Advancements in Detection Methods for Salmonella in Food: A Comprehensive Review
Abstract
1. Introduction
2. Traditional Culture-Based Methods
3. Immunological Techniques
4. Molecular Methods
5. Emerging Technologies
5.1. Electrochemical Aptasensors
5.2. Surface Plasmon Resonance
5.3. Surface Enhanced Raman Spectroscopy
5.4. Bacteriophages
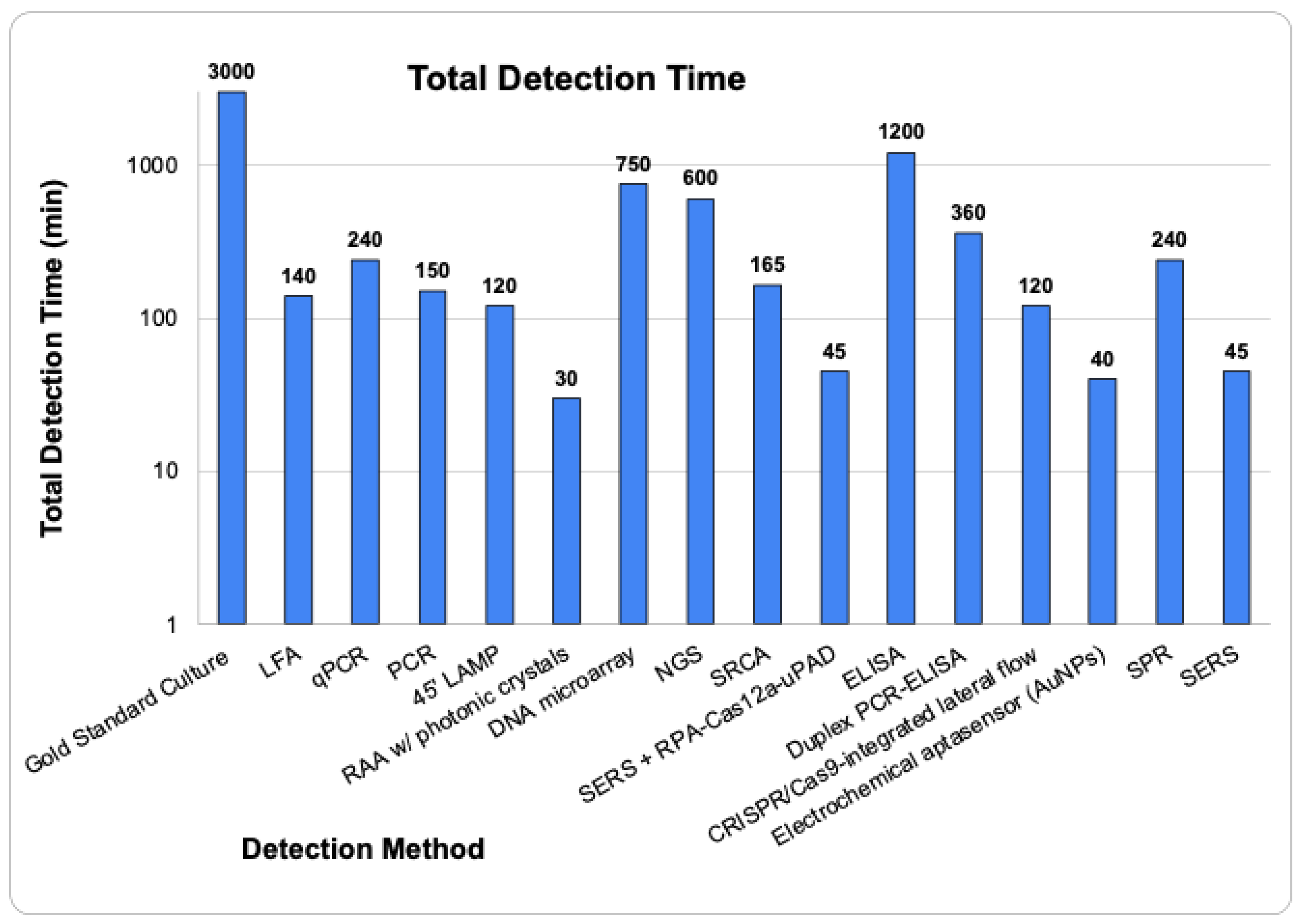
6. Challenges Associated with Detection Technologies: Limits of Detection and Enrichment Time
7. Future Directions
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- CDC. Salmonella Homepage. 2016. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 15 January 2021).
