Comparative Analysis of Immune Gene Transcription in Sea Bream (Sparus aurata) Challenged with RGNNV or RGNNV/SJNNV Betanodaviruses
Abstract
:1. Introduction
2. Material and Methods
2.1. Viral Propagation and Titration
2.2. Fish Trial
2.3. Sample Processing
2.4. Quantification of the RNA1 Viral Segment
2.5. Quantification of Immune Gene Transcription
| Gene | Primer | Sequence (5′-3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| RdRp | F | GGCTCAGATCTGGTAATGTTTCAA | 63 | [22] |
| R | CAAAGCCAAGGGAAGAAGCA | |||
| irf3 | F | TCAGAATGCCCCAAGAGATT | 107 | [23] |
| R | AGAGTCTCCGCCTTCAGATG | |||
| rtp3 | F | CAGGTGCAGCAAGTGTGGGA | 120 | This study |
| R | GTCTCACCTTGACCACGCCC | |||
| sacs | F | ACATCCGGACCTTGGTGCCT | 124 | This study |
| R | AGCGGTGGTGTAGTCTGTCCA | |||
| il-1β | F | AGCGCAGTAGAAGAGCGAAC | 117 | [24] |
| R | CACTCGGACTAAGTGCCTCTG | |||
| il-6 | F | CCAGATCCCCTCAAGATTCA | 144 | [24] |
| R | AAGGTGTCGGAGCTGTCG | |||
| il-10 | F | CAGGCCATGAACAACATCC | 143 | [24] |
| R | TGGACGTCAGATTTGAGCTG | |||
| tnf-α | F | TTCCGACTGGTGGACAATAAG | 143 | [24] |
| R | GAGATCCTGTGGCTGAGAGG | |||
| hsp70 | F | TGAGGTAAAGTCCACTGCCGGA | 132 | This study |
| R | AGCTCTCTTGTTGTCGCTGATGT | |||
| granb | F | GAAACAAAGGAACGGGTCAA | 128 | This study |
| R | GAGCTGTCCATCTTTTGCTTG | |||
| casp1 | F | TCGAAGAGACGGACAGTGTG | 123 | [24] |
| R | CGTTGATGGGGAACTCATCT | |||
| actb | F | ATTGTCAAACTGCACCCACA | 139 | [24] |
| R | GCTCAACAGCCTTGATGACA |
2.6. Statistical Analyses
3. Results
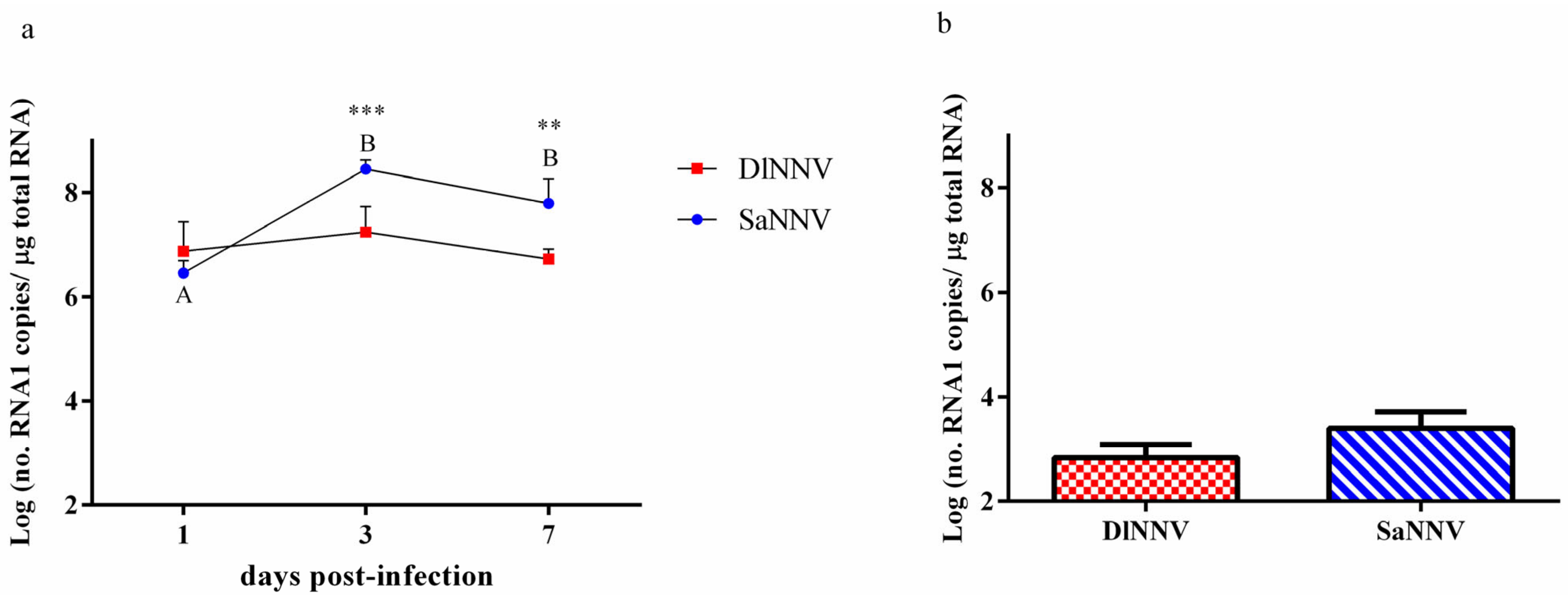
3.1. Fish Mortality and Viral Replication
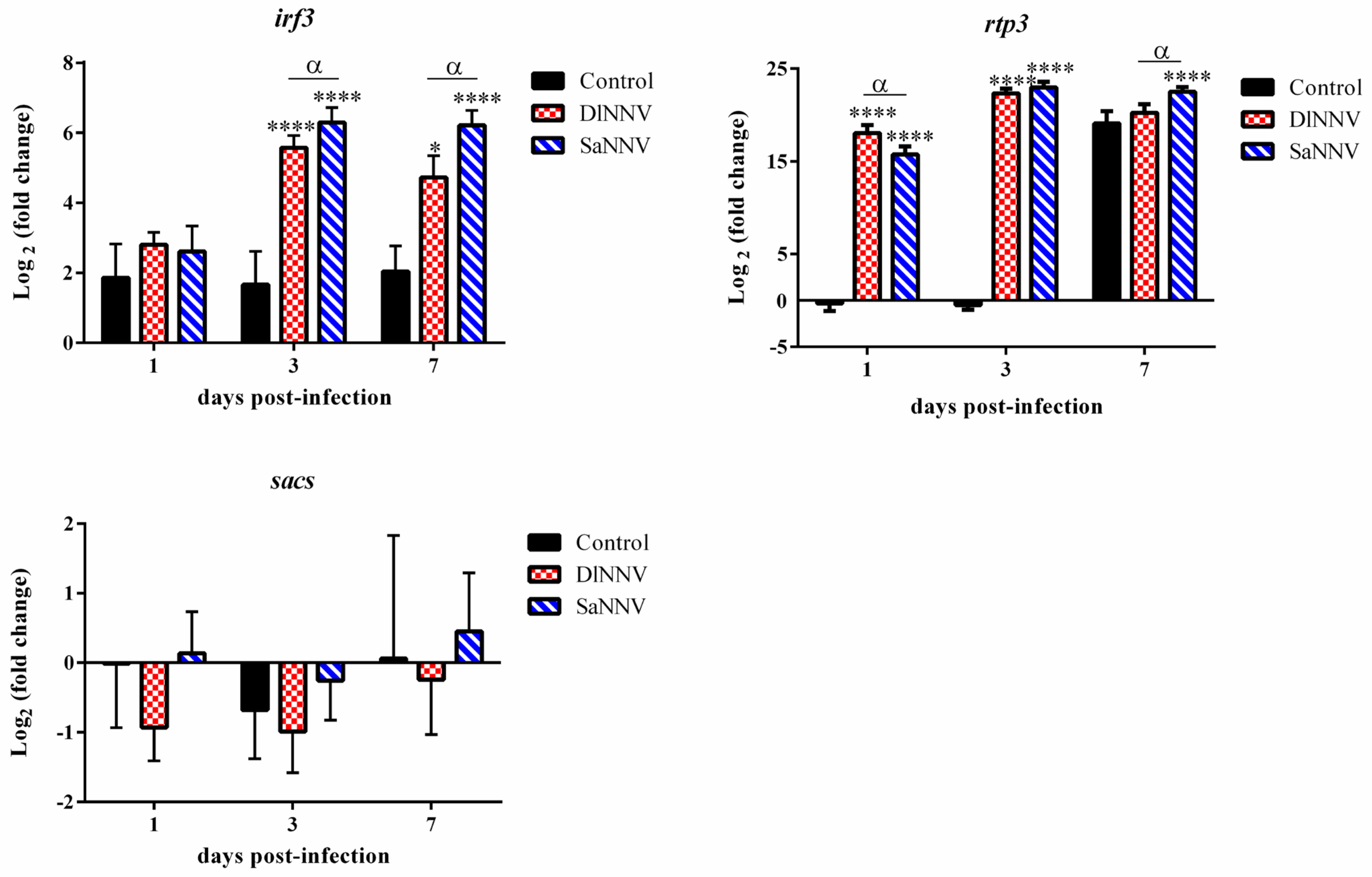
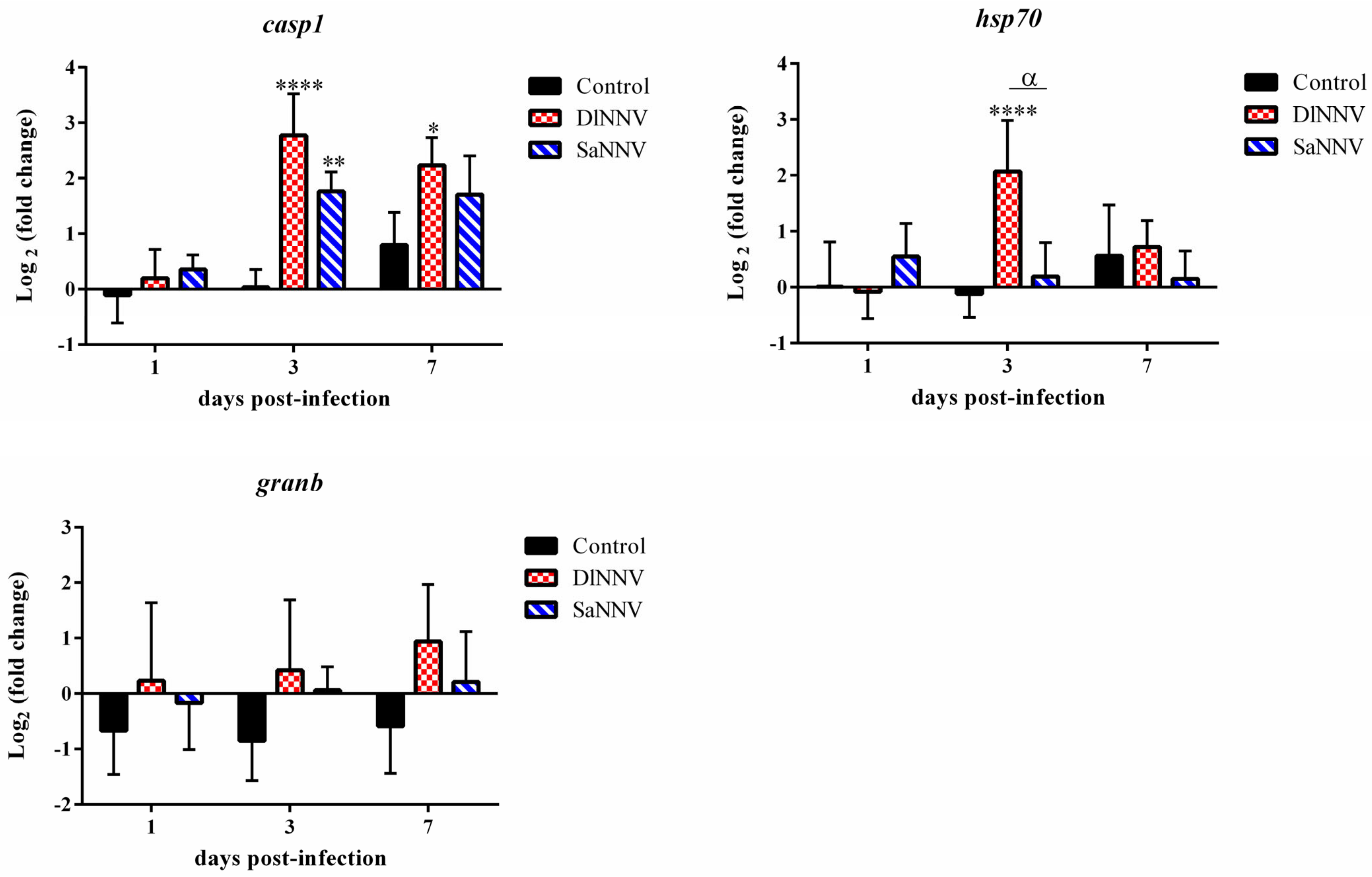
