Computational Workflow to Design Novel Vaccine Candidates and Small-Molecule Therapeutics for Schistosomiasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of Protein Targets
2.2. Prediction of Cytotoxic T Lymphocyte (CTL) and Helper T Lymphocyte (HTL) Epitopes
2.3. B-Cell Epitope Prediction
2.4. Selection of Overlapping Epitopes
2.5. Interferon Gamma (IFN-ɣ)-Inducing Epitopes
2.6. Antigenicity, Allergenicity, and Toxicity Analysis
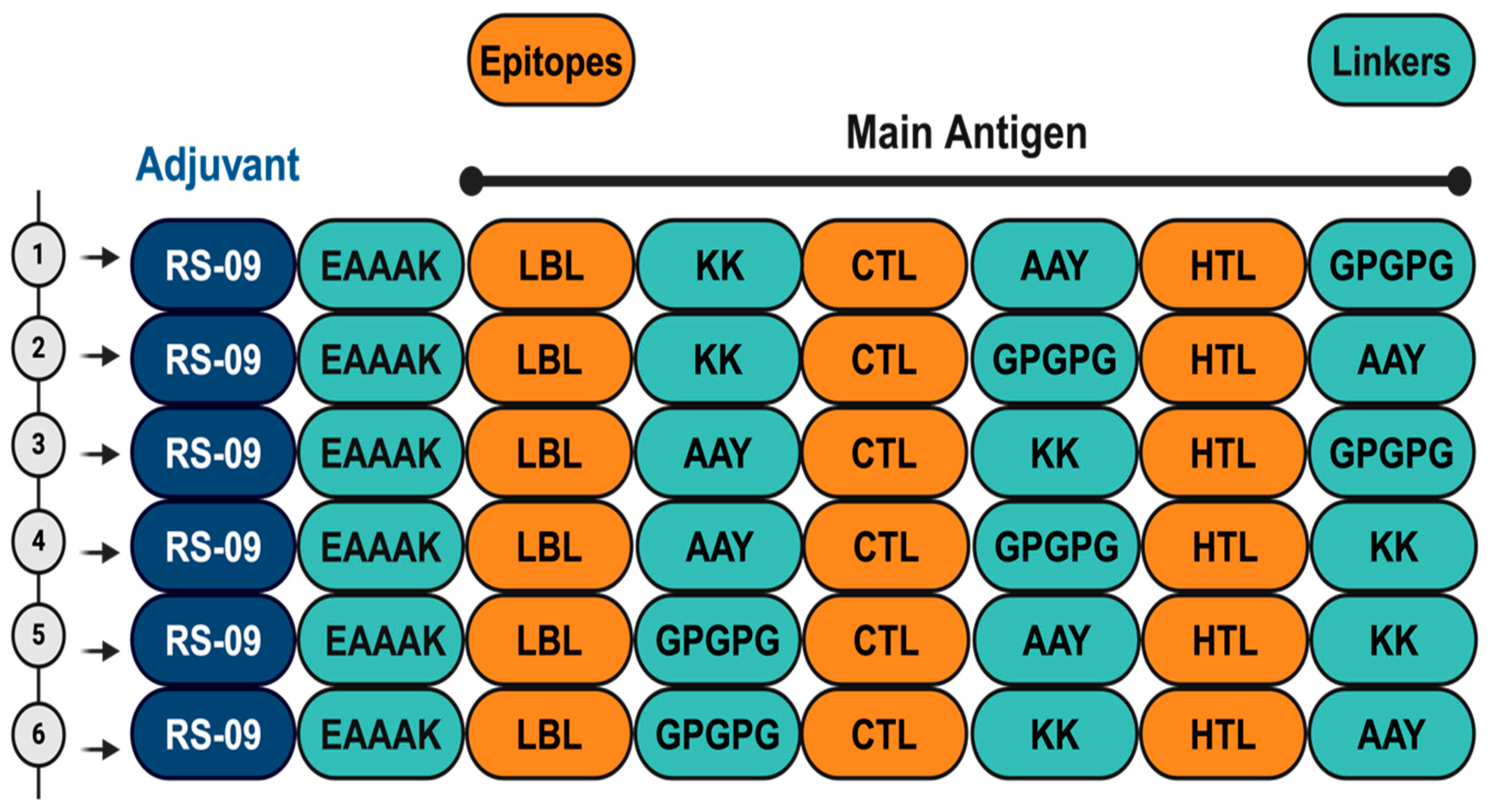
2.7. Construction of Vaccine Candidates
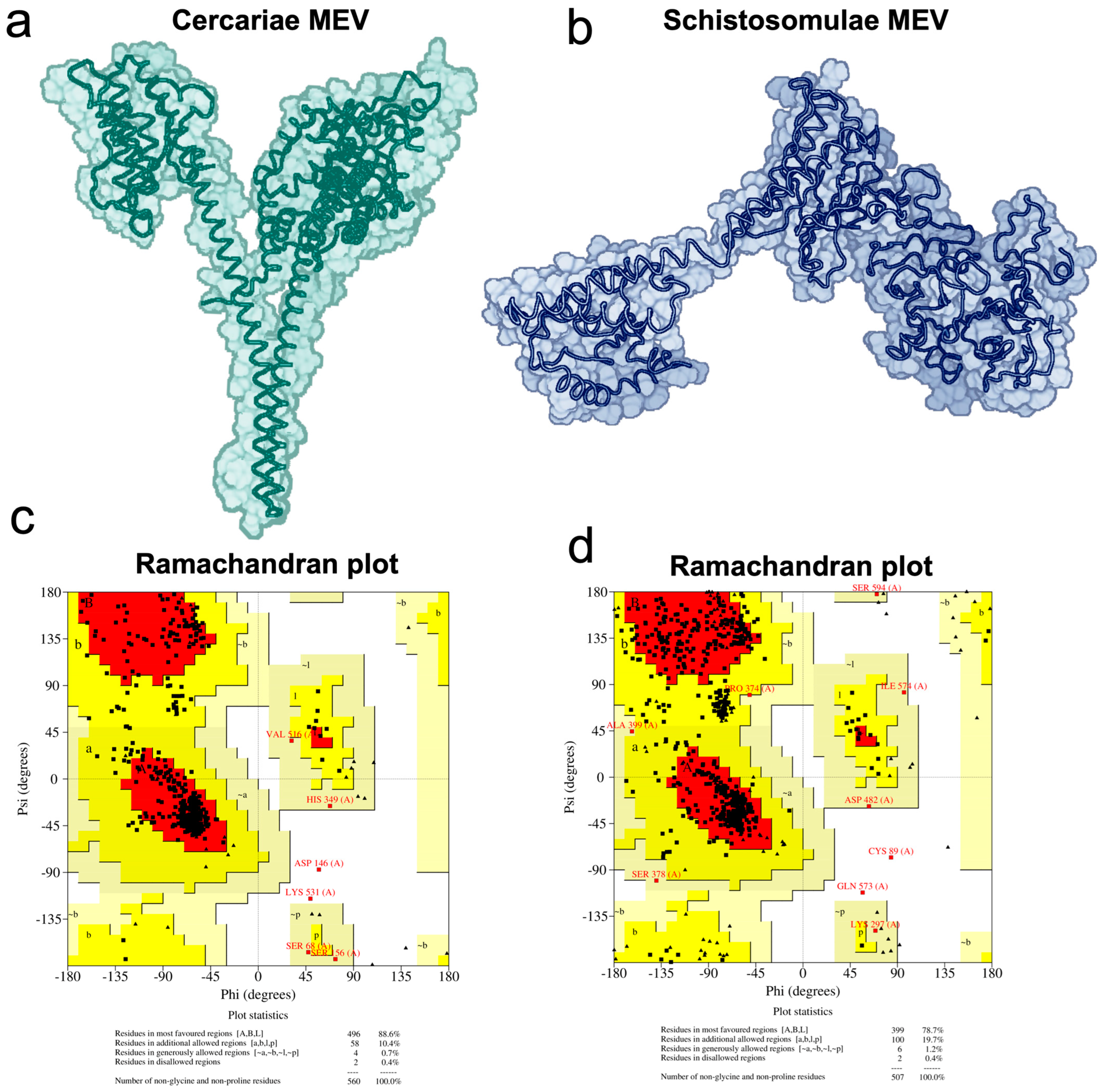
2.8. Vaccine 3D Structure Prediction and Physicochemical Properties Evaluation
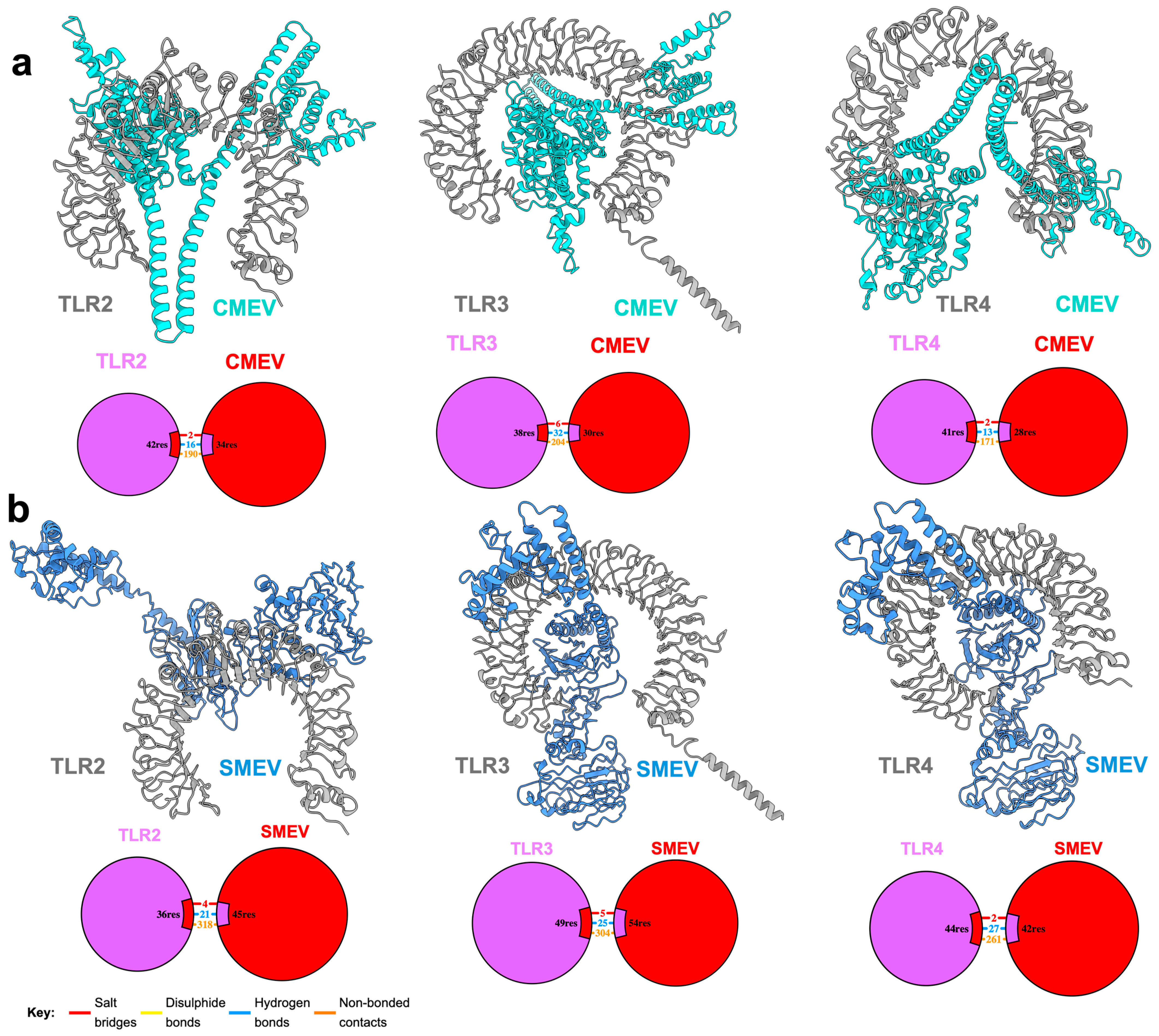
2.9. Molecular Docking of Construct with Immune Receptors
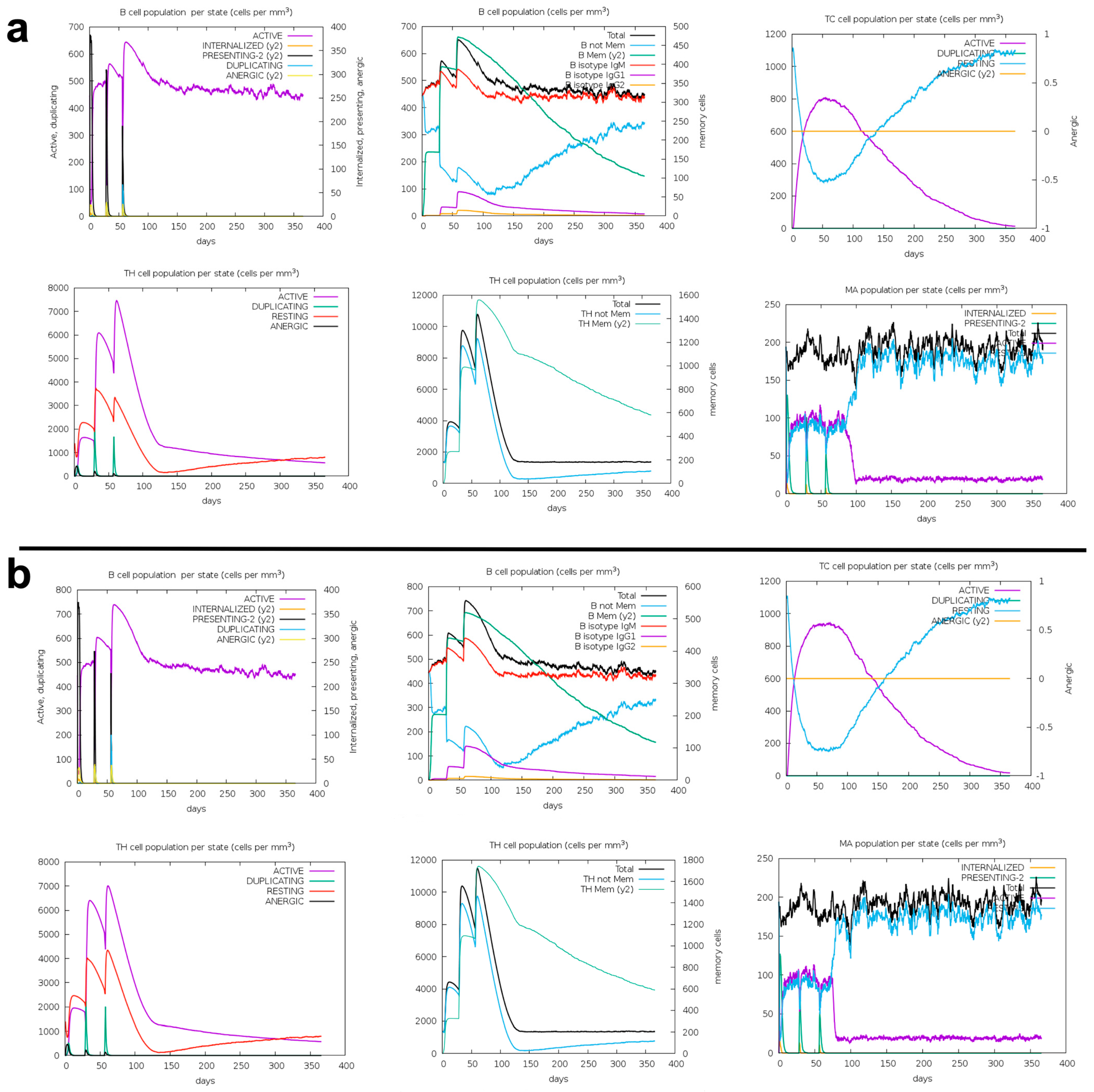
2.10. Immune Response Simulation
2.11. Selection of Cathepsin Drug Targets and Multiple Sequence Alignment
