Valp1, a Newly Identified Temperate Phage Facilitating Coexistence of Lysogenic and Non-Lysogenic Populations of Vibrio anguillarum
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Phage Isolation and Propagation
2.3. Phage Characterization
2.4. Isolation and Characterization of Lysogenic Strain P1.1
2.5. Co-Culture Experiments
2.6. Analysis and Statistics
3. Results
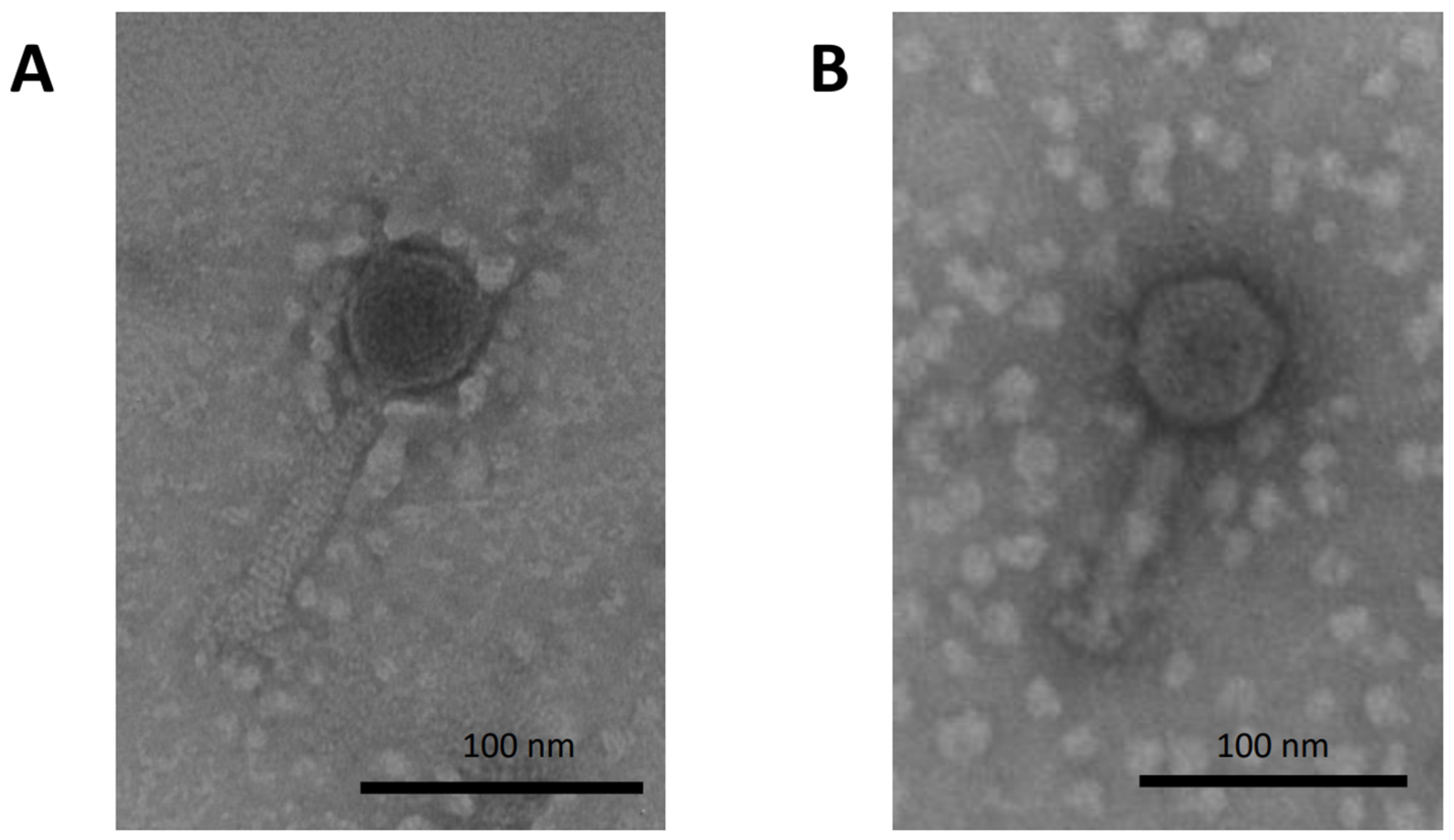
3.1. Phage Characterization and Isolation
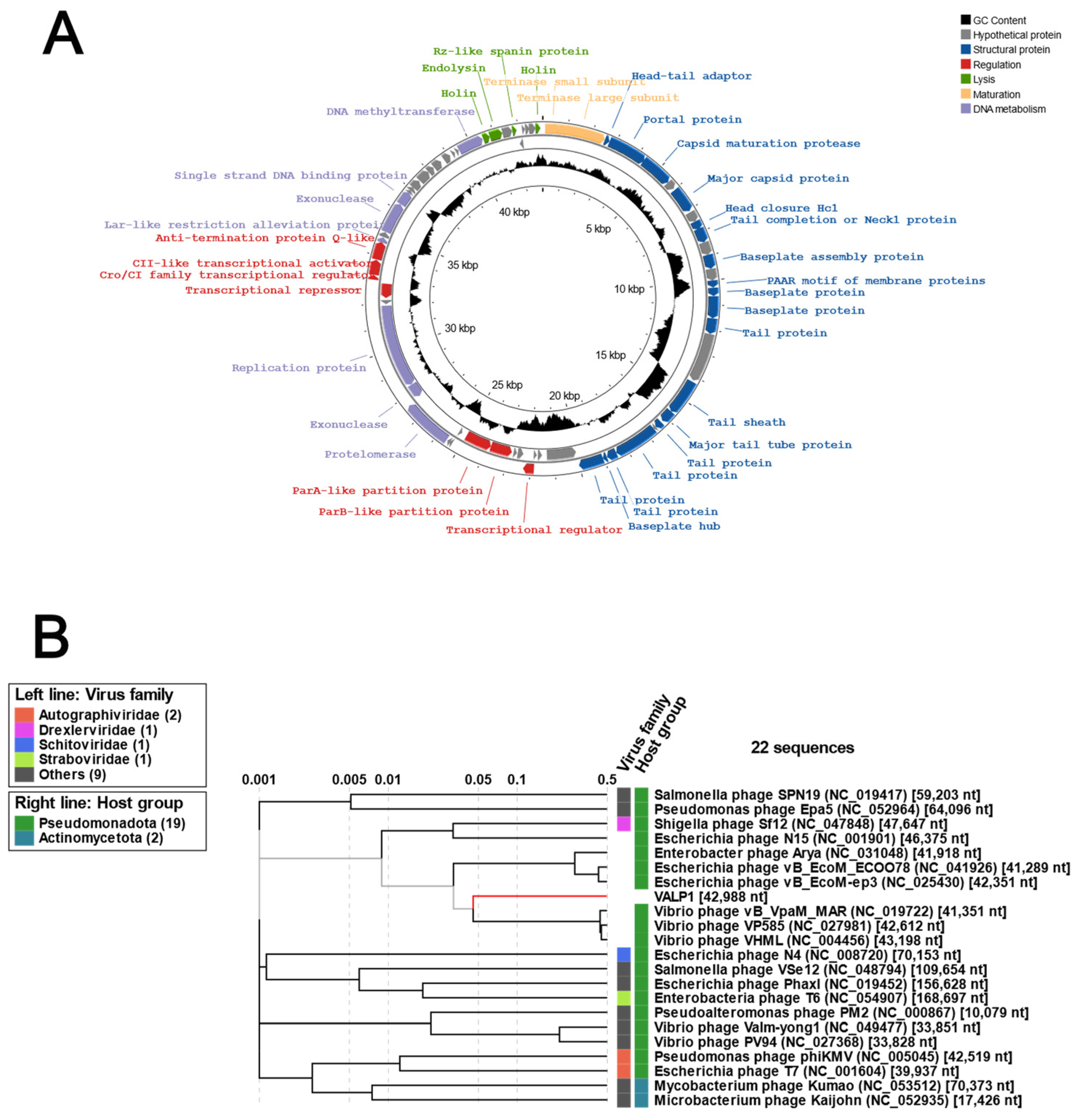
3.2. Genomic Characterization of Valp1 Phage
3.3. Infection of V. anguillarum with Valp1 Phage
3.4. Characterization of the V. anguillarum Lysogenic Strain P1.1
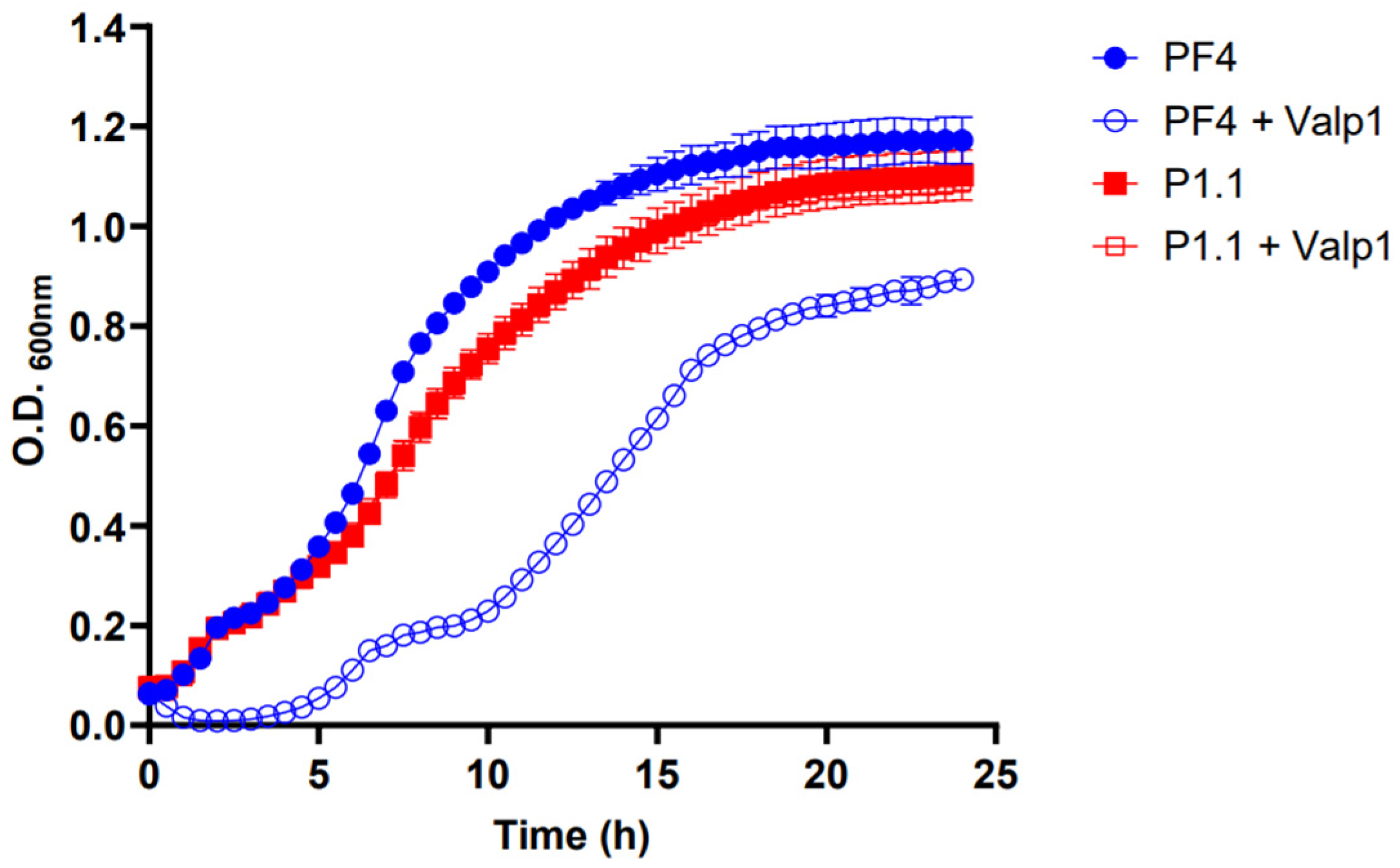
3.5. Coexistence between the V. anguillarum Strains PF4 and P1.1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grimes, D.J. The Vibrios: Scavengers, Symbionts, and Pathogens from the Sea. Microb. Ecol. 2020, 80, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.; Colwell, R.R. Vibrio Ecology, Pathogenesis, and Evolution. Front. Microbiol. 2014, 5, 256. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio Spp. Infections. Nat. Rev. Dis. Primers 2018, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.; Amal, M.N.A.; Yasin, I.S.M.; Zamri Saad, M.; Nasruddin, N.S.; Al-saari, N.; Mino, S.; Sawabe, T. Vibriosis in Cultured Marine Fishes: A Review. Aquaculture 2019, 512, 734289. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a Fish Pathogen: Virulence Factors, Diagnosis and Prevention: Pathogen Profile of Vibrio anguillarum. