Functional Analysis of Type III Effectors in Xanthomonas campestris pv. campestris Reveals Distinct Roles in Modulating Arabidopsis Innate Immunity
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Vector Constructions
2.3. Arabidopsis Protoplast Transient Expression
2.4. Gene Expression Analyses
2.5. Virulence Assays
2.6. Subcellular Localization
3. Results
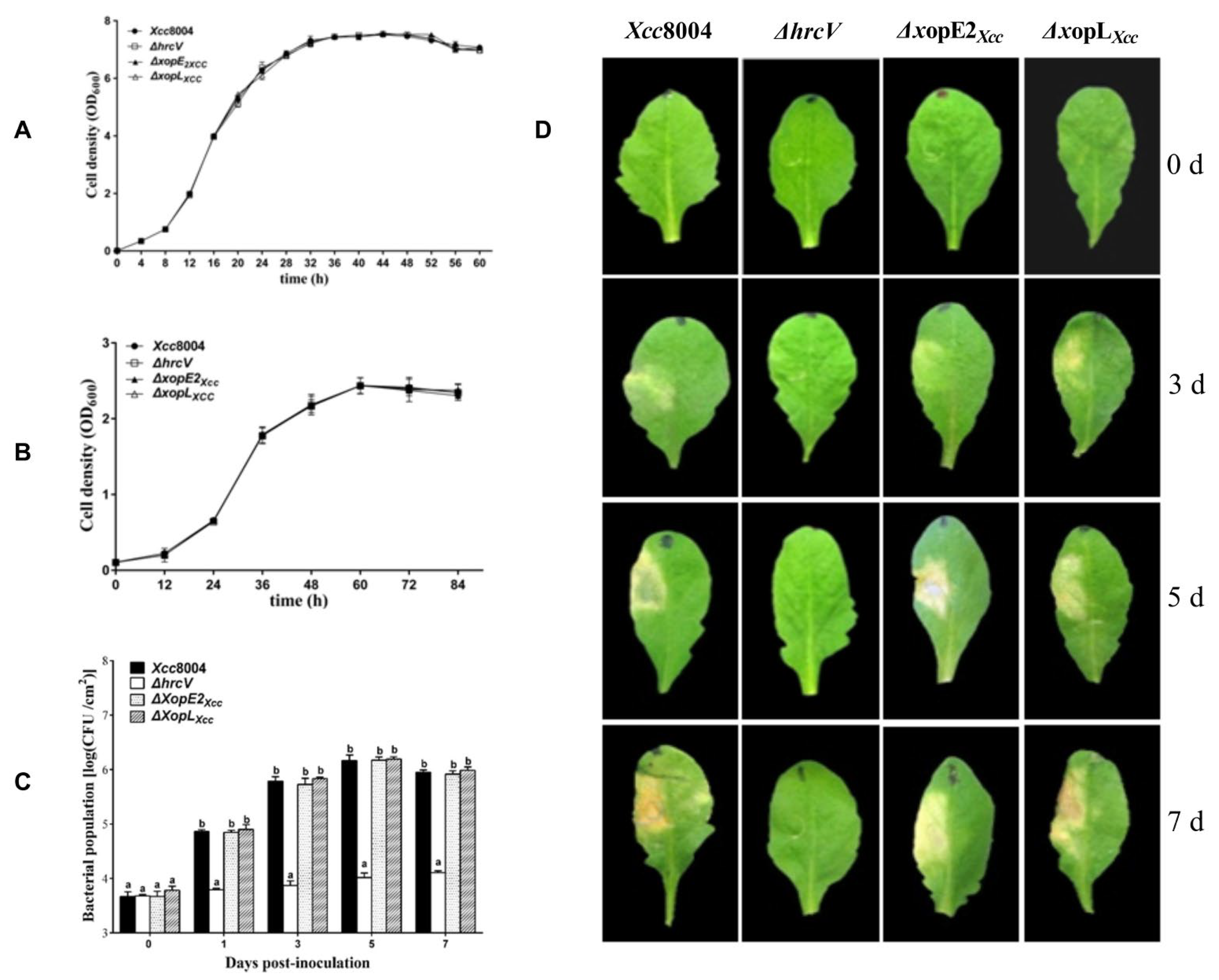
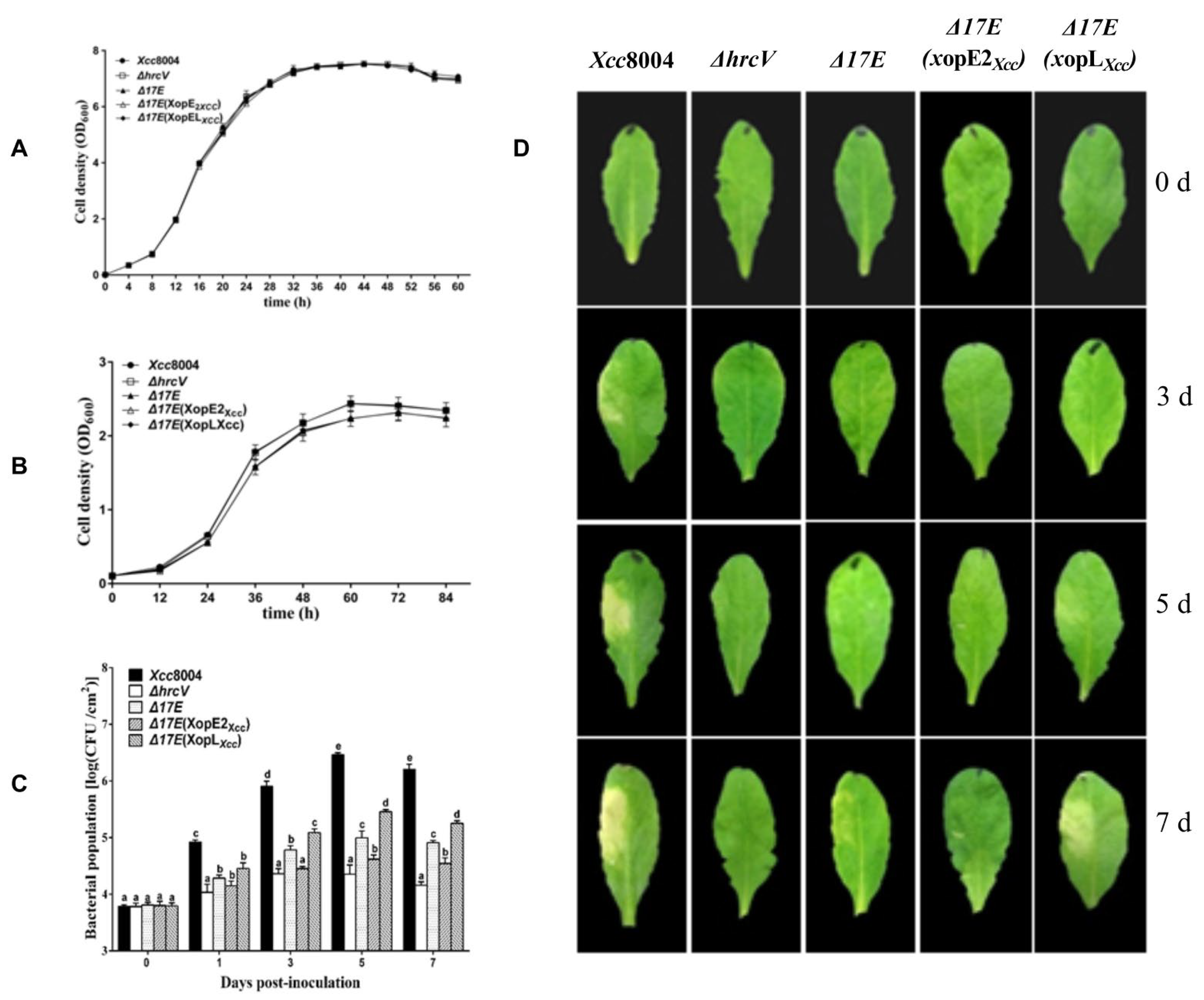
3.1. Distinct Contributions of XopE2Xcc and XopLXcc to Xcc 8004 Virulence on Arabidopsis
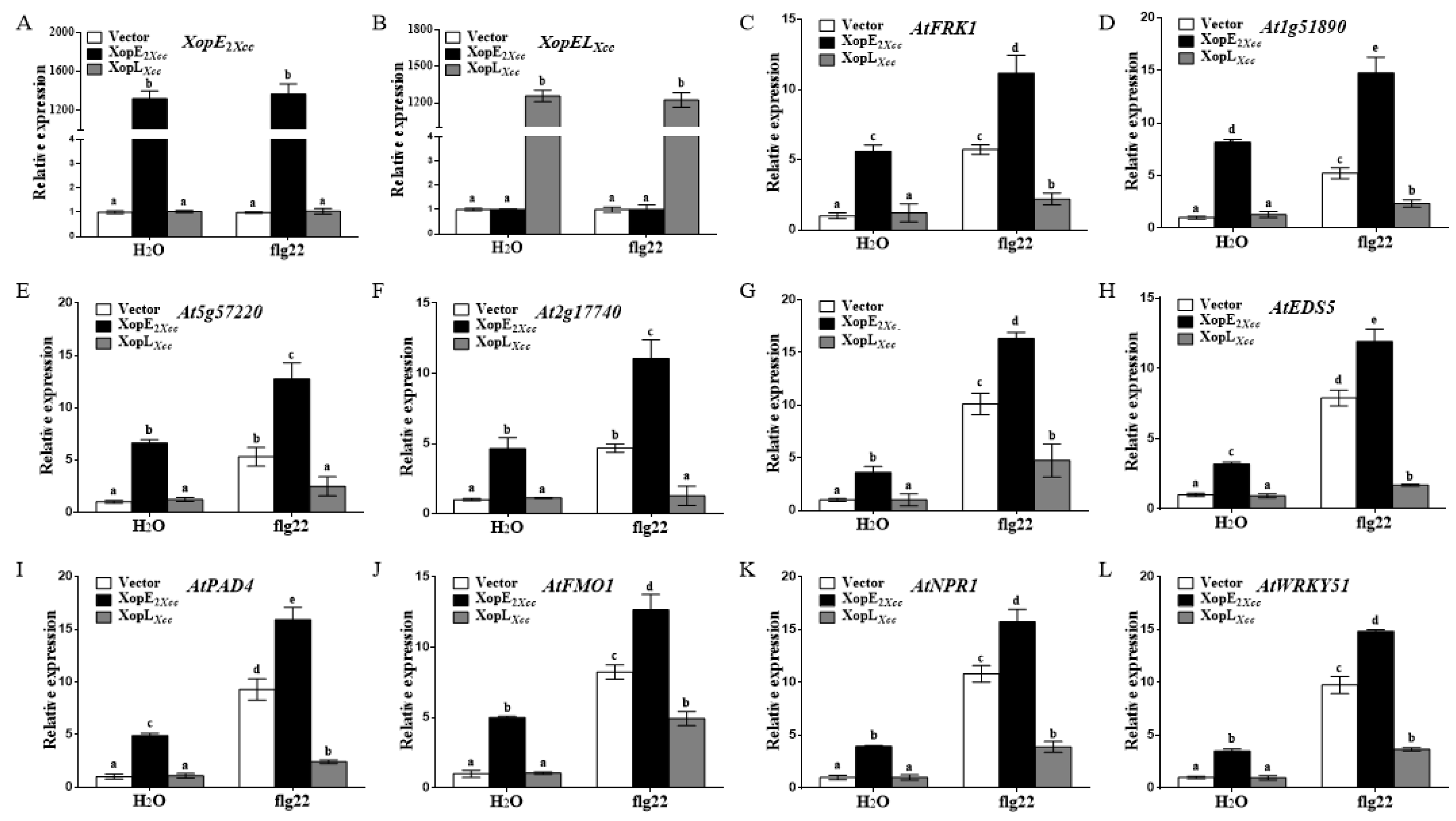
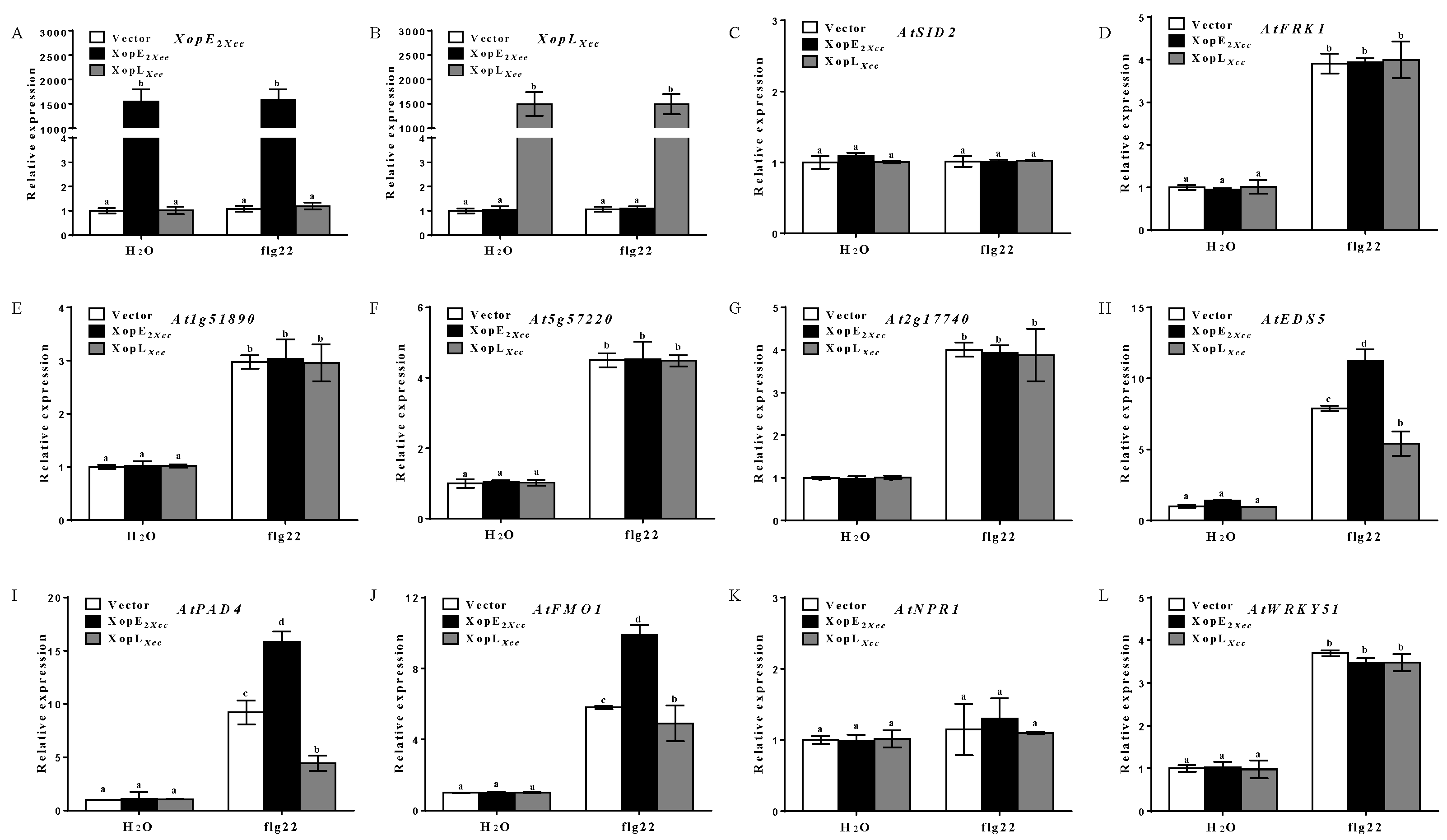
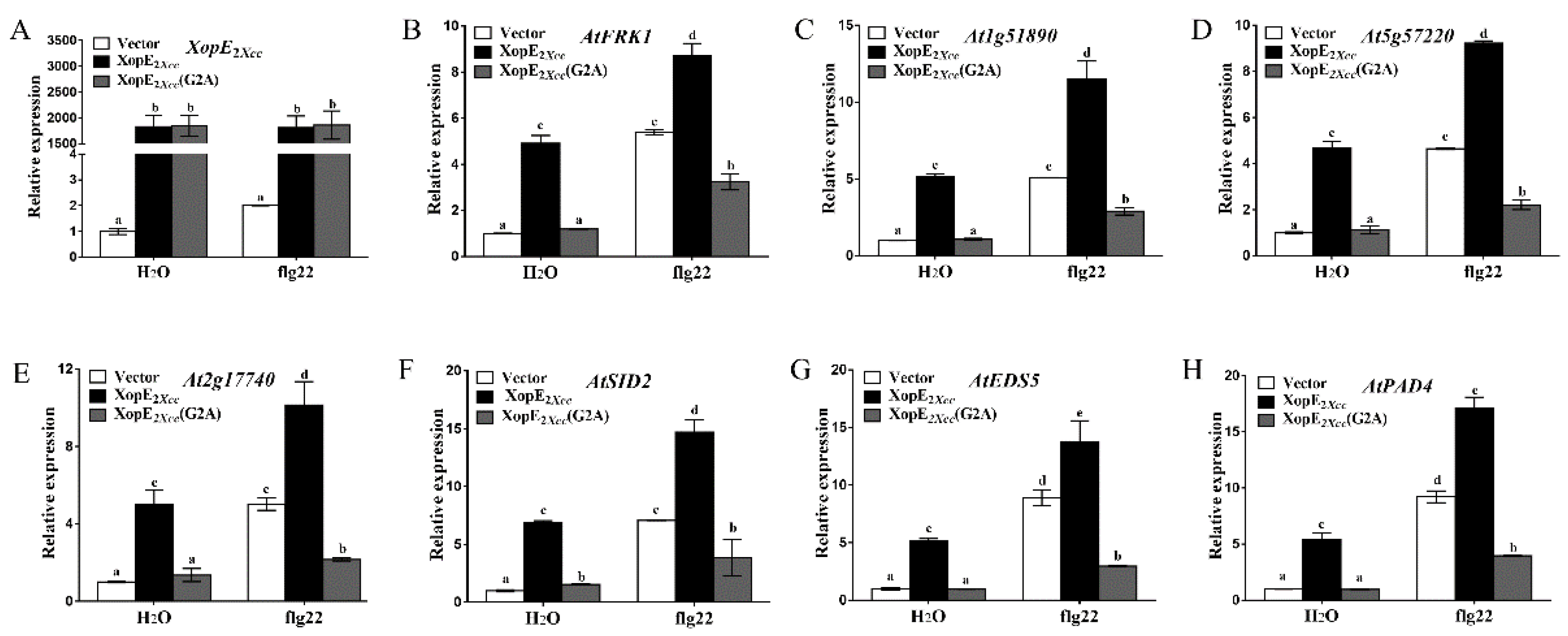
3.2. Effect of XopE2Xcc and XopLXcc on Defense Resistance in Arabidopsis
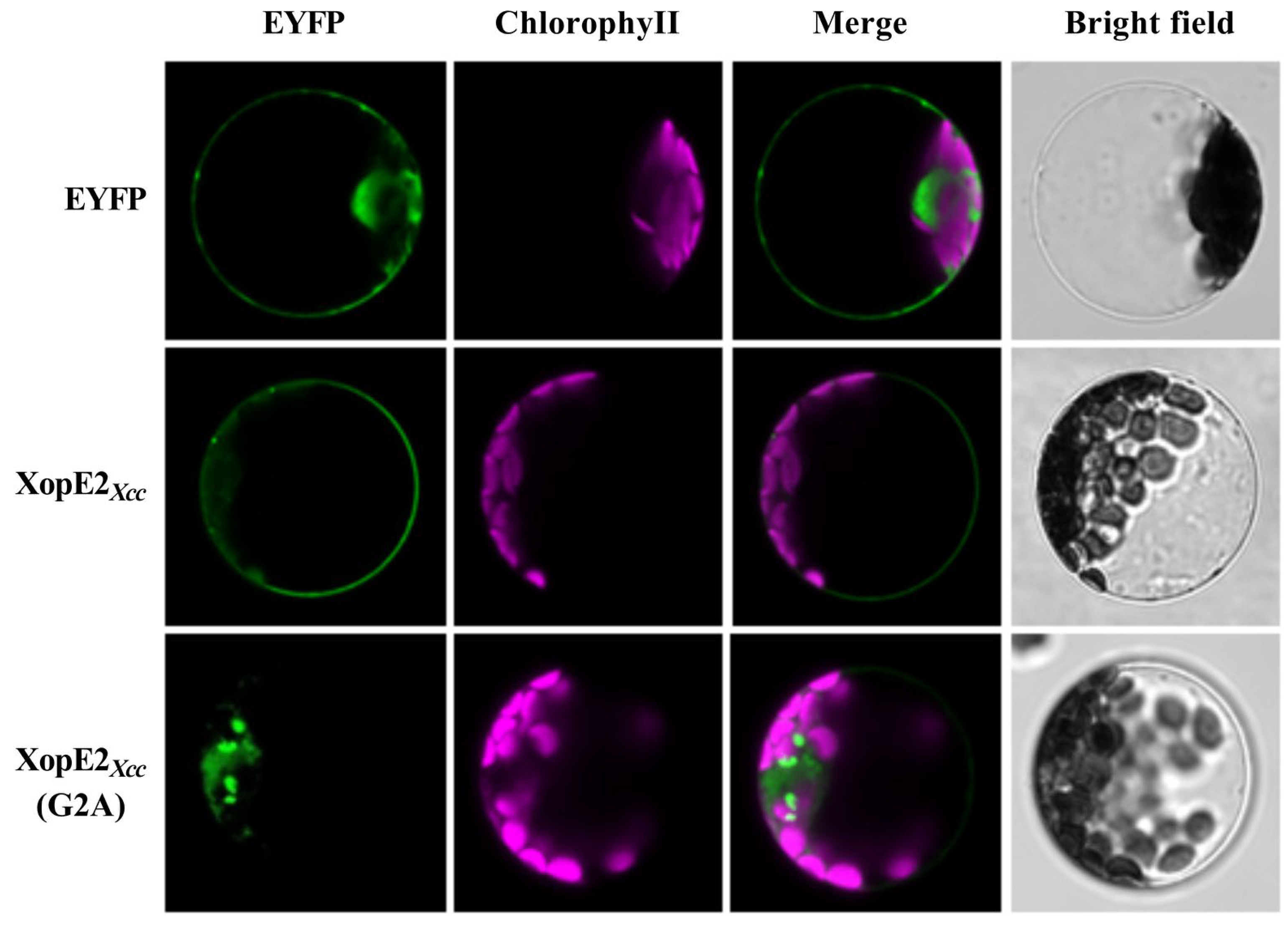
3.3. Subcellular Localization and Function Analysis of Mutant XopE2Xcc (G2A) Protein
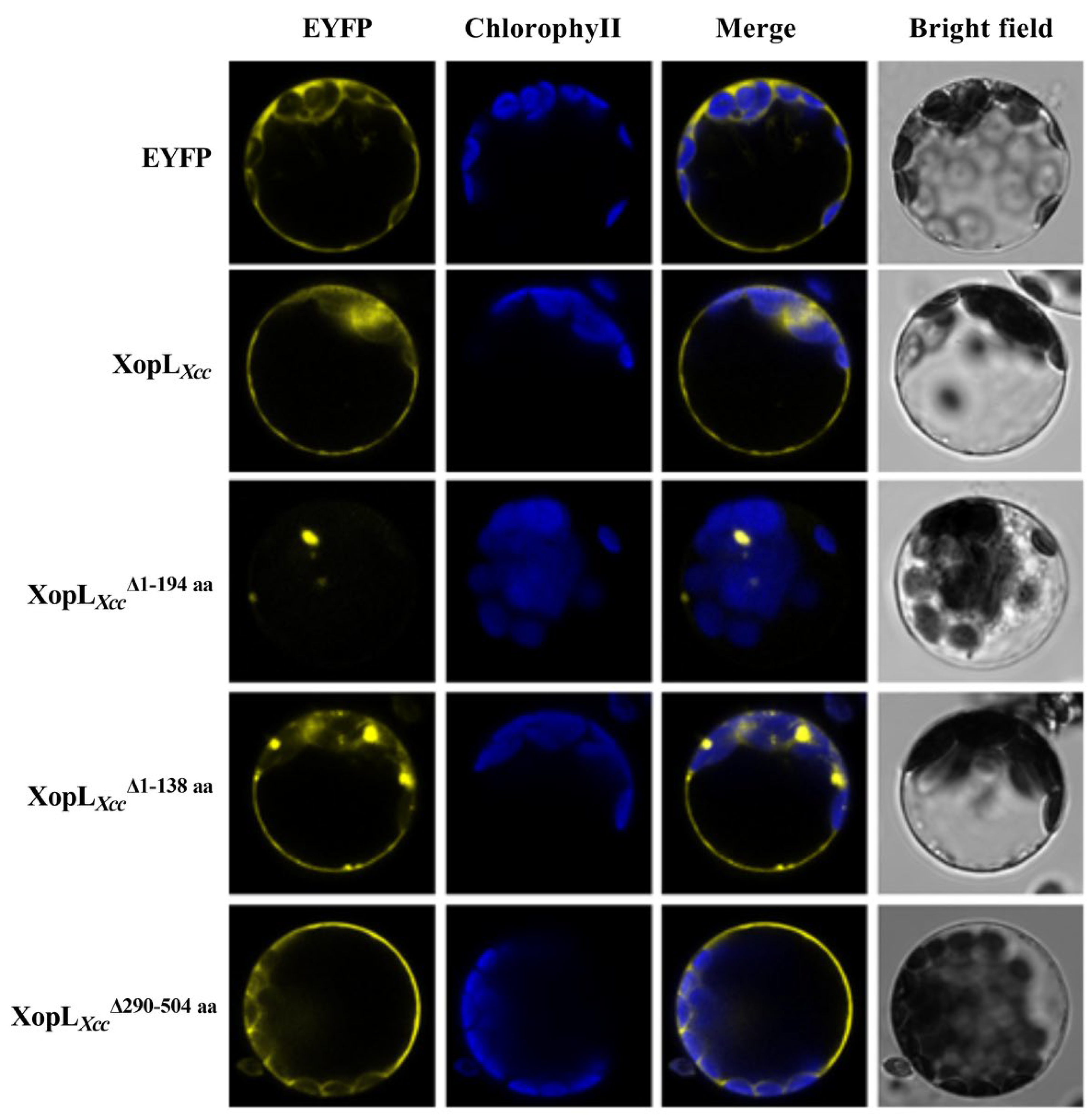
3.4. Subcellular Localization and Function Analysis of Various Mutant XopLXcc Proteins
4. Discussion
4.1. XopE2Xcc Triggers Avirulence in Arabidopsis via Upstream Activation of the SA Signaling Pathway