2.12. Molecular Modeling and Docking
3. Results
3.1. Selection of Extracellular Helices
3.2. Prediction of CTL, HTL, and B-Cell Epitopes
3.3. Antigenicity, Allergenicity, and Toxicity Analysis
3.4. Vaccine Candidates
3.5. Interaction of the Vaccine Candidates with Key Immune Receptors
3.6. Immune Response Simulation
3.7. Identification of Potential Inhibitors of Schistosoma Cathepsins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kokaliaris, C.; Garba, A.; Matuska, M.; Bronzan, R.N.; Colley, D.G.; Dorkenoo, A.M.; Ekpo, U.F.; Fleming, F.M.; French, M.D.; Kabore, A.; et al. Effect of Preventive Chemotherapy with Praziquantel on Schistosomiasis among School-Aged Children in Sub-Saharan Africa: A Spatiotemporal Modelling Study. Lancet Infect. Dis. 2022, 22, 136–149. [Google Scholar] [CrossRef]
- Sanches, R.C.O.; Tiwari, S.; Ferreira, L.C.G.; Oliveira, F.M.; Lopes, M.D.; Passos, M.J.F.; Maia, E.H.B.; Taranto, A.G.; Kato, R.; Azevedo, V.A.C.; et al. Immunoinformatics Design of Multi-Epitope Peptide-Based Vaccine against Schistosoma Mansoni Using Transmembrane Proteins as a Target. Front. Immunol. 2021, 12, 621706. [Google Scholar] [CrossRef]
- Becker, J.M.; Ganatra, A.A.; Kandie, F.; Mühlbauer, L.; Ahlheim, J.; Brack, W.; Torto, B.; Agola, E.L.; McOdimba, F.; Hollert, H.; et al. Pesticide Pollution in Freshwater Paves the Way for Schistosomiasis Transmission. Sci. Rep. 2020, 10, 3650. [Google Scholar] [CrossRef]
- White Bear, J.; Long, T.; Skinner, D.; McKerrow, J.H. Predictions of Novel Schistosoma Mansoni-Human Protein Interactions Consistent with Experimental Data. Sci. Rep. 2018, 8, 13092. [Google Scholar] [CrossRef]
- Schistosomiasis. Available online: https://www.who.int/news-room/fact-sheets/detail/schistosomiasis (accessed on 19 July 2024).