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.H.; Blackburn, N.; Larsen, J.L.; Olsen, J.E. Influences of Temperature, Salinity and Starvation on the Motility and Chemotactic Response of Vibrio anguillarum. Microbiology 2004, 150, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Higuera, G.; Bastías, R.; Tsertsvadze, G.; Romero, J.; Espejo, R.T. Recently Discovered Vibrio anguillarum Phages Can Protect against Experimentally Induced Vibriosis in Atlantic Salmon, Salmo salar. Aquaculture 2013, 392–395, 128–133. [Google Scholar] [CrossRef]
- Tan, D.; Gram, L.; Middelboe, M. Vibriophages and Their Interactions with the Fish Pathogen Vibrio anguillarum. Appl. Environ. Microbiol. 2014, 80, 3128–3140. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage Puppet Masters of the Marine Microbial Realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef]
- Brüssow, H.; Canchaya, C.; Hardt, W.-D. Phages and the Evolution of Bacterial Pathogens: From Genomic Rearrangements to Lysogenic Conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef]
- Fortier, L.-C.; Sekulovic, O. Importance of Prophages to Evolution and Virulence of Bacterial Pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Faruque, S.M.; Mekalanos, J.J. Phage-Bacterial Interactions in the Evolution of Toxigenic Vibrio cholerae. Virulence 2012, 3, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.; Oakey, J.; Bromage, E.; Owens, L. Experimental Bacteriophage-Mediated Virulence in Strains of Vibrio harveyi. Dis. Aquat. Org. 2003, 54, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Khemayan, K.; Prachumwat, A.; Sonthayanon, B.; Intaraprasong, A.; Sriurairatana, S.; Flegel, T.W. Complete Genome Sequence of Virulence-Enhancing Siphophage VHS1 from Vibrio harveyi. Appl. Environ. Microbiol. 