Comparative Study of T-Cell Repertoires after COVID-19 Immunization with Homologous or Heterologous Vaccine Booster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Total Antibody Detection against SARS-CoV-2
2.3. T-Cell Receptor Beta Sequencing and Bioinformatic Analysis
2.4. Statistical Methods
3. Results
3.1. Study Population and Reactogenicity
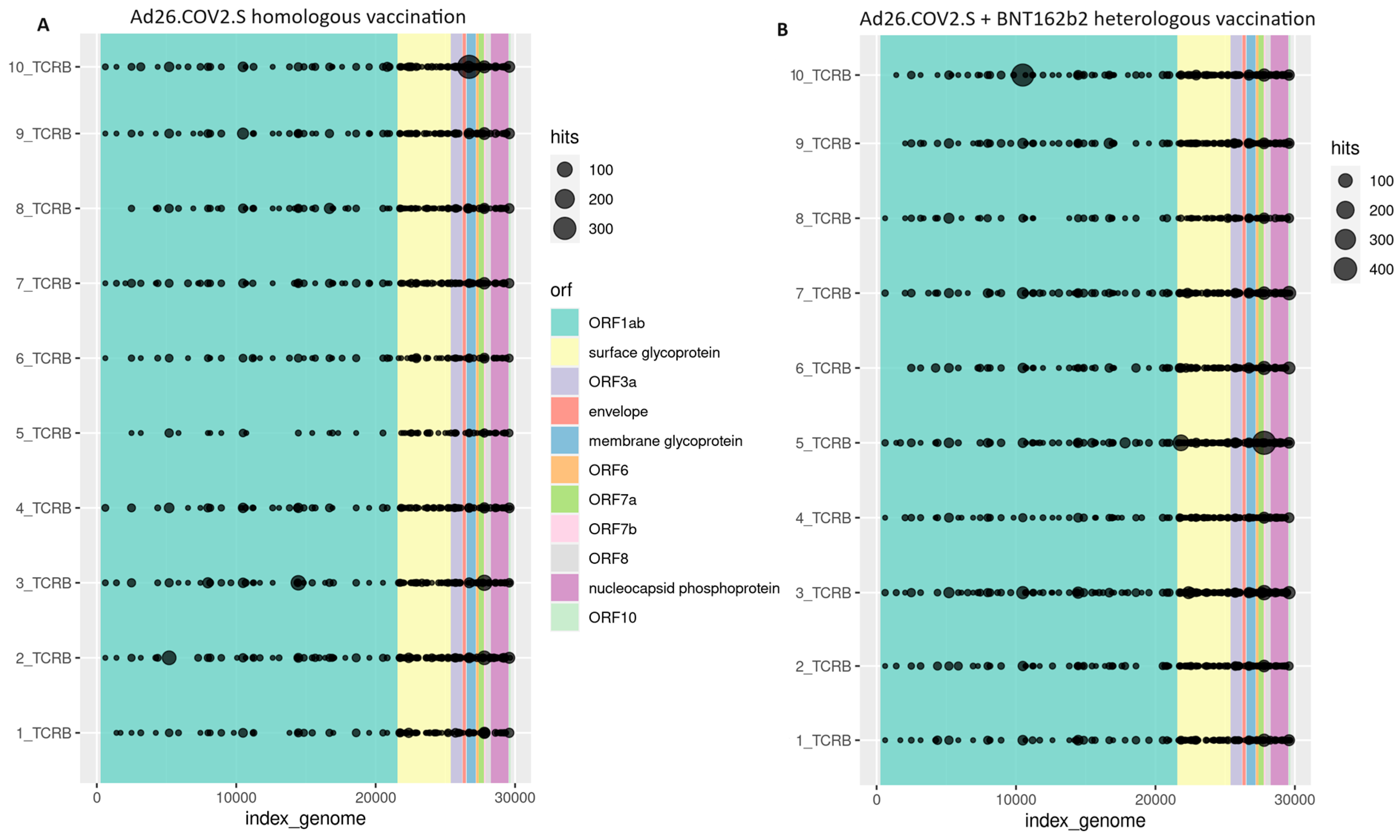
3.2. Cellular Immune Response
3.3. Humoral Immune Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tatsi, E.-B.; Filippatos, F.; Michos, A. SARS-CoV-2 Variants and Effectiveness of Vaccines: A Review of Current Evidence. Epidemiol. Infect. 2021, 149, e237. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Cari, L.; Naghavi Alhosseini, M.; Mencacci, A.; Migliorati, G.; Nocentini, G. Differences in the Expression Levels of SARS-CoV-2 Spike Protein in Cells Treated with MRNA-Based COVID-19 Vaccines: A Study on Vaccines from the Real World. Vaccines 2023, 11, 879. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef] [PubMed]
- Michos, A.; Tatsi, E.B.; Filippatos, F.; Dellis, C.; Koukou, D.; Efthymiou, V.; Kastrinelli, E.; Mantzou, A.; Syriopoulou, V. Association of Total and Neutralizing SARS-CoV-2 Spike -Receptor Binding Domain Antibodies with Epidemiological and Clinical Characteristics after Immunization with the 1st and 2nd Doses of the BNT162b2 Vaccine. Vaccine 2021, 39, 5963–5967. [Google Scholar] [CrossRef] [PubMed]
- Tatsi, E.B.; Filippatos, F.; Dellis, C.; Dourdouna, M.M.; Syriopoulou, V.; Michos, A. Kinetics of SARS-CoV-2 Spike Antibodies after the Second and Third Dose of the BNT162b2 COVID-19 Vaccine and Association with Epidemiological Characteristics and Breakthrough Infection in a Cohort Study of Healthcare Workers. Microorganisms 2023, 11, 2010. [Google Scholar] [CrossRef] [PubMed]
- Kanokudom, S.; Assawakosri, S.; Suntronwong, N.; Auphimai, C.; Nilyanimit, P.; Vichaiwattana, P.; Thongmee, T.; Yorsaeng, R.; Srimuan, D.; Thatsanatorn, T.; et al. Safety and Immunogenicity of the Third Booster Dose with Inactivated, Viral Vector, and MRNA COVID-19 Vaccines in Fully Immunized Healthy Adults with Inactivated Vaccine. Vaccines 2022, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Potesta, M.; Varchetta, S.; Fenoglio, D.; Iannetta, M.; Sarmati, L.; Mele, D.; Dentone, C.; Bassetti, M.; Montesano, C.; et al. Early and Polyantigenic CD4 T Cell Responses Correlate with Mild Disease in Acute COVID-19 Donors. Int. J. Mol. Sci. 2022, 23, 7155. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Rha, M.S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-Specific T Cell Memory Is Sustained in COVID-19 Convalescent Patients for 10 Months with Successful Development of Stem Cell-like Memory T Cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Dourdouna, M.M.; Tatsi, E.B.; Syriopoulou, V.; Michos, A. Evaluation of T Cell Responses with the QuantiFERON SARS-CoV-2 Assay in Individuals with 3 Doses of BNT162b2 Vaccine, SARS-CoV-2 Infection, or Hybrid Immunity. Diagn. Microbiol. Infect. Dis. 2023, 106, 115948. [Google Scholar] [CrossRef]
- Schwarz, M.; Mzoughi, S.; Lozano-Ojalvo, D.; Tan, A.T.; Bertoletti, A.; Guccione, E. T Cell Immunity Is Key to the Pandemic Endgame: How to Measure and Monitor It. Curr. Res. Immunol. 2022, 3, 215–221. [Google Scholar] [CrossRef]
- Sheridan, C. COVID-19 Testing Turns to T Cells. Nat. Biotechnol. 2021, 39, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Rossjohn, J.; Gras, S.; Miles, J.J.; Turner, S.J.; Godfrey, D.I.; McCluskey, J. T Cell Antigen Receptor Recognition of Antigen-Presenting Molecules. Annu. Rev. Immunol. 2015, 33, 169–200. [Google Scholar] [CrossRef] [PubMed]
- Robins, H.S.; Campregher, P.V.; Srivastava, S.K.; Wacher, A.; Turtle, C.J.; Kahsai, O.; Riddell, S.R.; Warren, E.H.; Carlson, C.S. Comprehensive Assessment of T-Cell Receptor Beta-Chain Diversity in Alphabeta T Cells. Blood 2009, 114, 4099–4107. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.; Bombardi, R.G.; Kozhevnikov, M.; Sinkovits, R.S.; Chen, E.C.; Branchizio, A.; Kose, N.; Day, S.B.; Pilkinton, M.; Gujral, M.; et al. High Frequency of Shared Clonotypes in Human T Cell Receptor Repertoires. Cell Rep. 2020, 32, 107882. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, W.S.; Emerson, R.O.; Lindau, P.; Vignali, M.; Snyder, T.M.; Desmarais, C.; Sanders, C.; Utsugi, H.; Warren, E.H.; McElrath, J.; et al. Dynamics of the Cytotoxic T Cell Response to a Model of Acute Viral Infection. J. Virol. 2015, 89, 4517–4526. [Google Scholar] [CrossRef] [PubMed]
- Herati, R.S.; Muselman, A.; Vella, L.; Bengsch, B.; Parkhouse, K.; Del Alcazar, D.; Kotzin, J.; Doyle, S.A.; Tebas, P.; Hensley, S.E.; et al. Successive Annual Influenza Vaccination Induces a Recurrent Oligoclonotypic Memory Response in Circulating T Follicular Helper Cells. Sci. Immunol. 2017, 2, eaag2152. [Google Scholar] [CrossRef] [PubMed]