- Galán-Relaño; Díaz, A.V.; Lorenzo, B.H.; Gómez-Gascón, L.; Rodríguez, M.M.; Jiménez, E.C.; Rodríguez, F.P.; Márquez, R.J.A. Salmonella and Salmonellosis: An Update on Public Health Implications and Control Strategies. Animals 2023, 13, 3666. [Google Scholar] [CrossRef] [PubMed]
- Grimont, P.A.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella. Institute Pasteur: Paris, France, 2007; pp. 1–166. [Google Scholar]
- Sanchez, S.; Hofacre, C.L.; Lee, M.D.; Maurer, J.J.; Doyle, M.P. Animal sources of salmonellosis in humans. J. Am. Veter- Med Assoc. 2002, 221, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Abulreesh, H.H. Salmonellae in the environment. In Salmonella—Distribution, Adaptation, Control Measures and Molecular Technologies; Bassam, A.A., Joshua, B.G., Eds.; IntechOpen: London, UK, 2012; pp. 22–31. [Google Scholar] [CrossRef]
- Rodriguez, A.; Pangloli, P.; Richards, H.A.; Mount, J.R.; Draughon, F.A. Prevalence of Salmonella in Diverse Environmental Farm Samples. J. Food Prot. 2006, 69, 2576–2580. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.F.; Wong, W.C.; Chai, L.C.; Nillian, E.; Ghazali, F.M.; Cheah, Y.K.; Nakaguchi, Y.; Nishibuchi, M.; Radu, S. Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control 2011, 22, 337–342. [Google Scholar] [CrossRef]
- Shen, X.; Yin, L.; Zhang, A.; Zhao, R.; Yin, D.; Wang, J.; Dai, Y.; Hou, H.; Pan, X.; Hu, X.; et al. Prevalence and Characterization of Salmonella Isolated from Chickens in Anhui, China. Pathogens 2023, 12, 465. [Google Scholar] [CrossRef]
- Institute of Medicine (US) and National Research Council (US) Committee on the Review of the Use of Scientific Criteria and Performance Standards for Safe Food. Scientific Criteria to Ensure Safe Food; National Academies Press (US): Washington, DC, USA, 2003. Available online: https://www.ncbi.nlm.nih.gov/books/NBK221565/ (accessed on 29 October 2024). [PubMed]
- He, Y.; Wang, J.; Zhang, R.; Chen, L.; Zhang, H.; Qi, X.; Chen, J. Epidemiology of foodborne diseases caused by Salmonella in Zhejiang Province, China, between 2010 and 2021. Front. Public Health 2023, 11, 1127925. [Google Scholar] [CrossRef]
- Huckstep, R.L. Typhoid Fever and Other Salmonella Infections. Am. J. Med Sci. 1963, 245, 154. [Google Scholar] [CrossRef]
- Acheson, D.; Hohmann, E.L. Nontyphoidal Salmonellosis. Clin. Infect. Dis. 2001, 32, 263–269. [Google Scholar] [CrossRef]
- Dhanoa, A.; Fatt, Q.K. Non-typhoidal Salmonella bacteraemia: Epidemiology, clinical characteristics and its’ association with severe immunosuppression. Ann. Clin. Microbiol. Antimicrob. 2009, 8, 15. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Crump, J.A.; Kretsinger, K.; Gay, K.; Hoekstra, R.M.; Vugia, D.J.; Hurd, S.; Segler, S.D.; Megginson, M.; Luedeman, L.J.; Shiferaw, B.; et al. Clinical Response and Outcome of Infection with Salmonella enterica Serotype Typhi with Decreased Susceptibility to Fluoroquinolones: A United States FoodNet Multicenter Retrospective Cohort Study. Antimicrob. Agents Chemother. 2008, 52, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Eikmeier, D.; Medus, C.; Smith, K. Incubation period for outbreak-associated, non-typhoidal salmonellosis cases, Minnesota, 2000–2015. Epidemiology Infect. 2018, 146, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Ghori, N.; Falkow, S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994, 180, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Zhang, P.; Piao, R.; Wang, Y. Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front. Cell. Infect. Microbiol. 2019, 9, 270. [Google Scholar] [CrossRef]
- Galán, J.E. Molecular and Cellular Bases of Salmonella Entry into Host Cells. Curr. Top. Microbiol. Immunol. 1996, 209, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-M.; Hobbie, S.; Galán, J.E. Requirement of CDC42 for Salmonella -Induced Cytoskeletal and Nuclear Responses. Science 1996, 274, 2115–2118. [Google Scholar] [CrossRef]
- Vassiloyanakopoulos, A.P.; Okamoto, S.; Fierer, J. The crucial role of polymorphonuclear leukocytes in resistance to Salmonella dublin infections in genetically susceptible and resistant mice. Proc. Natl. Acad. Sci. USA 1998, 95, 7676–7681. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Recalls, Market Withdrawals, & Safety Alerts. U.S. Department of Health and Human Services. 2024. Available online: https://www.fda.gov/safety/recalls-market-withdrawals-safety-alerts (accessed on 30 October 2024).
- Wang, M.; Zhang, Y.; Tian, F.; Liu, X.; Du, S.; Ren, G. Overview of Rapid Detection Methods for Salmonella in Foods: Progress and Challenges. Foods 2021, 10, 2402. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Guidance for Industry: Testing for Salmonella Species in Human Foods and Di-rect-Human-Contact Animal Foods. 2012. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/guidance-industry-testing-salmonella-species-human-foods-and-direct-human-contact-animal-foods (accessed on 30 October 2024).
- ISO 16140-2:2016; Microbiology of the Food Chain—Method Validation—Part 2: Protocol for the Validation of Alternative (Proprietary) Methods Against a Reference Method. International Organization for Standardization: Geneva, Switzerland, 2016. Available online: https://www.iso.org/obp/ui/en/#iso:std:iso:16140:-2:ed-1:v1:en (accessed on 29 October 2024).
- U.S. Food and Drug Administration Foods Program. Guidelines for the Validation of Analytical Methods for the Detection of Microbial Pathogens in Foods and Feeds, 3rd ed.; 2019. Available online: https://www.fda.gov/media/83812/download (accessed on 29 October 2024).
- European Union. Regulation (EU) 2017/625 of the European Parliament and of the Council of 15 March 2017 on Official Controls and Other Official Activities Carried Out to Ensure the Application of Food and Feed Law, Rules on Animal Health and Welfare, Plant Health and Plant Protection Products. EUR-Lex. 2017. Available online: http://data.europa.eu/eli/reg/2017/625/2022-01-28/eng (accessed on 29 October 2024).
- Neyaz, L.A.; Alghamdi, H.S.; Alghashmari, R.M.; Alswat, S.S.; Almaghrabi, R.O.; Bazaid, F.S.; Albarakaty, F.M.; Elbanna, K.; Abulreesh, H.H. A Comprehensive Review on the Current Status of Culture Media for Routine Standardized Isolation of Salmonella and Shigella Spp. from Contaminated Food. J. Umm Al-Qura Univ. Appll. Sci. 2024. [Google Scholar] [CrossRef]
- Mooijman, K.A. The new ISO 6579-1: A real horizontal standard for detection of Salmonella, at last! Food Microbiol. 2018, 71, 2–7. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. [WWW Document]. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/56712.html (accessed on 29 October 2024).