2012, 78, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- Argov, T.; Azulay, G.; Pasechnek, A.; Stadnyuk, O.; Ran-Sapir, S.; Borovok, I.; Sigal, N.; Herskovits, A.A. Temperate Bacteriophages as Regulators of Host Behavior. Curr. Opin. Microbiol. 2017, 38, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R. Bacteriophage Behavioral Ecology: How Phages Alter Their Bacterial Host’s Habits. Bacteriophage 2014, 4, e29866. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Andersen, N.; Kalatzis, P.G.; Middelboe, M. Large Phenotypic and Genetic Diversity of Prophages Induced from the Fish Pathogen Vibrio anguillarum. Viruses 2019, 11, 983. [Google Scholar] [CrossRef] [PubMed]
- Kalatzis, P.; Rørbo, N.; Castillo, D.; Mauritzen, J.; Jørgensen, J.; Kokkari, C.; Zhang, F.; Katharios, P.; Middelboe, M. Stumbling across the Same Phage: Comparative Genomics of Widespread Temperate Phages Infecting the Fish Pathogen Vibrio anguillarum. Viruses 2017, 9, 122. [Google Scholar] [CrossRef]
- Caruffo, M.; Navarrete, N.; Salgado, O.; Díaz, A.; López, P.; García, K.; Feijóo, C.G.; Navarrete, P. Potential Probiotic Yeasts Isolated from the Fish Gut Protect Zebrafish (Danio rerio) from a Vibrio anguillarum Challenge. Front. Microbiol. 2015, 6, 1093. [Google Scholar] [CrossRef]
- Castillo, D.; Alvise, P.D.; Xu, R.; Zhang, F.; Middelboe, M.; Gram, L. Comparative Genome Analyses of Vibrio anguillarum Strains Reveal a Link with Pathogenicity Traits. mSystems 2017, 2, e00001-17. [Google Scholar] [CrossRef]
- Silva-Rubio, A.; Avendaño-Herrera, R.; Jaureguiberry, B.; Toranzo, A.E.; Magariños, B. First Description of Serotype O3 in Vibrio anguillarum Strains Isolated from Salmonids in Chile. J. Fish Dis. 2008, 31, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 69–76. ISBN 978-1-60327-164-6. [Google Scholar]