- Alter, G.; Yu, J.; Liu, J.; Chandrashekar, A.; Borducchi, E.N.; Tostanoski, L.H.; McMahan, K.; Jacob-Dolan, C.; Martinez, D.R.; Chang, A.; et al. Immunogenicity of Ad26.COV2.S Vaccine against SARS-CoV-2 Variants in Humans. Nature 2021, 596, 268–272. [Google Scholar] [CrossRef]
- Dykema, A.G.; Zhang, B.; Woldemeskel, B.A.; Garliss, C.C.; Rashid, R.; Westlake, T.; Zhang, L.; Zhang, J.; Cheung, L.S.; Caushi, J.X.; et al. SARS-CoV-2 Vaccination Diversifies the CD4+ Spike-Reactive T Cell Repertoire in Patients with Prior SARS-CoV-2 Infection. eBioMedicine 2022, 80, 104048. [Google Scholar] [CrossRef]
- Yin, Q.; Luo, W.; Mallajosyula, V.; Bo, Y.; Guo, J.; Xie, J.; Sun, M.; Verma, R.; Li, C.; Constantz, C.M.; et al. A TLR7-Nanoparticle Adjuvant Promotes a Broad Immune Response against Heterologous Strains of Influenza and SARS-CoV-2. Nat. Mater. 2023, 22, 380–390. [Google Scholar] [CrossRef]
- Carlson, C.S.; Emerson, R.O.; Sherwood, A.M.; Desmarais, C.; Chung, M.W.; Parsons, J.M.; Steen, M.S.; LaMadrid-Herrmannsfeldt, M.A.; Williamson, D.W.; Livingston, R.J.; et al. Using Synthetic Templates to Design an Unbiased Multiplex PCR Assay. Nat. Commun. 2013, 4, 2680. [Google Scholar] [CrossRef] [PubMed]
- Robins, H.; Desmarais, C.; Matthis, J.; Livingston, R.; Andriesen, J.; Reijonen, H.; Carlson, C.; Nepom, G.; Yee, C.; Cerosaletti, K. Ultra-Sensitive Detection of Rare T Cell Clones. J. Immunol. Methods 2012, 375, 14–19. [Google Scholar] [CrossRef]
- Chiffelle, J.; Genolet, R.; Perez, M.A.; Coukos, G.; Zoete, V.; Harari, A. T-Cell Repertoire Analysis and Metrics of Diversity and Clonality. Curr. Opin. Biotechnol. 2020, 65, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.H.; Lee, J.H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [PubMed]
- Reuben, A.; Zhang, J.; Chiou, S.H.; Gittelman, R.M.; Li, J.; Lee, W.C.; Fujimoto, J.; Behrens, C.; Liu, X.; Wang, F.; et al. Comprehensive T Cell Repertoire Characterization of Non-Small Cell Lung Cancer. Nat. Commun. 2020, 11, 603. [Google Scholar] [CrossRef] [PubMed]
- Nolan, S.; Vignali, M.; Klinger, M.; Dines, J.N.; Kaplan, I.M.; Svejnoha, E.; Craft, T.; Boland, K.; Pesesky, M.; Gittelman, R.M.; et al. A Large-Scale Database of T-Cell Receptor Beta (TCRβ) Sequences and Binding Associations from Natural and Synthetic Exposure to SARS-CoV-2. Res. Sq. 2020, preprint. [Google Scholar] [CrossRef]