- Zhuang, L.; Gong, J.; Shen, Q.; Yang, J.; Song, C.; Liu, Q.; Zhao, B.; Zhang, Y.; Zhu, M. Advances in detection methods for viable Salmonella spp.: Current applications and challenges. Anal. Sci. 2023, 39, 1643–1660. [Google Scholar] [CrossRef] [PubMed]
- Oslan, S.N.H.; Yusof, N.Y.; Lim, S.J.; Ahmad, N.H. Rapid and sensitive detection of Salmonella in agro-Food and environmental samples: A review of advances in rapid tests and biosensors. J. Microbiol. Methods 2024, 219, 106897. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Esteve, P.; Tammela, P. Evaluation and validation of Biolog OmniLog(R) system for antibacterial activity assays. Lett. Appl. Microbiol. 2021, 72, 589–595. [Google Scholar] [CrossRef] [PubMed]
- ISO/TR No. 6579-3:2014; Microbiology of the food chain-horizontal method for the detection, enumeration and serotyping of Salmonella-Part 3: Guidelines for serotyping of Salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2014. Available online: https://www.iso.org/standard/56714.html (accessed on 29 October 2024).
- Wattiau, P.; Boland, C.; Bertrand, S. Methodologies for Salmonella enterica subsp. enterica Subtyping: Gold Standards and Alternatives. Appl. Environ. Microbiol. 2011, 77, 7877–7885. [Google Scholar] [CrossRef] [PubMed]
- Awang, M.S.; Bustami, Y.; Hamzah, H.H.; Zambry, N.S.; Najib, M.A.; Khalid, M.F.; Aziah, I.; Manaf, A.A. Advancement in Salmonella Detection Methods: From Conventional to Electrochemical-Based Sensing Detection. Biosensors 2021, 11, 346. [Google Scholar] [CrossRef]
- Tietjen, M.; Fung, D.Y.C. Salmonellae and Food Safety. Crit. Rev. Microbiol. 1995, 21, 53–83. [Google Scholar] [CrossRef]
- Mansfield, L.; Forsythe, S. The detection of Salmonella using a combined immunomagnetic separation and ELISA end-detection procedure. Lett. Appl. Microbiol. 2000, 31, 279–283. [Google Scholar] [CrossRef]
- Hu, J.; Huang, R.; Wang, Y.; Wei, X.; Wang, Z.; Geng, Y.; Jing, J.; Gao, H.; Sun, X.; Dong, C.; et al. Development of duplex PCR-ELISA for simultaneous detection of Salmonella spp. and Escherichia coli O157: H7 in food. J. Microbiol. Methods 2018, 154, 127–133. [Google Scholar] [CrossRef]
- Priyanka, B.; Patil, R.K.; Dwarakanath, S. A review on detection methods used for foodborne pathogens. Indian J. Med. Res. 2016, 144, 327–338. [Google Scholar] [CrossRef]
- Schenk, F.; Weber, P.; Vogler, J.; Hecht, L.; Dietzel, A.; Gauglitz, G. Development of a paper-based lateral flow immunoassay for simultaneous detection of lipopolysaccharides of Salmonella serovars. Anal. Bioanal. Chem. 2017, 410, 863–868. [Google Scholar] [CrossRef]
- Gao, P.; Wang, L.; He, Y.; Wang, Y.; Yang, X.; Fu, S.; Qin, X.; Chen, Q.; Man, C.; Jiang, Y. An Enhanced Lateral Flow Assay Based on Aptamer–Magnetic Separation and Multifold AuNPs for Ultrasensitive Detection of Salmonella Typhimurium in Milk. Foods 2021, 10, 1605. [Google Scholar] [CrossRef] [PubMed]
- Chirambo, A.C.; Nyirenda, T.S.; Jambo, N.; Msefula, C.; Kamng’Ona, A.; Molina, S.; Mandala, W.L.; Heyderman, R.S.; Iturizza-Gomara, M.; Henrion, M.Y.; et al. Performance of molecular methods for the detection of Salmonella in human stool specimens. Wellcome Open Res. 2021, 5, 237. [Google Scholar] [CrossRef] [PubMed]
- Aladhadh, M. A Review of Modern Methods for the Detection of Foodborne Pathogens. Microorganisms 2023, 11, 1111. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, L.; Wang, H.H. Critical Issues in Detecting Viable Listeria monocytogenes Cells by Real-Time Reverse Transcriptase PCR. J. Food Prot. 2012, 75, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Zendrini, A.; Carta, V.; Filipello, V.; Ragni, L.; Cosciani-Cunico, E.; Arnaboldi, S.; Bertasi, B.; Franceschi, N.; Ajmone-Marsan, P.; De Medici, D.; et al. One-Day Molecular Detection of Salmonella and Campylobacter in Chicken Meat: A Pilot Study. Foods 2021, 10, 1132. [Google Scholar] [CrossRef]
- Chen, M.; Lan, X.; Zhu, L.; Ru, P.; Xu, W.; Liu, H. PCR Mediated Nucleic Acid Molecular Recognition Technology for Detection of Viable and Dead Foodborne Pathogens. Foods 2022, 11, 2675. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, 63. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, N.; Zheng, L.; Cai, G.; Lin, J. A lab-on-chip device for the sample-in-result-out detection of viable Salmonella using loop-mediated isothermal amplification and real-time turbidity monitoring. Lab A Chip 2020, 20, 2296–2305. [Google Scholar] [CrossRef]
- Li, M.; Luan, Z.; Liu, Y.; Yang, C.; Wang, Y.; Ma, C.; Shi, C. Ultrafast bacterial cell lysis using a handheld corona treater and loop-mediated isothermal amplification for rapid detection of foodborne pathogens. Food Control. 2021, 128, 108178. [Google Scholar] [CrossRef]
- Williams, M.R.; Telli, A.E.; Telli, N.; Islam, D.T.; Hashsham, S.A. Direct or DNA Extraction-Free Amplification and Quantification of Foodborne Pathogens. Methods Mol. Biol. 2019, 2852, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Ndraha, N.; Lin, H.-Y.; Wang, C.-Y.; Hsiao, H.-I.; Lin, H.-J. Rapid detection methods for foodborne pathogens based on nucleic acid amplification: Recent advances, remaining challenges, and possible opportunities. Food Chem. Mol. Sci. 2023, 7, 100183. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Zhou, D.; Xie, G.; Liu, J.; Xiong, Q.; Feng, X.; Xu, H. The fluorescent probe-based recombinase-aided amplification for rapid detection of Escherichia coli O157:H7. Mol. Cell. Probes 2021, 60, 101777. [Google Scholar] [CrossRef] [PubMed]
- Milton, A.A.P.; Momin, K.M.; Priya, G.B.; Ghatak, S.; Das, S.; Gandhale, P.N.; Angappan, M.; Sen, A. Development of novel visual detection methodology for Salmonella in meat using saltatory rolling circle amplification. J. Appl. Microbiol. 2021, 131, 2361–2371. [Google Scholar] [CrossRef] [PubMed]
- Delgado, E.; Katchman, B.; Stice, S.; Calle, A. Independent evaluation of a DNA microarray system for Salmonella detection in ground beef. Food Microbiol. 2023, 118, 104406. [Google Scholar] [CrossRef]
- Katchman, B.A.; Sinton, M. DNA Microarrays: A Powerful Tool for Pathogen Detection; PathogenDx, 2019; Available online: https://pathogendx.com/wp-content/uploads/2019/12/What-is-a-DNA-Microarray_BAK-072419_1.pdf (accessed on 29 October 2024).