- Kutter, E. Phage Host Range and Efficiency of Plating. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 141–149. ISBN 978-1-60327-164-6. [Google Scholar]
- Kalatzis, P.G.; Bastías, R.; Kokkari, C.; Katharios, P. Isolation and Characterization of Two Lytic Bacteriophages, φSt2 and φGrn1; Phage Therapy Application for Biological Control of Vibrio alginolyticus in Aquaculture Live Feeds. PLoS ONE 2016, 11, e0151101. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.; Rasche, H.; Maughmer, C.; Criscione, A.; Mijalis, E.; Liu, M.; Hu, J.C.; Young, R.; Gill, J.J. Galaxy and Apollo as a Biologist-Friendly Interface for High-Quality Cooperative Phage Genome Annotation. PLoS Comput. Biol. 2020, 16, e1008214. [Google Scholar] [CrossRef] [PubMed]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A Fast Scalable Bacteriophage Annotation Tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Shang, J.; Peng, C.; Liao, H.; Tang, X.; Sun, Y. PhaBOX: A Web Server for Identifying and Characterizing Phage Contigs in Metagenomic Data. Bioinform. Adv. 2023, 3, vbad101. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhao, H.; Xian, Y.; Hussain, M.A.; Wu, X. Multiplex PCR Assays for the Detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an Internal Amplification Control. Diagn. Microbiol. Infect. Dis. 2014, 79, 115–118. [Google Scholar] [CrossRef]
- León, M.; Kokkari, C.; García, K.; Castillo, D.; Katharios, P.; Bastías, R. Diversification of Vibrio anguillarum Driven by the Bacteriophage CHOED. Front. Microbiol. 2019, 10, 1396. [Google Scholar] [CrossRef]
- Oyarbide, U.; Iturria, I.; Rainieri, S.; Pardo, M.A. Use of Gnotobiotic Zebrafish to Study Vibrio anguillarum Pathogenicity. Zebrafish 2015, 12, 71–80. [Google Scholar] [CrossRef]