3.2. Kinetics of Immune Gene Transcription
3.3. Principal Component Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Padros, F.; Caggiano, M.; Toffan, A.; Constenla, M.; Zarza, C.; Ciulli, S. Integrated management strategies for viral nervous necrosis (VNN) disease control in marine fish farming in the Mediterranean. Pathogens 2022, 11, 330. [Google Scholar] [CrossRef]
- Doan, Q.K.; Vandeputte, M.; Chatain, B.; Morin, T.; Allal, F. Viral encephalopathy and retinopathy in aquaculture: A review. J. Fish Dis. 2017, 40, 717–742. [Google Scholar] [CrossRef]
- Sahul Hammed, A.S.; Ninawe, A.S.; Nakai, Y.; Chi, S.C.; Johnson, K.L. ICTV virus taxonomy profile: Nodaviridae. J. Gen. Virol. 2019, 100, 3–4. [Google Scholar] [CrossRef]
- Ariff, N.; Abdullah, A.; Amal-Azmai, M.N.; Musa, N.; Zainathan, S.C. Risk factors associated with viral nervous necrosis in hybrid groupers in Malaysia and the high similarity of its causative agent nervous necrosis virus to reassortant red-spotted grouper nervous necrosis virus/striped jack nervous necrosis virus strains. Vet. World 2019, 12, 1273–1284. [Google Scholar] [CrossRef]
- Bitchava, K.; Xylouri, E.; Fragkiadaki, E.; Athanassopoulou, F.; Papanastassopoulou, M.; Sabatakou, O. First incidence of clinical signs of nodavirus infection in sea bream, Sparus aurata L. Isr. J. Aquac. 2007, 59, 3–9. [Google Scholar]
- Chérif, N.; El Jeni, R.; Amdouni, F.; Zreilli, S.; Djabou, H.; Khemiri, S.; Tliba, I.; Bouhaouala-Zahar, B.; Maatoug, K.; Zaafran, S.; et al. Phylogeography of betanodavirus genotypes circulating in Tunisian aquaculture sites, 2012–2019. Dis. Aquat. Org. 2021, 146, 53–63. [Google Scholar] [CrossRef]