- Catalano, S.; Sène, M.; Diouf, N.D.; Fall, C.B.; Borlase, A.; Léger, E.; Bâ, K.; Webster, J.P. Rodents as Natural Hosts of Zoonotic Schistosoma Species and Hybrids: An Epidemiological and Evolutionary Perspective from West Africa. J. Infect. Dis. 2018, 218, 429–433. [Google Scholar] [CrossRef]
- Adenowo, A.F.; Oyinloye, B.E.; Ogunyinka, B.I.; Kappo, A.P. Impact of Human Schistosomiasis in Sub-Saharan Africa. Braz. J. Infect. Dis. 2015, 19, 196. [Google Scholar] [CrossRef]
- Andrews, P.; Thomas, H.; Pohlke, R.; Seubert, J. Praziquantel. Med. Res. Rev. 1983, 3, 147–200. [Google Scholar] [CrossRef]
- Senghor, B.; Diaw, O.T.; Doucoure, S.; Sylla, S.N.; Seye, M.; Talla, I.; Bâ, C.T.; Diallo, A.; Sokhna, C. Efficacy of Praziquantel against Urinary Schistosomiasis and Reinfection in Senegalese School Children Where There Is a Single Well-Defined Transmission Period. Parasit. Vectors 2015, 8, 362. [Google Scholar] [CrossRef]
- Dejon-Agobé, J.C.; Edoa, J.R.; Honkpehedji, Y.J.; Zinsou, J.F.; Adégbitè, B.R.; Ngwese, M.M.; Mangaboula, A.; Lell, B.; Grobusch, M.P.; Mordmüller, B.; et al. Schistosoma Haematobium Infection Morbidity, Praziquantel Effectiveness and Reinfection Rate among Children and Young Adults in Gabon. Parasit. Vectors 2019, 12, 577. [Google Scholar] [CrossRef]
- Caffrey, C.R. Schistosomiasis and Its Treatment. Future Med. Chem. 2015, 7, 675–676. [Google Scholar] [CrossRef]
- Caffrey, C.R.; El-Sakkary, N.; Mäder, P.; Krieg, R.; Becker, K.; Schlitzer, M.; Drewry, D.H.; Vennerstrom, J.L.; Grevelding, C.G. Drug Discovery and Development for Schistosomiasis. In Neglected Tropical Diseases: Drug Discovery and Development; Wiley: Hoboken, NJ, USA, 2019; pp. 187–225. [Google Scholar] [CrossRef]
- Sabah, A.A.; Fletcher, C.; Webbe, G.; Doenhoff, M.J. Schistosoma Mansoni: Chemotherapy of Infections of Different Ages. Exp. Parasitol. 1986, 61, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Kovač, J.; Vargas, M.; Keiser, J. In Vitro and in Vivo Activity of R- and S- Praziquantel Enantiomers and the Main Human Metabolite Trans-4-Hydroxy-Praziquantel against Schistosoma haematobium. Parasit. Vectors 2017, 10, 365. [Google Scholar] [CrossRef]
- Friedrich, L.; Park, S.K.; Ballard, P.; Ho Baeurle, T.H.; Maillard, D.; Bödding, M.; Keiser, J.; Marchant, J.S.; Spangenberg, T. Metabolism of (R)-Praziquantel versus the Activation of a Parasite Transient Receptor Potential Melastatin Ion Channel. ChemMedChem 2023, 18, e202300140. [Google Scholar] [CrossRef]
- Onile, O.S.; Fadahunsi, A.I.; Adekunle, A.A.; Oyeyemi, B.F.; Anumudu, C.I. An Immunoinformatics Approach for the Design of a Multi-Epitope Subunit Vaccine for Urogenital Schistosomiasis. PeerJ 2020, 8, e8795. [Google Scholar] [CrossRef]
- Al-Naseri, A.; Al-Absi, S.; El Ridi, R.; Mahana, N. A Comprehensive and Critical Overview of Schistosomiasis Vaccine Candidates. J. Parasit. Dis. 2021, 45, 557–580. [Google Scholar] [CrossRef]
- Arumugam, S. In-Silico Design of Envelope Based Multi-Epitope Vaccine Candidate against Kyasanur Forest Disease Virus. Sci. Rep. 2021, 11, 17118. [Google Scholar] [CrossRef]
- Chao, P.; Zhang, X.; Zhang, L.; Yang, A.; Wang, Y.; Chen, X. Proteomics-Based Vaccine Targets Annotation and Design of Multi-Epitope Vaccine against Antibiotic-Resistant Streptococcus gallolyticus. Sci. Rep. 2024, 14, 4836. [Google Scholar] [CrossRef]
- Danazumi, A.U.; Iliyasu Gital, S.; Idris, S.; BS Dibba, L.; Balogun, E.O.; Górna, M.W. Immunoinformatic Design of a Putative Multi-Epitope Vaccine Candidate against Trypanosoma brucei Gambiense. Comput. Struct. Biotechnol. J. 2022, 20, 5574–5585. [Google Scholar] [CrossRef]
- Danazumi, A.U.; Adepoju, O.A.; Dibba, L.B.; Ibrahim, B.; Gital, S.I.; Joseph, G.I.; Chang, G.; Balogun, E.O. Probing the Proteome of Mpox Virus for in Silicodesign of a Multiepitope Vaccine. Future Drug Discov. 2023, 5, FDD86. [Google Scholar] [CrossRef]
- Mahmud, S.; Rafi, M.O.; Paul, G.K.; Promi, M.M.; Shimu, M.S.S.; Biswas, S.; Bin Emran, T.; Dhama, K.; Alyami, S.A.; Moni, M.A.; et al. Designing a Multi-Epitope Vaccine Candidate to Combat MERS-CoV by Employing an Immunoinformatics Approach. Sci. Rep. 2021, 11, 15431. [Google Scholar] [CrossRef] [PubMed]
- Naz, A.; Shahid, F.; Butt, T.T.; Awan, F.M.; Ali, A.; Malik, A. Designing Multi-Epitope Vaccines to Combat Emerging Coronavirus Disease 2019 (COVID-19) by Employing Immuno-Informatics Approach. Front. Immunol. 2020, 11, 548248. [Google Scholar] [CrossRef] [PubMed]
- Rcheulishvili, N.; Mao, J.; Papukashvili, D.; Feng, S.; Liu, C.; Yang, X.; Lin, J.; He, Y.; Wang, P.G. Development of a Multi-Epitope Universal MRNA Vaccine Candidate for Monkeypox, Smallpox, and Vaccinia Viruses: Design and In Silico Analyses. Viruses 2023, 15, 1120. [Google Scholar] [CrossRef] [PubMed]