- Frans, I.; Dierckens, K.; Crauwels, S.; Van Assche, A.; Leisner, J.; Larsen, M.H.; Michiels, C.W.; Willems, K.A.; Lievens, B.; Bossier, P.; et al. Does Virulence Assessment of Vibrio anguillarum Using Sea Bass (Dicentrarchus labrax) Larvae Correspond with Genotypic and Phenotypic Characterization? PLoS ONE 2013, 8, e70477. [Google Scholar] [CrossRef] [PubMed]
- Vidgen, M.; Carson, J.; Higgins, M.; Owens, L. Changes to the Phenotypic Profile of Vibrio harveyi When Infected with the Vibrio harveyi Myovirus-like (VHML) Bacteriophage. J. Appl. Microbiol. 2006, 100, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Zabala, B.; Hammerl, J.A.; Espejo, R.T.; Hertwig, S. The Linear Plasmid Prophage Vp58.5 of Vibrio parahaemolyticus Is Closely Related to the Integrating Phage VHML and Constitutes a New Incompatibility Group of Telomere Phages. J. Virol. 2009, 83, 9313–9320. [Google Scholar] [CrossRef] [PubMed]
- Oakey, H.J.; Cullen, B.R.; Owens, L. The Complete Nucleotide Sequence of the Vibrio harveyi Bacteriophage VHML. J. Appl. Microbiol. 2002, 93, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V. N15: The Linear Phage–Plasmid. Plasmid 2011, 65, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Mobberley, J.M.; Authement, R.N.; Segall, A.M.; Paul, J.H. The Temperate Marine Phage ΦHAP-1 of Halomonas aquamarina Possesses a Linear Plasmid-Like Prophage Genome. J. Virol. 2008, 82, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhong, L.; Shen, M.; Fang, P.; Qin, Z. Characterization of Streptomyces Plasmid-Phage pFP4 and Its Evolutionary Implications. Plasmid 2012, 68, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Berryhill, B.A.; Garcia, R.; McCall, I.C.; Manuel, J.A.; Chaudhry, W.; Petit, M.-A.; Levin, B.R. The Book of Lambda Does Not Tell Us That Naturally Occurring Lysogens of Escherichia coli Are Likely to Be Resistant as Well as Immune. Proc. Natl. Acad. Sci. USA 2023, 120, e2212121120. [Google Scholar] [CrossRef]