- Kaplan, M.; Pekmez, K.; Cagirgan, A.A.; Arslan, F.; Özkan, B.; Kalayci, G. The first detection of betanodavirus reassortant genotype (RGNNV/SJNNV) isolated from gilthead sea bream (Sparus aurata) in the Turkish coastlines: The importance of screening and monitoring studies for identifying the source of infection. J. Fish Dis. 2022, 45, 783–793. [Google Scholar] [CrossRef]
- Olveira, J.G.; Souto, S.; Dopazo, C.P.; Thiery, R.; Barja, J.L.; Bandin, I. Comparative analysis of both genomic segments of betanodaviruses isolated from epizootic outbreaks in farmed fish species provides evidence for genetic reassortment. J. Gen. Virol. 2009, 90, 2940–2951. [Google Scholar] [CrossRef]
- Panzarin, V.; Fusaro, A.; Monne, I.; Cappellozza, E.; Patarnello, P.; Bovo, G.; Capua, I.; Holmes, E.C.; Cattoli, G. Molecular epidemiology and evolutionary dynamics of betanodavirus in southern Europe. Infect. Genet. Evol. 2012, 12, 63–70. [Google Scholar] [CrossRef]
- Toffan, A.; Pascoli, F.; Pretto, T.; Panzarin, V.; Abbadi, M.; Buratin, A.; Quartesan, R.; Gijon, D.; Padros, F. Viral nervous necrosis in gilthead sea bream (Sparus aurata) caused by reassortant betanodavirus RGNNV/SJNNV: An emerging threat for Mediterranean aquaculture. Sci. Rep. 2017, 7, 46755. [Google Scholar] [CrossRef]
- Toffolo, V.; Negrisolo, E.; Maltese, C.; Bovo, G.; Belvedere, P.; Colombo, I.; Dalla Valle, L. Phylogeny of betanodaviruses and molecular evolution of their RNA polymerase and coat proteins. Mol. Phylogenet. Evol. 2007, 43, 298–308. [Google Scholar] [CrossRef]