- U.S. Food and Safety Inspection Service. Foodborne Pathogen Test Kits Validated by Independent Organizations. 2024. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/documents/Validated-Test-Kit.pdf (accessed on 29 October 2024).
- Den Bakker, H.C.; Allard, M.W.; Bopp, D.; Brown, E.W.; Fontana, J.; Iqbal, Z.; Kinney, A.; Limberger, R.; Musser, K.; Shudt, M.; et al. Rapid Whole-Genome Sequencing for Surveillance of Salmonella enterica Serovar Enteritidis. Emerg. Infect. Dis. 2014, 20, 1306–1314. [Google Scholar] [CrossRef]
- Nafea, A.M.; Wang, Y.; Wang, D.; Salama, A.M.; Aziz, M.A.; Xu, S.; Tong, Y. Application of next-generation sequencing to identify different pathogens. Front. Microbiol. 2024, 14, 1329330. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Whole Genome Sequencing (WGS) Program. U.S. Department of Health and Human Services. 2024. Available online: https://www.fda.gov/food/microbiology-research-food/whole-genome-sequencing-wgs-program (accessed on 29 October 2024).
- U.S. Food and Drug Administration. GenomeTrakr Fast Facts. U.S. Department of Health and Human Services. 2024. Available online: https://www.fda.gov/food/whole-genome-sequencing-wgs-program/genometrakr-fast-facts (accessed on 29 October 2024).
- Lin, A.; Singh, A.; Allred, A.; Allard, M.; Waltman, D.; Imanian, B.; Ng, J.H.; Sanahmadi, Y.; Khaksar, R. Targeted Next-Generation Sequencing Assay for Direct Detection and Serotyping of Salmonella from Enrichment. J. Food Prot. 2024, 87, 100256. [Google Scholar] [CrossRef]
- Persad, A.K.; Fahmy, H.A.; Anderson, N.; Cui, J.; Topalcengiz, Z.; Jeamsripong, S.; Spanninger, P.M.; Buchanan, R.L.; Kniel, K.E.; Jay-Russell, M.T.; et al. Identification and Subtyping of Salmonella Isolates Using Matrix-Assisted Laser Desorption–Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF). Microorganisms 2022, 10, 688. [Google Scholar] [CrossRef]
- Kliem, M.; Sauer, S. The essence on mass spectrometry based microbial diagnostics. Curr. Opin. Microbiol. 2012, 15, 397–402. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom. Rev. 2012, 32, 188–217. [Google Scholar] [CrossRef] [PubMed]
- Bruker. Microbial Analysis by MALDI-TOF. 2024. Available online: https://www.bruker.com/en/products-and-solutions/microbiology-and-diagnostics/microbial-identification.html (accessed on 30 October 2024).
- Mangmee, S.; Reamtong, O.; Kalambaheti, T.; Roytrakul, S.; Sonthayanon, P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Control. 2020, 113, 107188. [Google Scholar] [CrossRef]