- Dines, J.N.; Manley, T.J.; Svejnoha, E.; Simmons, H.M.; Taniguchi, R.; Klinger, M.; Baldo, L.; Robins, H. The ImmuneRACE Study: A Prospective Multicohort Study of Immune Response Action to COVID-19 Events with the ImmuneCODETM Open Access Database. medRxiv 2020, preprint. [Google Scholar]
- Warren, R.L.; Freeman, J.D.; Zeng, T.; Choe, G.; Munro, S.; Moore, R.; Webb, J.R.; Holt, R.A. Exhaustive T-Cell Repertoire Sequencing of Human Peripheral Blood Samples Reveals Signatures of Antigen Selection and a Directly Measured Repertoire Size of at Least 1 Million Clonotypes. Genome Res. 2011, 21, 790–797. [Google Scholar] [CrossRef]
- Shen, Y.; Voigt, A.; Leng, X.; Rodriguez, A.A.; Nguyen, C.Q. A Current and Future Perspective on T Cell Receptor Repertoire Profiling. Front. Genet. 2023, 14, 1159109. [Google Scholar] [CrossRef]
- Freeman, J.D.; Warren, R.L.; Webb, J.R.; Nelson, B.H.; Holt, R.A. Profiling the T-Cell Receptor Beta-Chain Repertoire by Massively Parallel Sequencing. Genome Res. 2009, 19, 1817–1824. [Google Scholar] [CrossRef]
- Sacco, K.; Castagnoli, R.; Vakkilainen, S.; Liu, C.; Delmonte, O.M.; Oguz, C.; Kaplan, I.M.; Alehashemi, S.; Burbelo, P.D.; Bhuyan, F.; et al. Immunopathological Signatures in Multisystem Inflammatory Syndrome in Children and Pediatric COVID-19. Nat. Med. 2022, 28, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Mazouz, S.; Boisvert, M.; Abdel-Hakeem, M.S.; Khedr, O.; Bruneau, J.; Shoukry, N.H. Expansion of Unique Hepatitis C Virus–Specific Public CD8+ T Cell Clonotypes during Acute Infection and Reinfection. J. Immunol. 2021, 207, 1180–1193. [Google Scholar] [CrossRef] [PubMed]
- Musvosvi, M.; Huang, H.; Wang, C.; Xia, Q.; Rozot, V.; Krishnan, A.; Acs, P.; Cheruku, A.; Obermoser, G.; Leslie, A.; et al. T Cell Receptor Repertoires Associated with Control and Disease Progression Following Mycobacterium Tuberculosis Infection. Nat. Med. 2023, 29, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Bunse, L.; Wick, A.; Bunse, T.; Le Cornet, L.; Harting, I.; Sahm, F.; Sanghvi, K.; Tan, C.L.; Poschke, I.; et al. A Vaccine Targeting Mutant IDH1 in Newly Diagnosed Glioma. Nature 2021, 592, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Lyke, K.E.; Deming, M.E.; Jackson, L.A.; Branche, A.R.; El