee, S.; Manisegaran, M.; Campanile, M.; Del Vecchio, P.; Petraccone, L.; Winter, R. Binding Properties of RNA Quadruplex of SARS-CoV-2 to Berberine Compared to Telomeric DNA Quadruplex. Int. J. Mol. Sci. 2022, 23, 5690. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Zhang, F.-T.; Cai, L.-Y.; Zhou, Y.-L.; Buurma, N.J.; Zhang, X.-X. Investigation of the interactions between methylene blue and intramolecular G-quadruplexes: An explicit distinction in electrochemical behavior. Analyst 2017, 142, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, T.S.; Pereira, R.O.; de Mello, H.L.; Costa, J. Methemoglobinemia: From diagnosis to treatment. Rev. Bras. Anestesiol. 2008, 58, 651–664. [Google Scholar] [CrossRef]
- Bojadzic, D.; Alcazar, O.; Buchwald, P. Methylene Blue Inhibits the SARS-CoV-2 Spike–ACE2 Protein-Protein Interaction–a Mechanism that can Contribute to its Antiviral Activity Against COVID-19. Brief Research Report. Front. Pharmacol. 2021, 11, 600372. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Zhao, C.; Yang, J.; Wang, Z.; Ren, J.; Qu, X. Unlocking G-Quadruplexes as Targets and Tools against COVID-19. Chin. J. Chem. 2023, 41, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Y.; Wang, Y.; Feng, Y.; Li, D.; Li, S.; Qin, P.; Yang, X.; Chen, L.; Zhao, J.; et al. Characterization of RNA G-quadruplexes in porcine epidemic diarrhea virus genome and the antiviral activity of G-quadruplex ligands. Int. J. Biol. Macromol. 2023, 231, 123282. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef] [PubMed]
- Butovskaya, E.; Heddi, B.; Bakalar, B.; Richter, S.N.; Phan, A.T. Major G-Quadruplex Form of HIV-1 LTR Reveals a (3 + 1) Folding Topology Containing a Stem-Loop. J. Am. Chem. Soc. 2018, 140, 13654–13662. [Google Scholar] [CrossRef] [PubMed]
- De Nicola, B.; Lech, C.J.; Heddi, B.; Regmi, S.; Frasson, I.; Perrone, R.; Richter, S.N.; Phan, A.T. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 2016, 44, 6442–6451. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zareie, A.R.; Dabral, P.; Verma, S.C. G-Quadruplexes in the Regulation of Viral Gene Expressions and Their Impacts on Controlling Infection. Pathogens 2024, 13, 60. https://doi.org/10.3390/pathogens13010060
Zareie AR, Dabral P, Verma SC. G-Quadruplexes in the Regulation of Viral Gene Expressions and Their Impacts on Controlling Infection. Pathogens. 2024; 13(1):60. https://doi.org/10.3390/pathogens13010060
Chicago/Turabian StyleZareie, Andrew R., Prerna Dabral, and Subhash C. Verma. 2024. "G-Quadruplexes in the Regulation of Viral Gene Expressions and Their Impacts on Controlling Infection" Pathogens 13, no. 1: 60. https://doi.org/10.3390/pathogens13010060