- Yang, S.-M.; Kim, E.; Kim, D.; Baek, J.; Yoon, H.; Kim, H.-Y. Rapid Detection of Salmonella Enteritidis, Typhimurium, and Thompson by Specific Peak Analysis Using Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Foods 2021, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Q.; Zhou, M.; Li, C.; Yan, C.; Huang, L.; Qin, P. Development of a CRISPR/Cas9-integrated lateral flow strip for rapid and accurate detection of Salmonella. Food Control. 2022, 142. [Google Scholar] [CrossRef]
- Guo, W.; Yang, Q.; Liu, J.; Chen, X.; Zhang, Y.; Zhang, W. Multiple fluorescent saltatory rolling circle amplification (SRCA) for simultaneous and sensitive detection of Salmonella spp. and Shigella spp. in food. LWT 2022, 168. [Google Scholar] [CrossRef]
- Huang, L.; Yuan, N.; Guo, W.; Zhang, Y.; Zhang, W. An electrochemical biosensor for the highly sensitive detection of Staphylococcus aureus based on SRCA-CRISPR/Cas12a. Talanta 2022, 252, 123821. [Google Scholar] [CrossRef]
- Cirocka, A.; Zarzeczańska, D.; Wcisło, A. Good Choice of Electrode Material as the Key to Creating Electrochemical Sensors—Characteristics of Carbon Materials and Transparent Conductive Oxides (TCO). Materials 2021, 14, 4743. [Google Scholar] [CrossRef]
- Zambry, N.S.; Najib, M.A.; Awang, M.S.; Selvam, K.; Khalid, M.F.; Bustami, Y.; Hamzah, H.H.; Ozsoz, M.; Manaf, A.A.; Aziah, I. Aptamer-Based Electrochemical Biosensors for the Detection of Salmonella: A Scoping Review. Diagnostics 2022, 12, 3186. [Google Scholar] [CrossRef]
- Fathi, S.; Saber, R.; Adabi, M.; Rasouli, R.; Douraghi, M.; Morshedi, M.; Farid-Majidi, R. Novel Competitive Voltammetric Aptasensor Based on Electrospun Carbon Nanofibers-Gold Nanoparticles Modified Graphite Electrode for Salmonella enterica serovar Detection. Biointerface Res. Appl. Chem. 2020, 11, 8702–8715. [Google Scholar] [CrossRef]
- Ma, X.; Jiang, Y.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Methods 2014, 98, 94–98. [Google Scholar] [CrossRef]
- Ge, C.; Yuan, R.; Yi, L.; Yang, J.; Zhang, H.; Li, L.; Nian, W.; Yi, G. Target-induced aptamer displacement on gold nanoparticles and rolling circle amplification for ultrasensitive live Salmonella typhimurium electrochemical biosensing. J. Electroanal. Chem. 2018, 826, 174–180. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Appaturi, J.N.; Thong, K.L.; Lai, C.W.; Ibrahim, F.; Leo, B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 2019, 127, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Hasan, R.; Pulingam, T.; Appaturi, J.N.; Zifruddin, A.N.; Teh, S.J.; Lim, T.W.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 2018, 554, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.; Chen, F.-C.; Bridgman, R.C. Detection of Salmonella Typhimurium in Romaine Lettuce Using a Surface Plasmon Resonance Biosensor. Biosensors 2019, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.; Chen, F.-C.; Bridgman, R.C. Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce. Sensors 2022, 22, 475. [Google Scholar] [CrossRef]