- Lennerz, V.; Gross, S.; Gallerani, E.; Sessa, C.; Mach, N.; Boehm, S.; Hess, D.; Von Boehmer, L.; Knuth, A.; Ochsenbein, A.F.; et al. Immunologic Response to the Survivin-Derived Multi-Epitope Vaccine EMD640744 in Patients with Advanced Solid Tumors. Cancer Immunol. Immunother. 2014, 63, 381–394. [Google Scholar] [CrossRef]
- Caffrey, C.R.; Lima, A.P.; Steverding, D. Cysteine Peptidases of Kinetoplastid Parasites. Adv. Exp. Med. Biol. 2011, 712, 84–99. [Google Scholar] [CrossRef]
- Caffrey, C.R.; Salter, J.P.; Lucas, K.D.; Khiem, D.; Hsieh, I.; Lim, K.C.; Ruppel, A.; McKerrow, J.H.; Sajid, M. SmCB2, a Novel Tegumental Cathepsin B from Adult Schistosoma mansoni. Mol. Biochem. Parasitol. 2002, 121, 49–61. [Google Scholar] [CrossRef] [PubMed]
- McKerrow, J.H.; Engel, J.C.; Caffrey, C.R. Cysteine Protease Inhibitors as Chemotherapy for Parasitic Infections. Bioorg. Med. Chem. 1999, 7, 639–644. [Google Scholar] [CrossRef]
- Brady, C.P.; Dowd, A.J.; Brindley, P.J.; Ryan, T.; Day, S.R.; Dalton, J.P. Recombinant Expression and Localization of Schistosoma Mansoni Cathepsin L1 Support Its Role in the Degradation of Host Hemoglobin. Infect. Immun. 1999, 67, 368–374. [Google Scholar] [CrossRef]
- Jílková, A.; Rubešová, P.; Fanfrlík, J.; Fajtová, P.; Řezáčová, P.; Brynda, J.; Lepšík, M.; Mertlíková-Kaiserová, H.; Emal, C.D.; Renslo, A.R.; et al. Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors. ACS Infect. Dis. 2021, 7, 1077–1088. [Google Scholar] [CrossRef]
- Morales, M.E.; Rinaldi, G.; Gobert, G.N.; Kines, K.J.; Tort, J.F.; Brindley, P.J. RNA Interference of Schistosoma Mansoni Cathepsin D, the Apical Enzyme of the Hemoglobin Proteolysis Cascade. Mol. Biochem. Parasitol. 2008, 157, 160–168. [Google Scholar] [CrossRef]
- Štefanic, S.; Dvořák, J.; Horn, M.; Braschi, S.; Sojka, D.; Ruelas, D.S.; Brian, S.; Lim, K.C.; Hopkins, S.D.; McKerrow, J.H.; et al. RNA Interference in Schistosoma Mansoni Schistosomula: Selectivity, Sensitivity and Operation for Larger-Scale Screening. PLoS Negl. Trop. Dis. 2010, 4, e850. [Google Scholar] [CrossRef]
- Cogo, R.M.; Pavani, T.F.A.; Mengarda, A.C.A.; Cajas, R.A.; Teixeira, T.R.; Fukui-Silva, L.; Sun, Y.U.; Liu, L.J.; Amarasinghe, D.K.; Yoon, M.C.; et al. Pharmacophore Virtual Screening Identifies Riboflavin as an Inhibitor of the Schistosome Cathepsin B1 Protease with Antiparasitic Activity. ACS Omega 2024, 9, 25356–25369. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.F.; Senger, M.R.; Moreira-Filho, J.T.; Júnior, F.J.d.V.; Dantas, R.F.; Owens, R.; Andrade, C.H.; Neves, B.J.; Silva-Junior, F.P. Discovery of New Schistosoma Mansoni Aspartyl Protease Inhibitors by Structure-Based Virtual Screening. Mem. Inst. Oswaldo Cruz 2023, 118. [Google Scholar] [CrossRef]
- Spiwoková, P.; Horn, M.; Fanfrlík, J.; Jílková, A.; Fajtová, P.; Leontovyč, A.; Houštecká, R.; Bieliková, L.; Brynda, J.; Chanová, M.; et al. Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis. ACS Infect. Dis. 2024, 10, 1935–1948. [Google Scholar] [CrossRef]
- Jílková, A.; Řezáčová, P.; Lepšík, M.; Horn, M.; Váchová, J.; Fanfrlík, J.; Brynda, J.; McKerrow, J.H.; Caffrey, C.R.; Mareš, M. Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma Mansoni. J. Biol. Chem. 2011, 286, 35770–35781. [Google Scholar] [CrossRef] [PubMed]
- About the Global Health Priority Box|Medicines for Malaria Venture. Available online: https://www.mmv.org/mmv-open/global-health-priority-box/about-global-health-priority-box (accessed on 18 July 2024).
- Hallgren, J.; Tsirigos, K.D.; Damgaard Pedersen, M.; Juan, J.; Armenteros, A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of Protein Subcellular Localization Using Deep Learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Andreatta, M.; Nielsen, M. Gapped Sequence Alignment Using Artificial Neural Networks: Application to the MHC Class i System. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef]
- Calis, J.J.A.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved Methods for Predicting Peptide Binding Affinity to MHC Class II Molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef]
- Clifford, J.N.; Høie, M.H.; Deleuran, S.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-3.0: Improved B-Cell Epitope Prediction Using Protein Language Models. Protein Sci. 2022, 31, e4497. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving Sequence-Based B-Cell Epitope Prediction Using Conformational Epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- NPS@: MULTALIN Multiple Alignment. Available online: https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_multalin.html (accessed on 16 August 2021).