- Stewart, F.M.; Levin, B.R. The Population Biology of Bacterial Viruses: Why Be Temperate. Theor. Popul. Biol. 1984, 26, 93–117. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arce, M.; Venegas, G.; Paez, K.; Latz, S.; Navarrete, P.; Caruffo, M.; Feijoo, C.; García, K.; Bastías, R. Valp1, a Newly Identified Temperate Phage Facilitating Coexistence of Lysogenic and Non-Lysogenic Populations of Vibrio anguillarum. Pathogens 2024, 13, 285. https://doi.org/10.3390/pathogens13040285
Arce M, Venegas G, Paez K, Latz S, Navarrete P, Caruffo M, Feijoo C, García K, Bastías R. Valp1, a Newly Identified Temperate Phage Facilitating Coexistence of Lysogenic and Non-Lysogenic Populations of Vibrio anguillarum. Pathogens. 2024; 13(4):285. https://doi.org/10.3390/pathogens13040285
Chicago/Turabian StyleArce, Manuel, Guillermo Venegas, Karla Paez, Simone Latz, Paola Navarrete, Mario Caruffo, Carmen Feijoo, Katherine García, and Roberto Bastías. 2024. "Valp1, a Newly Identified Temperate Phage Facilitating Coexistence of Lysogenic and Non-Lysogenic Populations of Vibrio anguillarum" Pathogens 13, no. 4: 285. https://doi.org/10.3390/pathogens13040285
APA StyleArce, M., Venegas, G., Paez, K., Latz, S., Navarrete, P., Caruffo, M., Feijoo, C., García, K., & Bastías, R. (2024). Valp1, a Newly Identified Temperate Phage Facilitating Coexistence of Lysogenic and Non-Lysogenic Populations of Vibrio anguillarum. Pathogens, 13(4), 285. https://doi.org/10.3390/pathogens13040285