- Oh, S.Y.; Heo, N.S.; Shukla, S.; Cho, H.J.; Vilian, A.E.; Kim, J.; Lee, S.Y.; Han, Y.-K.; Yoo, S.M.; Huh, Y.S. Development of gold nanoparticle-aptamer-based LSPR sensing chips for the rapid detection of Salmonella typhimurium in pork meat. Sci. Rep. 2017, 7, 10130. [Google Scholar] [CrossRef]
- Liang, F.; Li, Y.; Hu, M.; Shi, C.; Cui, Y.; Young, G.M.; Yang, M.M.; Zhang, J. Nanoparticle-Enhanced SPR Immunobiosensor for Rapid Detection of Salmonella Typhimurium by Immunomagnetic Separation (SSRN Scholarly Paper 4680036). 2024. Available online: https://ssrn.com/abstract=4680036 (accessed on 3 December 2024).
- Asgari, S.; Dhital, R.; Mustapha, A.; Lin, M. Duplex detection of foodborne pathogens using a SERS optofluidic sensor coupled with immunoassay. Int. J. Food Microbiol. 2022, 383, 109947. [Google Scholar] [CrossRef]
- Chuesiang, P.; Ryu, V.; Siripatrawan, U.; He, L.; McLandsborough, L. Aptamer-based surface enhanced Raman spectroscopy (SERS) for the rapid detection of Salmonella Enteritidis contaminated in ground beef. LWT 2021, 150. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Li, J.; Tu, Z.; Gu, B.; Wang, S. Ultrasensitive and multiplex detection of four pathogenic bacteria on a bi-channel lateral flow immunoassay strip with three-dimensional membrane-like SERS nanostickers. Biosens. Bioelectron. 2022, 214, 114525. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhao, Z.; Lian, K.; Yin, L.; Wang, J.; Man, S.; Liu, G.; Ma, L. SERS-based CRISPR/Cas assay on microfluidic paper analytical devices for supersensitive detection of pathogenic bacteria in foods. Biosens. Bioelectron. 2022, 207, 114167. [Google Scholar] [CrossRef]
- Jia, F.; Li, B.; He, Y.; Shen, Y.; Chen, J.; Li, X.; Li, Y. An amplification-free CRISPR-SERS biosensor for specific, sensitive and rapid detection of Salmonella Typhimurium in poultry. LWT 2023, 189. [Google Scholar] [CrossRef]
- Zheng, S.; Xiao, J.; Zhang, J.; Sun, Q.; Liu, D.; Liu, Y.; Gao, X. Python-assisted detection and photothermal inactivation of Salmonella typhimurium and Staphylococcus aureus on a background-free SERS chip. Biosens. Bioelectron. 2023, 247, 115913. [Google Scholar] [CrossRef] [PubMed]