4.2. XopLXcc Suppresses PTI-Related and SA-Related Genes in Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ryan, R.P.; Vorhölter, F.J.; Potnis, N.; Jones, J.B.; Van Sluys, M.A.; Bogdanove, A.J.; Dow, J.M. Pathogenomics of Xanthomonas: Understanding bacterium-plant interactions. Nat. Rev. Microbiol. 2011, 9, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Gürlebeck, D.; Thieme, F.; Bonas, U. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant. Physiol. 2006, 163, 233–255. [Google Scholar] [CrossRef] [PubMed]
- White, F.F.; Potnis, N.; Jones, J.B.; Koebnik, R. The type III effectors of Xanthomonas. Mol. Plant. Pathol. 2009, 10, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant. Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.; Cascón, T.; Vicedo, B.; García-Agustín, P.; Hamberg, M.; Castresana, C. Role of 9-lipoxygenase and α-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol. Plant. 2012, 5, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Jia, Y.T.; Ren, S.X.; He, Y.Q.; Feng, J.X.; Lu, L.F.; Sun, Q.H.; Ying, G.; Tang, D.J.; Tang, H.; et al. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 2005, 15, 757–767. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Q.; Zhang, L.; Jiang, B.L.; Zhang, Z.C.; Xu, R.Q.; Tang, D.J.; Qin, J.; Jiang, W.; Zhang, X.; Liao, J.; et al. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 2007, 8, R218. [Google Scholar] [CrossRef] [PubMed]