- Vazquez-Salgado, L.; Olveira, J.G.; Dopazo, C.P.; Bandin, I. Detection of different Betanodavirus genotypes in wild fish from Spanish Atlantic coastal waters (Galicia, northwestern Spain). J. Aquat. Anim. Health 2024, 36, 57–69. [Google Scholar] [CrossRef]
- Volpe, E.; Gustinelli, A.; Caffara, M.; Errani, F.; Quaglio, F.; Fioravanti, M.L.; Ciulli, S. Viral nervous necrosis outbreaks caused by the RGNNV/SJNNV reassortant betanodavirus in Gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax). Aquaculture 2020, 523, 735155. [Google Scholar]
- Carballo, C.; Garcia-Rosado, E.; Borrego, J.J.; Alonso, M.C. SJNNV down-regulates RGNNV replication in European sea bass by the induction of the type I interferon system. Vet. Res. 2016, 47, 6. [Google Scholar] [CrossRef]
- Souto, S.; Lopez-Jimena, B.; Alonso, M.C.; Garcia-Rosado, E.; Bandin, I. Experimental susceptibility of European sea bass and Senegalese ole to different betanodavirus isolates. Vet. Microbiol. 2015, 177, 53–61. [Google Scholar] [CrossRef]
- Vendramin, N.; Toffan, A.; Mancin, M.; Cappellozza, E.; Panzarin, V.; Bovo, G.; Catolli, G.; Capua, I.; Terregino, C. Comparative pathogenicity study of ten different betanodavirus strains in experimentally infected European sea, Dicentrarchus labrax (L.). J. Fish Dis. 2014, 37, 271–383. [Google Scholar] [CrossRef]
- Toffan, A.; Biasini, L.; Pretto, T.; Abbadi, M.; Buratin, A.; Franch, R.; Dalla Rovere, G.; Panzarin, V.M.; Marsella, A.; Bargelloni, L.; et al. Age dependency of RGNNV/SJNNV viral encephalo-retinopathy in Gilthead sea bream (Sparus aurata). Aquaculture 2021, 539, 736605. [Google Scholar]
- Castric, J.; Thiery, R.; Jeffroy, J.; de Kinkelin, P.; Raymond, J. Sea bream Sparus aurata, an asymptomatic contagious fish host for nodavirus. Dis. Aquat. Organ. 2001, 47, 33–38. [Google Scholar] [CrossRef]