- Speziali, E.; Bethony, J.; Martins-Filho, O.; Fraga, L.A.O.; Lemos, D.S.; Souza, L.J.; Correa-Oliveira, R.; Faria, A.M.C. Production of Interferon-Gamma by Natural Killer Cells and Aging in Chronic Human Schistosomiasis. Mediat. Inflamm. 2004, 13, 327. [Google Scholar] [CrossRef] [PubMed]
- Brito, C.F.A.; Caldas, I.R.; Coura Filho, P.; Correa-Oliveira, R.; Oliveira, S.C. CD4+ T Cells of Schistosomiasis Naturally Resistant Individuals Living in an Endemic Area Produce Interferon-γ and Tumour Necrosis Factor-α in Response to the Recombinant 14kdaSchistosoma Mansoni Fatty Acid-Binding Protein. Scand. J. Immunol. 2000, 51, 595–601. [Google Scholar] [CrossRef]
- IFNepitope: A Server for Predicting and Designing IFN-Gamma Inducing Epitopes. Available online: https://webs.iiitd.edu.in/raghava/ifnepitope/application.php (accessed on 5 September 2021).
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Dimitrov, I.; Flower, D.R.; Doytchinova, I. AllerTOP-a Server for in Silico Prediction of Allergens. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef]
- Rathore, A.S.; Arora, A.; Choudhury, S.; Tijare, P.; Raghava, G.P.S. ToxinPred 3.0: An Improved Method for Predicting the Toxicity of Peptides. Comput. Biol. Med. 2024, 179, 108926. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Dustin Schaeffer, R.; et al. Accurate Prediction of Protein Structures and Interactions Using a 3-Track Neural Network. Science 2021, 373, 871. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- ClusPro 2.0: Protein-Protein Docking. Available online: https://cluspro.bu.edu/help.php (accessed on 7 September 2021).
- Rapin, N.; Lund, O.; Castiglione, F. Immune System Simulation Online. Bioinformatics 2011, 27, 2013–2014. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Dunkel, M.; Fullbeck, M.; Neumann, S.; Preissner, R. SuperNatural: A Searchable Database of Available Natural Compounds. Nucleic Acids Res. 2006, 34, D678–D683. [Google Scholar] [CrossRef] [PubMed]
- Rappé, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a Full Periodic Table Force Field for Molecular Mechanics and Molecular Dynamics Simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Wardi, Y. A Stochastic Steepest-Descent Algorithm. J. Optim. Theory Appl. 1988, 59, 307–323. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Saad, A.E.; Ashour, D.S.; Osman, E.M. Different Panel of Toll-like Receptors Expression during Chronic Schistosoma Mansoni Infection in Experimental Animals. Exp. Parasitol. 2022, 239, 108317. [Google Scholar] [CrossRef]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional Classification of Class II Human Leukocyte Antigen (HLA) Molecules Reveals Seven Different Supertypes and a Surprising Degree of Repertoire Sharing across Supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef]