- Guy, E.; Lautier, M.; Chabannes, M.; Roux, B.; Lauber, E.; Arlat, M.; Noël, L.D. XopAC-triggered immunity against Xanthomonas depends on arabidopsis receptor-Like cytoplasmic kinase genes PBL2 and RIPK. PLoS ONE 2013, 8, e73469. [Google Scholar] [CrossRef]
- Wang, L.F.; Tang, X.Y.; He, C.Z. The bifunctional effector AvrXccC of Xanthomonas campestris pv. campestris requires plasma membrane-anchoring for host recognition. Mol. Plant. Pathol. 2007, 8, 491–501. [Google Scholar] [CrossRef]
- Tan, L.T.; Rong, W.; Luo, H.L.; Chen, Y.H.; He, C.Z. The Xanthomonas campestris effector protein XopDxcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 2014, 204, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.L.; Li, C.A.X.; Ma, Z.M.; Han, X.; Luo, Y.Y.; Yang, L.; Yu, J.; Miao, Y.S. Xanthomonas effector XopR hijacks host actin cytoskeleton via complex coacervation. Nat. Commun. 2021, 12, 4064. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.L.; He, Y.Q.; Cen, W.J.; Wei, H.Y.; Jiang, G.F.; Jiang, W.; Hang, X.H.; Feng, J.X.; Lu, G.T.; Tang, D.H.; et al. The type III secretion effector xopXccN of Xanthomonas campestris pv. campestris is required for full virulence. Res. Microbiol. 