- Iwamoto, T.; Nakai, T.; Mori, K.; Arimoto, M.; Furusawa, I. Cloning of the fish cell line SSN-1 for piscine nodavirus. Dis. Aquat. Org. 2000, 43, 81–89. [Google Scholar] [CrossRef]
- Reed, L.J.; Müench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lopez-Jimena, B.; Garcia-Rosado, E.; Infante, C.; Adams, A.; Infante, C.; Castro, D.; Borrego, J.J.; Alonso, M.C. Effect of the coexistence on the replication of striped jack nervous necrosis virus (SJNNV) and red-spotted grouper nervous necrosis virus (RGNNV) using an in vitro approach. J. Appl. Ichthyol. 2014, 30, 916–922. [Google Scholar] [CrossRef]
- Lopez-Jimena, B.; Alonso, M.C.; Thompson, K.D.; Adams, A.; Infante, C.; Castro, D.; Borrego, J.J.; Garcia-Rosado, E. Tissue distribution of Red Spotted Grouper Nervous Necrosis Virus (RGNNV) genome in experimentally infected European seabass (Dicentrarchus labrax). Vet. Immunol. 2011, 154, 86–95. [Google Scholar] [CrossRef]
- Valero, Y.; Morcillo, P.; Meseguer, J.; Buonocore, F.; Esteban, M.A.; Chaves-Pozo, E.; Cuesta, A. Characterization of the IFN pathway in the teleost fish gonad against vertically transmitted viral nervous necrosis virus. J. Gen. Virol. 2015, 96, 2176–2187. [Google Scholar] [CrossRef]
- Leiva-Rebollo, R.; Labella, A.M.; Borrego, J.J.; Castro, D. Immune gene expression in gilthead seabream (Sparus aurata) after lymphocystis disease virus (LCDV-Sa) challenge resulting in asymptomatic infection. J. Appl. Microbiol. 2020, 128, 41–53. [Google Scholar]