- Jung, L.-S.; Ding, T.; Ahn, J. Evaluation of lytic bacteriophages for control of multidrug-resistant Salmonella Typhimurium. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Thanki, A.M.; Brown, N.; Millard, A.D.; Clokie, M.R.J. Genomic Characterization of Jumbo Salmonella Phages That Effectively Target United Kingdom Pig-Associated Salmonella Serotypes. Front. Microbiol. 2019, 10, 1491. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Vinner, G.K.; Lopez, V.C.; Thanki, A.M.; Phothaworn, P.; Thiennimitr, P.; Garcia, A.; AbuOun, M.; Anjum, M.F.; Korbsrisate, S.; et al. An Optimized Bacteriophage Cocktail Can Effectively Control Salmonella in vitro and in Galleria mellonella. Front. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Lopez-Garcia, A.V.; AbuOun, M.; Nunez-Garcia, J.; Nale, J.Y.; Gaylov, E.E.; Phothaworn, P.; Sukjoi, C.; Thiennimitr, P.; Malik, D.J.; Korbsrisate, S.; et al. Pathogen genomics and phage-based solutions for accurately identifying and controlling Salmonella pathogens. Front. Microbiol. 2023, 14. [Google Scholar] [CrossRef]
- Cho, I.-H.; Ku, S. Current Technical Approaches for the Early Detection of Foodborne Pathogens: Challenges and Opportunities. Int. J. Mol. Sci. 2017, 18, 2078. [Google Scholar] [CrossRef]
- Quintela, I.A.; Vasse, T.; Lin, C.-S.; Wu, V.C.H. Advances, applications, and limitations of portable and rapid detection technologies for routinely encountered foodborne pathogens. Front. Microbiol. 2022, 13, 1054782. [Google Scholar] [CrossRef]
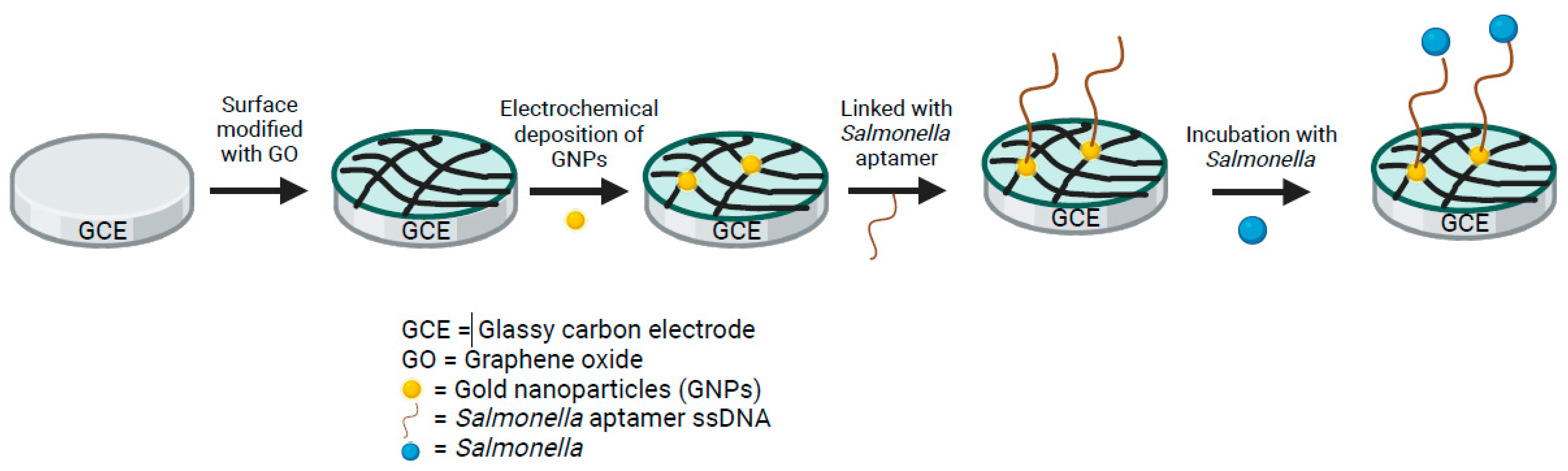
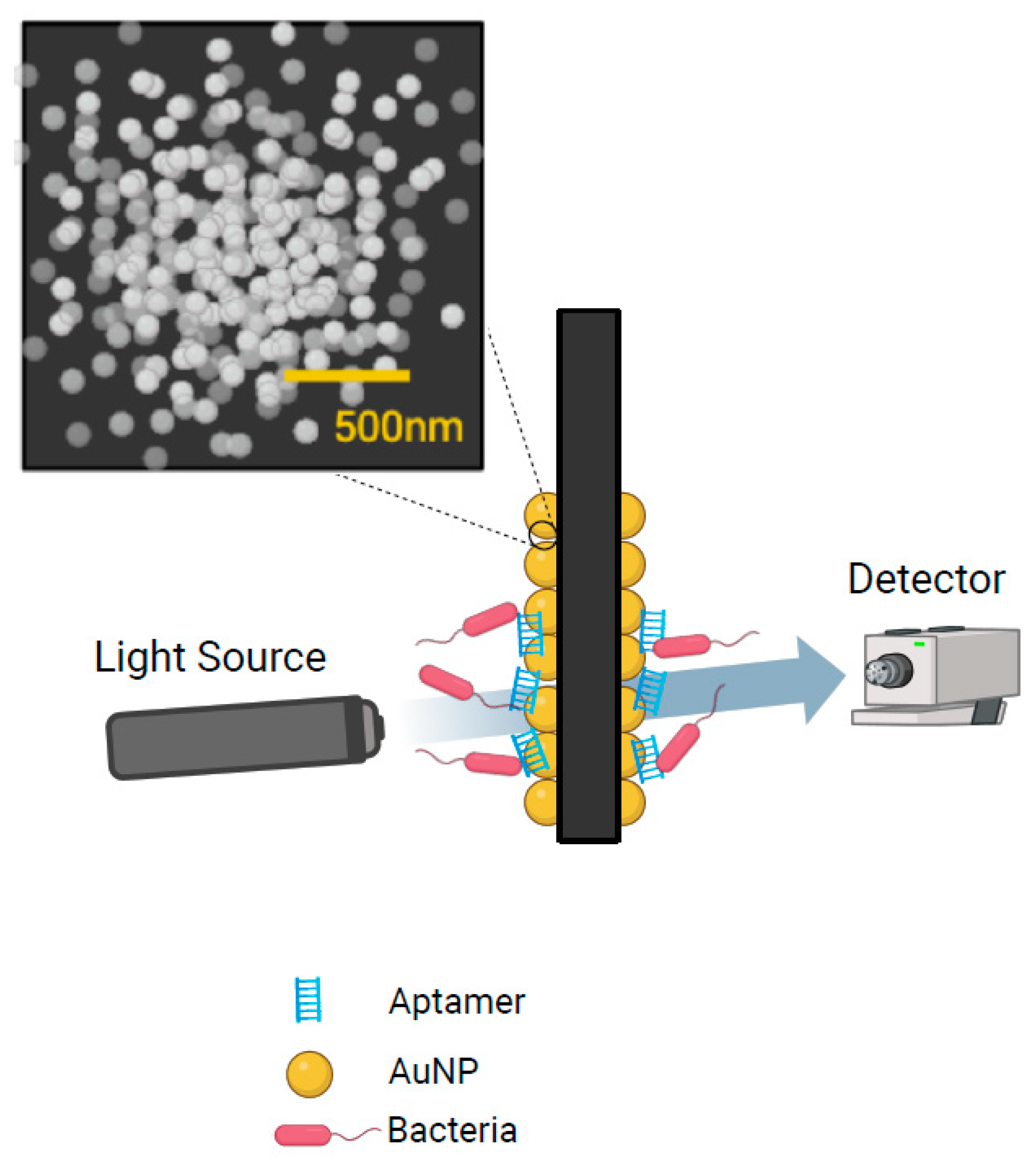
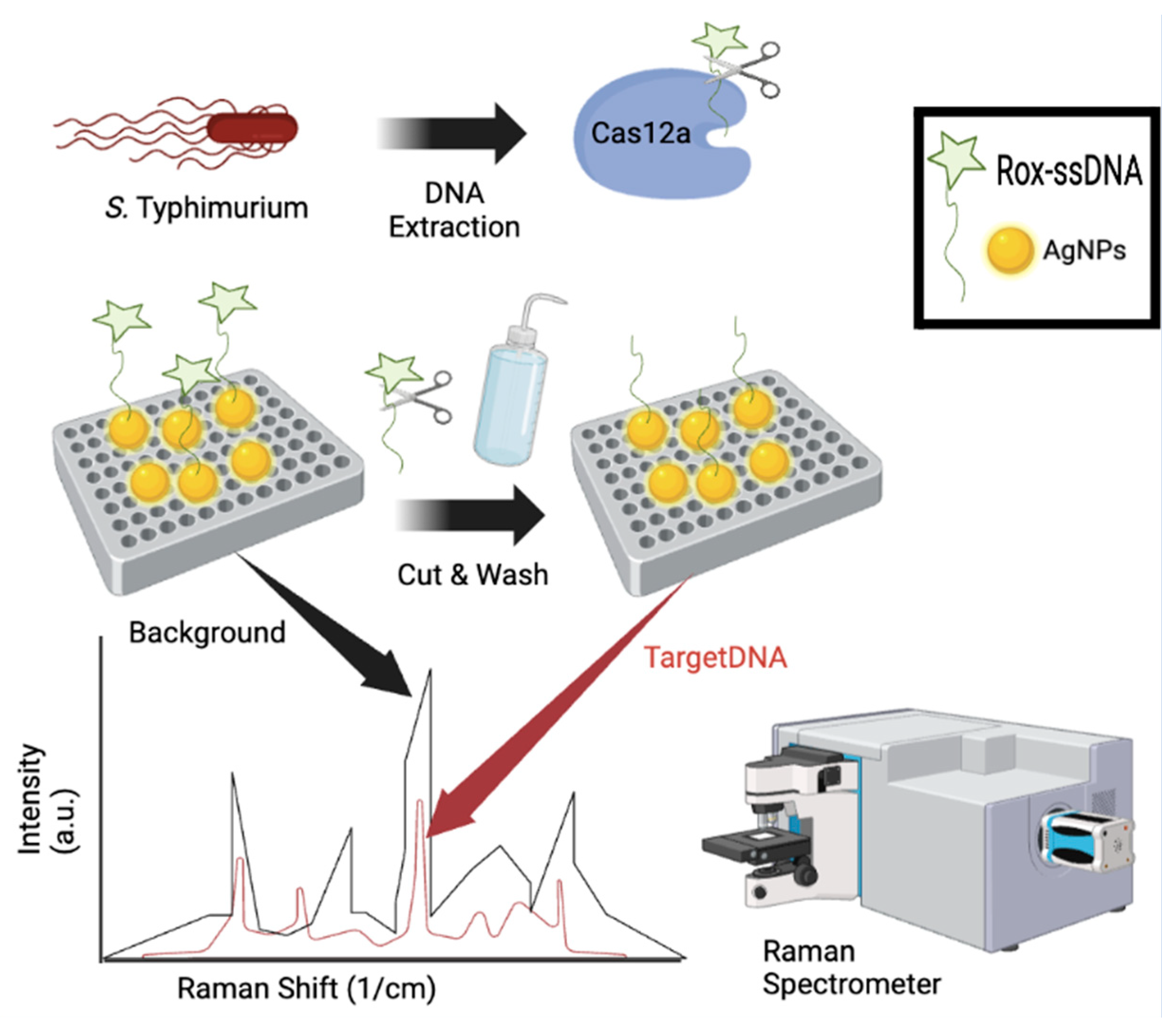
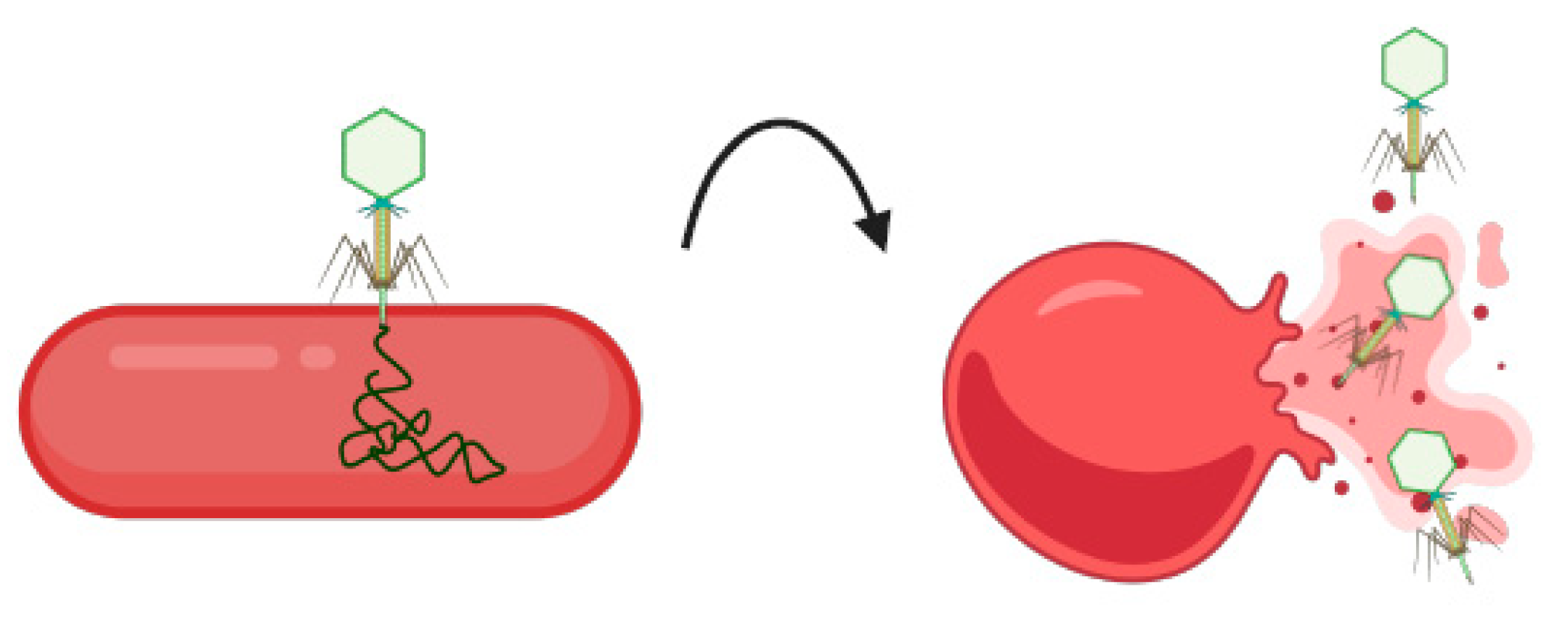


| Emerging Technologies: Electrochemical Aptasensors | ||||
|---|---|---|---|---|
| Sensor Material | Limit of Detection (CFU/mL) | Detection Time (Min) | Source | Author |
| Gold nanoparticle (AuNPs)/Graphite electrode (GE) | 1 | 40 | S. enterica in milk | [75] |
| Glassy Carbon Electrode (GCE)/Graphene oxide (GO) | 3 | 35 | Salmonella spp. in pork | [76] |
| Gold (Au)/AuNPs | 16 | 60 | S. Typhimurium in milk | [77] |
| Reduced Graphene oxide-titanium dioxide (rGO-TiO2) | 10 | 5 | S. Typhimurium in Chicken meat | [78] |
| Multi-walled carbon nanotubes (MWCNTs) | 55 (S. Enteritidis); 67 (S. Typhimurium) | 20 | S. Enteritidis + Typhimurium in chicken meat | [79] |
| Emerging Technologies: Surface Plasmon Resonance (SPR) | ||||
|---|---|---|---|---|
| Sensor | Limit of Detection | Detection Time | Source | Author |
| SPR Biosensor | 0.9 log CFU/g; 5.7 log CFU/g | 24 h enrichment; <2 min detection | S. Typhimurium in romaine lettuce | [80] |
| Magnetic Nanoparticles Enhanced SPR | 5.2 log CFU/g | 4 h (3.9 h to prep, <2 min detection) | S. Typhimurium in romaine lettuce | [81] |
| Gold nanoparticle aptamer-based localized SPR | 4 log CFU/mL | 35 min | S. Typhimurium in pork meat | [82] |
| Sandwich Antibody-AuNP SPR | 42 CFU/mL | <50 min | S. Typhimurium in milk | [83] |