2008, 159, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Yang, F.; Rong, W.; Wu, X.G.; Zhang, J.; Chen, S.; He, C.Z.; Zhou, J.M. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 2012, 485, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Tao, J.; Luo, H.L.; Tan, L.T.; Rong, W.; Li, H.P.; He, C.Z. A type III effector XopLxc8004 is vital for Xanthomonas campestris pathovar campestris to regulate plant immunity. Res. Microbiol. 2019, 170, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Üstün, S.; Bartetzko, V.; Börnke, F. The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 2013, 9, e1003427. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.Q.; Li, X.Z.; Wei, H.Y.; Jiang, B.; Li, K.; He, Y.Q.; Feng, J.X.; Tang, J.L. Regulation of eight avr genes by hrpG and hrpX in Xanthomonas campestris pv. campestris and their role in pathogenicity. Prog. Nat. Sci. Mater. 2006, 16, 1288–1294. [Google Scholar]
- Jiang, W.; Jiang, B.L.; Xu, R.Q.; Huang, J.D.; Wei, H.Y.; Jiang, G.F.; Cen, W.J.; Liu, J.; Ge, Y.Y.; Li, G.H.; et al. Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant. Microbe Interact. 2009, 22, 1401–1411. [Google Scholar] [CrossRef]
- Chen, S.B.; Songkumarn, P.; Liu, J.L.; Wang, G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant. Physiol. 2009, 150, 1111–1121. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, Y.; Peng, J.S.; Zhong, C.; Yi, H.Y.; Ow, D.W.; Gong, J.M. Fission yeast HMT1 lowers seed cadmium through phytochelatin-dependent vacuolar sequestration in arabidopsis. Plant. Physiol. 2012, 158, 1779–1788. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Lauber, E.; Roby, D.; Arlat, M.; Kroj, T. Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant. Pathol. 2005, 6, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Thieme, F.; Szczesny, R.; Urban, A.; Kirchner, O.; Hause, G.; Bonas, U. New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant. Microbe Interact. 2007, 20, 1250–1261. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1999, 1451, 1–16. [Google Scholar] [CrossRef]
- Nimchuk, Z.L.; Fisher, E.J.; Desveaux, D.; Chang, J.H.; Dangl, J.L. The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol. Plant. Microbe Interact. 2007, 20, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.G.; Holub, E.B. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant. Pathol. 2013, 14, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Kvitko, B.H.; Park, D.H.; Velásquez, A.C.; Wei, C.F.; Russell, A.B.; Martin, G.B.; Schneider, D.J.; Collmer, A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009, 5, e1000388. [Google Scholar] [CrossRef]