- Gadelha, C.; Zhang, W.; Chamberlain, J.W.; Chait, B.T.; Wickstead, B.; Field, M.C. Architecture of a Host-Parasite Interface: Complex Targeting Mechanisms Revealed through Proteomics. Mol. Cell Proteom. 2015, 14, 1911–1926. [Google Scholar] [CrossRef]
- De Gregorio, E.; Rappuoli, R. Vaccines for the Future: Learning from Human Immunology. Microb. Biotechnol. 2012, 5, 149. [Google Scholar] [CrossRef] [PubMed]
- Pandya, N.; Kumar, A. Immunoinformatics Analysis for Design of Multi-Epitope Subunit Vaccine by Using Heat Shock Proteins against Schistosoma mansoni. J. Biomol. Struct. Dyn. 2023, 41, 1859–1878. [Google Scholar] [CrossRef] [PubMed]
- Parker-Manuel, S.J.; Ivens, A.C.; Dillon, G.P.; Wilson, R.A. Gene Expression Patterns in Larval Schistosoma Mansoni Associated with Infection of the Mammalian Host. PLoS Negl. Trop. Dis. 2011, 5, e1274. [Google Scholar] [CrossRef] [PubMed]
- Majer, P.; Collins, J.R.; Gulnik, S.V.; Erickson, J.W. Structure-Based Subsite Specificity Mapping of Human Cathepsin D Using Statine-Based Inhibitors. Protein Sci. 1997, 6, 1458–1466. [Google Scholar] [CrossRef]
- Riva, D.; Barison, A.; Stefanello, M.É.A.; Poliquesi, C.B.; Ruiz, A.L.T.G.; De Carvalho, J.E.; Salvador, M.J. Estudo Químico de Sinningia Allagophylla Guiado Por Testes de Atividade Antiproliferativa. Quim. Nova 2012, 35, 974–977. [Google Scholar] [CrossRef]
- Scharf, D.R.; Verdan, M.H.; Ribeiro, M.A.; Simionatto, E.L.; Sá, E.L.; Salvador, M.J.; Barison, A.; Stefanello, M.E.A. Naphthochromenes and Related Constituents from the Tubers of Sinningia Allagophylla. J. Nat. Prod. 2016, 79, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Winiewski, V.; Verdan, M.H.; Santos Oliveira, C.; Eloyane Barreto Rodrigues, T.; Salvador, M.J.; Alves Stefanello, M.É. Two New Naphthoquinone Derivatives from Sinningia conspicua (Gesneriaceae). Nat. Prod. Res. 2023, 1–6. [Google Scholar] [CrossRef]
- Serain, A.F.; Buitrago-Mejia, A.J.; de Souza, G.C.A.; Corrêa, W.R.; Stefanello, M.E.A.; Salvador, M.J. Antitumoral Photoinduced Effects of Crude Extract, Fractions, and Naphthoquinones from Sinningia magnifica (Otto & A. Dietr.) Wiehler (Gesneriaceae) in a Bioguided Study. Photochem. Photobiol. 2024, 100, 190–203. [Google Scholar] [CrossRef]
- Lira, R.; Nascimento, D.V.; Lopes, K.C.; Soares, M.R.S.; Torres, J.B. Assessment of Boll Weevil Susceptibility to Isocycloseram and Ethiprole and Differential Toxicity to Natural Enemies. Neotrop. Entomol. 2024, 53, 682–693. [Google Scholar] [CrossRef]
- Hirashima, S.; Amimoto, T.; Iwamoto, Y.; Takeda, K. Photodegradation of the Phenylpyrazole Insecticide Ethiprole in Aquatic Environments and a Comparison with Fipronil. Environ. Sci. Pollut. Res. 2024, 31, 53447–53457. [Google Scholar] [CrossRef]