| Emerging Technologies: Surface Enhanced Raman Spectroscopy (SERS) | ||||
|---|---|---|---|---|
| Sensor | Limit of Detection | Total time of Detection | Source | Author |
| SERS optofluidic sensor coupled with immunoassay | 10 CFU/200g | 2 h (w/15 min enrichment) | S. enterica + E. coli strains in lettuce and packed salad samples | [84] |
| SERS + short-chain adenine and fluorescein molecule | 4 log | 4 h | S. Enteritidis and S. Gaminara in ground beef | [85] |
| GO@Au/Ag-based SERS-LFA | 9 cells/mL | 20 min | S. Typhimurium, E. coli, S. aureus, L. monocytogenes in human urine + blood | [86] |
| RPA-Cas12a-μPAD SERS | 3–4 CFU/mL | 45 min | S. Typhimurium in meat and milk | [87] |
| CRISPR-SERS | 110 CFU/mL | 2 h | S. Typhimurium in minced poultry | [88] |
| Background-free SERS w/sandwich configuration (+ 100% photothermal inactivation of all bacteria) | 10 CFU/mL | 90 min | S. Typhimurium & S. aureus in human blood | [89] |
| Emerging Technologies: Bacteriophages [93] | |||
|---|---|---|---|
| Phage | Sensitivity for S. Enteritidis | Sensitivity for S. Typhimurium | Detection Time (Hours) |
| SPFM1 | 76.19% | 33.33% | 18 |
| SPFM2 | 76.19% | 40.00% | 18 |
| SPFM3 | 76.19% | 26.67% | 18 |
| SPFM4 | 66.67% | 33.33% | 18 |
| SPFM5 | 85.71% | 33.33% | 18 |
| SPFM6 | 76.19% | 33.33% | 18 |
| SPFM7 | 66.67% | 33.33% | 18 |
| SPFM8 | 80.95% | 33.33% | 18 |
| SPFM9 | 80.95% | 20.00% | 18 |
| SPFM10 | 14.29% | No effect | 18 |
| SPFM11 | 66.67% | 20.00% | 18 |
| SPFM12 | 14.29% | No effect | 18 |
| SPFM13 | 85.71% | 40.00% | 18 |
| SPFM14 | 61.90% | 26.67% | 18 |
| SPFM15 | 71.43% | 26.67% | 18 |
| SPFM16 | 80.95% | 26.67% | 18 |
| SPFM17 | 79.19% | 26.67% | 18 |
| SPFM19 | 14.29% | No effect | 18 |
| SPFM20 | 71.43% | 33.33% | 18 |
| SPFM21 | 71.43% | 26.67% | 18 |
| STW-77 | 80.95% | 66.67% | 18 |
| SEW-109 | 42.86% | 40.00% | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, A.; Wolfram, A.; Desin, T.S. Advancements in Detection Methods for Salmonella in Food: A Comprehensive Review. Pathogens 2024, 13, 1075. https://doi.org/10.3390/pathogens13121075
Patel A, Wolfram A, Desin TS. Advancements in Detection Methods for Salmonella in Food: A Comprehensive Review. Pathogens. 2024; 13(12):1075. https://doi.org/10.3390/pathogens13121075
Chicago/Turabian StylePatel, Aayushi, Andrew Wolfram, and Taseen S. Desin. 2024. "Advancements in Detection Methods for Salmonella in Food: A Comprehensive Review" Pathogens 13, no. 12: 1075. https://doi.org/10.3390/pathogens13121075
APA StylePatel, A., Wolfram, A., & Desin, T. S. (2024). Advancements in Detection Methods for Salmonella in Food: A Comprehensive Review. Pathogens, 13(12), 1075. https://doi.org/10.3390/pathogens13121075