- He, P.; Shan, L.; Lin, N.C.; Martin, G.B.; Kemmerling, B.; Nürnberger, T.; Sheen, J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 2006, 125, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Akimoto-Tomiyama, C.; Furutani, A.; Tsuge, S.; Washington, E.J.; Nishizawa, Y.; Minami, E.; Ochiai, H. XopR, a type III effector secreted by Xanthomonas oryzae pv. oryzae, suppresses microbe-associated molecular pattern-triggered immunity in arabidopsis thaliana. Mol. Plant. Microbe Interact. 2012, 25, 505–514. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network properties of robust immunity in plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.Q.; Blanvillain, S.; Feng, J.X.; Jiang, B.L.; Li, X.Z.; Wei, H.Y.; Kroj, T.; Lauber, E.; Roby, D.; Chen, B.; et al. AvrAC, a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype col-0. J. Bacteriol. 2008, 190, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Stork, W.; Kim, J.G.; Mudgett, M.B. Functional analysis of plant defense suppression and activation by the Xanthomonas core type III effector XopX. Mol. Plant. Microbe Interact. 2015, 28, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, A.; Reddy, J.D.; El-Yacoubi, B.; Gabriel, D.W. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one gene affected race specificity. Mol. Plant. Microbe Interact. 2005, 18, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.P.; Dong, X.N. Perception of the plant immune signal salicylic acid. Curr. Opin. Plant. Biol. 2014, 20, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.U.; Schulze, S.; Skarina, T.; Xu, X.H.; Cui, H.; Eschen-Lippold, L.; Egler, M.; Srikumar, T.; Raught, B.; Lee, J.; et al. A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 2013, 9, e1003121. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.L.; Adlung, N.; Lampe, C.; Bonas, U.; Schattat, M.H. The Xanthomonas effector XopL uncovers the role of microtubules in stromule extension and dynamics in Nicotiana benthamiana. Plant J. 2018, 93, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Soni, M.; Mondal, K.K. Xanthomonas axonopodis pv. punicae uses XopL effector to suppress pomegranate immunity. J. Integr. Plant. Biol. 2018, 60, 341–357. [Google Scholar] [CrossRef]