- Li, L.; Zhao, X.; Hu, K.; Xu, W.; Wang, M.; Wang, H. Enantioselective Toxicity and Potential Endocrine-Disruptive Effects of the Insecticides Flufiprole and Ethiprole on Danio Rerio. J. Agric. Food Chem. 2024, 72, 1509–1515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Z.; Li, Q.X.; Hua, R.; Wu, X. Enantioselective Metabolism of Phenylpyrazole Insecticides by Rat Liver Microsomal CYP3A1, CYP2E1 and CYP2D2. Pestic. Biochem. Physiol. 2021, 176, 1509–15153. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Inomata, A. Reproductive and Neurobehavioral Effects of Ethiprole Administered to Mice in the Diet. Birth Defects Res. 2017, 109, 1568–1585. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, F.; Wang, P.; Jiang, W.; Zhang, Z.; Liu, D.; Zhou, Z. Enantioselective Toxic Effects and Environmental Behavior of Ethiprole and Its Metabolites against Chlorella pyrenoidosa. Environ. Pollut. 2019, 244, 757–765. [Google Scholar] [CrossRef]

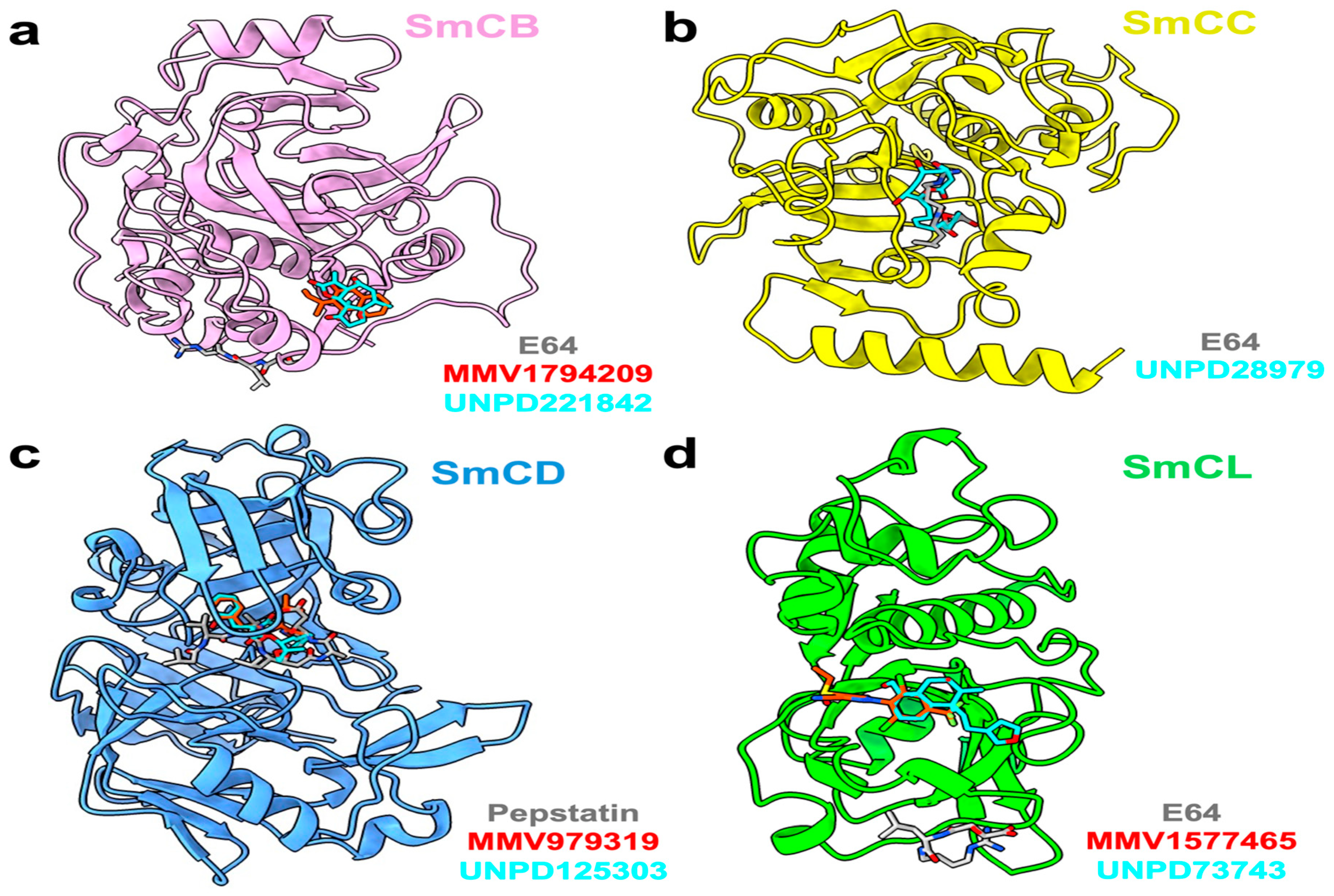
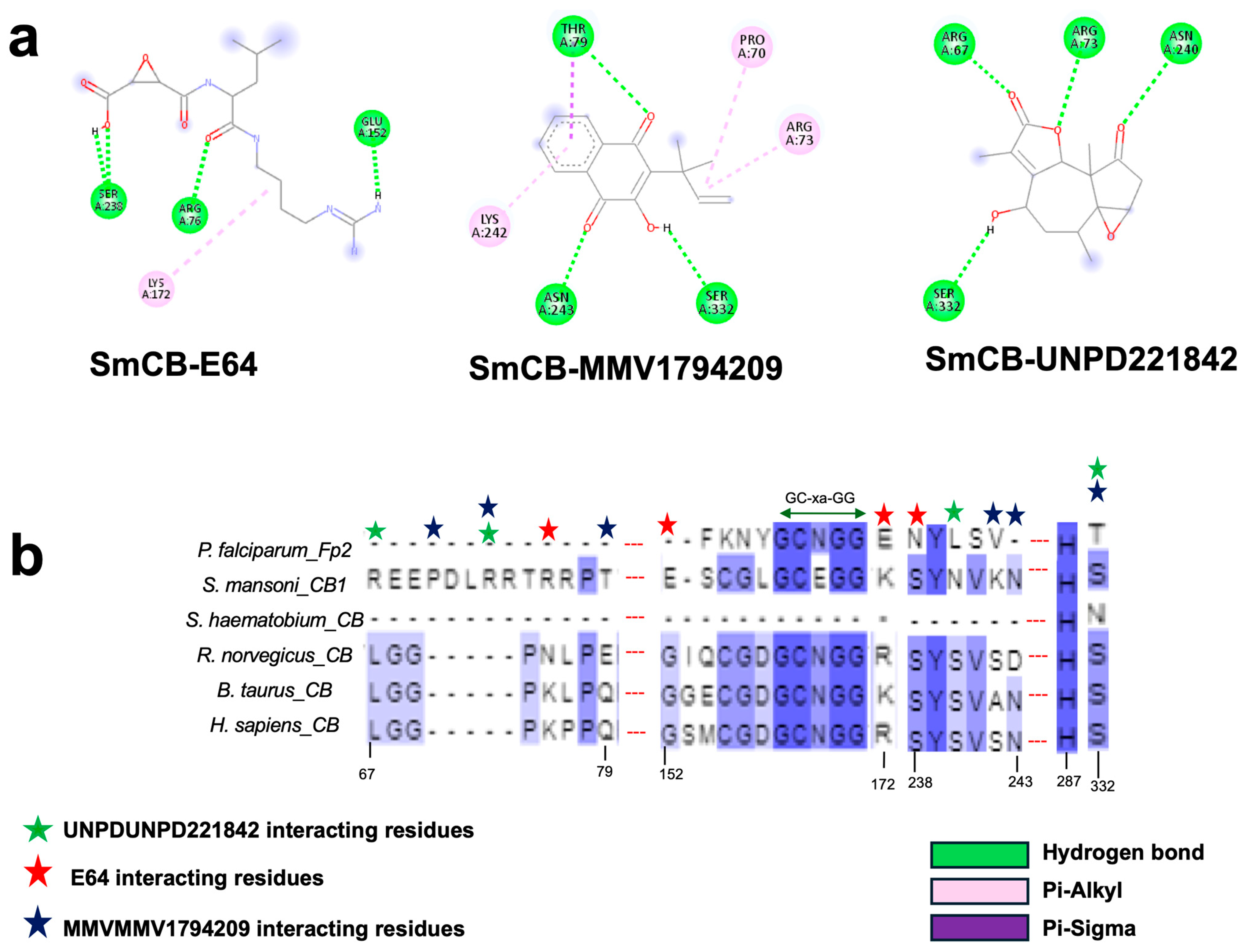
| Target Cathepsin | Hit ID | Hit Structure | Binding Energy (kcal/mol) |
|---|---|---|---|
| B | UNPD221842 | 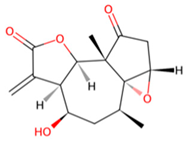 | −7.6 |
| MMV1794209 | 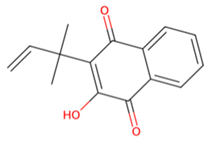 | −6.7 | |
| C | UNPD28979 | 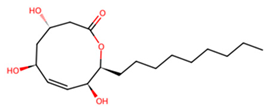 | −6.7 |
| D | UNPD125303 | 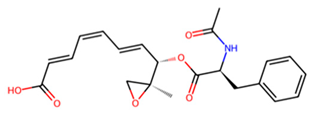 | −7.6 |
| MMV979319 | 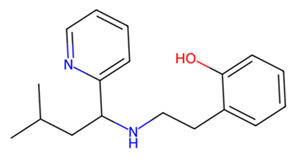 | −7.2 | |
| L | UNPD73743 | 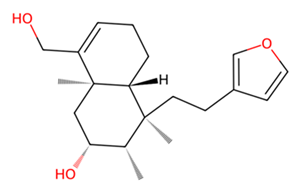 | −7.4 |
| MMV1577465 | 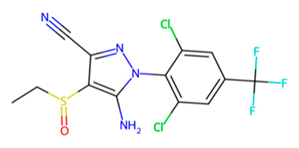 | −6.8 | |
| B C L | E-64 | 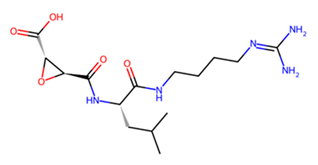 | −6 −6.7 −5.3 |
| D | Pepstatin | 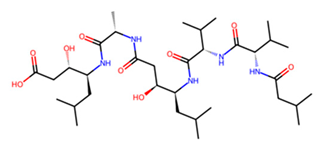 | −7.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balogun, E.O.; Joseph, G.I.; Olabode, S.C.; Dayaso, N.A.; Danazumi, A.U.; Bashford-Rogers, R.; Mckerrow, J.H.; Jeelani, G.; Caffrey, C.R. Computational Workflow to Design Novel Vaccine Candidates and Small-Molecule Therapeutics for Schistosomiasis. Pathogens 2024, 13, 850. https://doi.org/10.3390/pathogens13100850
Balogun EO, Joseph GI, Olabode SC, Dayaso NA, Danazumi AU, Bashford-Rogers R, Mckerrow JH, Jeelani G, Caffrey CR. Computational Workflow to Design Novel Vaccine Candidates and Small-Molecule Therapeutics for Schistosomiasis. Pathogens. 2024; 13(10):850. https://doi.org/10.3390/pathogens13100850
Chicago/Turabian StyleBalogun, Emmanuel Oluwadare, Gideon Ibrahim Joseph, Samuel Charles Olabode, Naziru Abdulkadir Dayaso, Ammar Usman Danazumi, Rachael Bashford-Rogers, James H. Mckerrow, Ghulam Jeelani, and Conor R. Caffrey. 2024. "Computational Workflow to Design Novel Vaccine Candidates and Small-Molecule Therapeutics for Schistosomiasis" Pathogens 13, no. 10: 850. https://doi.org/10.3390/pathogens13100850
APA StyleBalogun, E. O., Joseph, G. I., Olabode, S. C., Dayaso, N. A., Danazumi, A. U., Bashford-Rogers, R., Mckerrow, J. H., Jeelani, G., & Caffrey, C. R. (2024). Computational Workflow to Design Novel Vaccine Candidates and Small-Molecule Therapeutics for Schistosomiasis. Pathogens, 13(10), 850. https://doi.org/10.3390/pathogens13100850