Sahly, H.M.; Rostad, C.A.; Martin, J.M.; Johnston, C.; Rupp, R.E.; et al. Homologous and Heterologous Covid-19 Booster Vaccinations. N. Engl. J. Med. 2022, 386, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Soni, M.; Migliori, E.; Fu, J.; Assal, A.; Chan, H.T.; Pan, J.; Khatiwada, P.; Ciubotariu, R.; May, M.S.; Pereira, M.; et al. The Prospect of Universal Coronavirus Immunity: A Characterization of Reciprocal and Non-Reciprocal T Cell Responses against SARS-CoV2 and Common Human Coronaviruses. Biorxiv Prepr. Serv. Biol. 2023, preprint. [Google Scholar] [CrossRef] [PubMed]
- Dykema, A.G.; Zhang, B.; Woldemeskel, B.A.; Garliss, C.C.; Cheung, L.S.; Choudhury, D.; Zhang, J.; Aparicio, L.; Bom, S.; Rashid, R.; et al. Functional Characterization of CD4+ T Cell Receptors Crossreactive for SARS-CoV-2 and Endemic Coronaviruses. J. Clin. Investig. 2021, 131, e146922. [Google Scholar] [CrossRef]
- Corpas, M.; de Mendoza, C.; Moreno-Torres, V.; Pintos, I.; Seoane, P.; Perkins, J.R.; Ranea, J.A.G.; Fatumo, S.; Korcsmaros, T.; Martín-Villa, J.M.; et al. Genetic Signature Detected in T Cell Receptors from Patients with Severe COVID-19. iScience 2023, 26, 107735. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Elyanow, R.; Snyder, T.M.; Dalai, S.C.; Gittelman, R.M.; Boonyaratanakornkit, J.; Wald, A.; Selke, S.; Wener, M.H.; Morishima, C.; Greninger, A.L.; et al. T Cell Receptor Sequencing Identifies Prior SARS-CoV-2 Infection and Correlates with Neutralizing Antibodies and Disease Severity. JCI Insight 2022, 7, e150070. [Google Scholar] [CrossRef]


| Booster | Samples | Total Rearrangements (Productive and Non-Productive) | Total Productive Rearrangements (TPRs) | COVID-19 TPRs | Spike COVID-19 TPRs | Non-Spike COVID-19 TPRs | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | n | % | n | % | n | % | n | % | ||
| Homologous (Ad26.COV2.S) | 1 | 40,539 | 32,584 | 80.4 | 280 | 0.86 | 64 | 22.9 | 216 | 77.1 |
| 2 | 67,002 | 54,912 | 81.9 | 491 | 0.89 | 94 | 19.1 | 397 | 80.9 | |
| 3 | 39,855 | 31,820 | 79.8 | 273 | 0.86 | 50 | 18.3 | 223 | 81.7 | |
| 4 | 49,515 | 39,218 | 79.2 | 320 | 0.82 | 55 | 17.2 | 265 | 82.8 | |
| 5 | 17,275 | 14,297 | 82.8 | 120 | 0.84 | 23 | 19.2 | 97 | 80.8 | |
| 6 | 29,311 | 24,534 | 83.7 | 192 | 0.78 | 30 | 15.6 | 162 | 84.4 | |
| 7 | 45,995 | 37,872 | 82.3 | 310 | 0.82 | 56 | 18.1 | 254 | 81.9 | |
| 8 | 36,642 | 30,725 | 83.9 | 267 | 0.87 | 44 | 16.5 | 223 | 83.5 | |