- Bengtsson, E.; Lindblom, K.; Tillgren, V.; Aspberg, A. The leucine-rich repeat protein PRELP binds fibroblast cell-surface proteoglycans and enhances focal adhesion formation. Biochem. J. 2016, 473, 1153–1164. [Google Scholar] [CrossRef]
- Nitipan, S.; Sritrakul, T.; Kunjantarachot, A.; Prapong, S. Identification of epitopes in Leptospira borgpetersenii leucine-rich repeat proteins. Infect. Genet. Evol. 2013, 14, 46–57. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhou, H.; Zhou, M.; Li, N.; Jiang, B.; He, Y. Functional Analysis of Type III Effectors in Xanthomonas campestris pv. campestris Reveals Distinct Roles in Modulating Arabidopsis Innate Immunity. Pathogens 2024, 13, 448. https://doi.org/10.3390/pathogens13060448
Huang J, Zhou H, Zhou M, Li N, Jiang B, He Y. Functional Analysis of Type III Effectors in Xanthomonas campestris pv. campestris Reveals Distinct Roles in Modulating Arabidopsis Innate Immunity. Pathogens. 2024; 13(6):448. https://doi.org/10.3390/pathogens13060448
Chicago/Turabian StyleHuang, Jing, Hao Zhou, Min Zhou, Nana Li, Bole Jiang, and Yongqiang He. 2024. "Functional Analysis of Type III Effectors in Xanthomonas campestris pv. campestris Reveals Distinct Roles in Modulating Arabidopsis Innate Immunity" Pathogens 13, no. 6: 448. https://doi.org/10.3390/pathogens13060448