| 9 | 46,544 | 37,636 | 80.9 | 354 | 0.94 | 51 | 14.4 | 303 | 85.6 | |
| 10 | 54,074 | 45,556 | 84.3 | 376 | 0.83 | 62 | 16.5 | 314 | 83.5 | |
| Mean (±SD) | 42,675.2 (±13,610.4) | 34,915.4 (±11,121.6) | 81.9 (±1.8) | 298.3 (±101.1) | 0.85 (±0.1) | 52.9 (±19.5) | 17.8 (±2.3) | 245.4 (±83.2) | 82.2 (±2.3) | |
| Heterologous (BNT162b2) | 1 | 76,222 | 60,980 | 80.0 | 478 | 0.78 | 83 | 17.36 | 395 | 82.6 |
| 2 | 72,380 | 57,184 | 79.0 | 430 | 0.75 | 68 | 15.81 | 362 | 84.2 | |
| 3 | 118,297 | 95,538 | 80.8 | 903 | 0.95 | 163 | 18.05 | 740 | 82.0 | |
| 4 | 55,435 | 45,147 | 81.4 | 385 | 0.85 | 68 | 17.66 | 317 | 82.3 | |
| 5 | 104,044 | 86,987 | 83.6 | 668 | 0.77 | 112 | 16.77 | 556 | 83.2 | |
| 6 | 70,600 | 59,007 | 83.6 | 430 | 0.73 | 76 | 17.67 | 354 | 82.3 | |
| 7 | 71,065 | 60,089 | 84.6 | 591 | 0.98 | 90 | 15.23 | 501 | 84.8 | |
| 8 | 40,028 | 32,497 | 81.2 | 249 | 0.77 | 36 | 14.46 | 213 | 85.5 | |
| 9 | 74,774 | 60,613 | 81.1 | 529 | 0.87 | 83 | 15.69 | 446 | 84.3 | |
| 10 | 92,306 | 73,476 | 79.6 | 562 | 0.76 | 123 | 21.89 | 439 | 78.1 | |
| Mean (±SD) | 77,515.1 (±22,616.4) | 63,151.8 (±18,441.5) | 81.5 (±1.9) | 522.5 (±178.0) | 0.82 (±0.1) | 90.2 (±35.1) | 17.06 (±2.1) | 432.3 (±144.8) | 82.9 (±2.1) | |
| p-value | 0.001 | 0.001 | 0.600 | 0.003 | 0.351 | 0.009 | 0.479 | 0.003 | 0.480 | |
| Vaccine Booster | Tabs-S (U/mL) |
|---|---|
| Homologous (Ad26.COV2.S) n = 10 | 1781.0 (1314.0) |
| Heterologous (BNT162b2) n = 10 | 10,115.0 (6993.0) |
| p-value | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatsi, E.-B.; Filippatos, F.; Bello, T.; Syriopoulou, V.; Michos, A. Comparative Study of T-Cell Repertoires after COVID-19 Immunization with Homologous or Heterologous Vaccine Booster. Pathogens 2024, 13, 284. https://doi.org/10.3390/pathogens13040284
Tatsi E-B, Filippatos F, Bello T, Syriopoulou V, Michos A. Comparative Study of T-Cell Repertoires after COVID-19 Immunization with Homologous or Heterologous Vaccine Booster. Pathogens. 2024; 13(4):284. https://doi.org/10.3390/pathogens13040284
Chicago/Turabian StyleTatsi, Elizabeth-Barbara, Filippos Filippatos, Thomas Bello, Vasiliki Syriopoulou, and Athanasios Michos. 2024. "Comparative Study of T-Cell Repertoires after COVID-19 Immunization with Homologous or Heterologous Vaccine Booster" Pathogens 13, no. 4: 284. https://doi.org/10.3390/pathogens13040284