- Esteban, M.A.; Meseguer, J.; Tafalla, C.; Cuesta, A. NK-like and oxidative burst activities are the main early cellular innate immune responses activated after virus inoculation in reservoir fish. Fish Shellfish Immunol. 2008, 25, 433–438. [Google Scholar] [CrossRef]
- Perez-Prieto, S.I.; Garcia-Rosado, E.; Rodriguez, S.; Castro, D.; Borrego, J.J. Antigenic properties and experimental transmission to several fish species of a marine birnavirus isolated from sole (Solea senegalensis). Vet. Microbiol. 2001, 82, 11–25. [Google Scholar] [CrossRef]
- Garcia-Rosado, E.; Castro, D.; Rodriguez, S.; Perez-Prieto, S.I.; Borrego, J.J. Isolation and characterization of lymphocystis virus (FLDV) from gilthead seabream (Sparus aurata L.) using a new homologous cell line. Bull. Eur. Assoc. Fish Pathol. 1999, 19, 53–56. [Google Scholar]
- Chaves-Pozo, E.; Guardiola, F.A.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Nodavirus infection induces a great innate cell-mediated cytotoxic activity in resistant, gilthead seabream, and susceptible, European sea bass, teleost fish. Fish Shellfish Immunol. 2012, 33, 1159–1166. [Google Scholar] [CrossRef]
- Lopez-Muñoz, A.; Sepulcre, M.P.; Garcia-Moreno, D.; Fuentes, I.; Bejar, J.; Manchado, M.; Alvarez, M.C.; Meseguer, J.; Mulero, V. Viral nervous necrosis virus persistently replicates in the central nervous system of asymptomatic gilthead seabream and promotes a transient inflammatory response followed by the infiltration of IgM+ B lymphocytes. Dev. Comp. Immunol. 2012, 37, 429–437. [Google Scholar] [CrossRef]
- Cherif, N.; Thiery, R.; Castric, J.; Biacchesi, S.; Bremont, M.; Thabti, F.; Limem, L.; Hammami, S. Viral encephalopathy and retinopathy of Dicentrarchus labrax and Sparus aurata farmed in Tunisia. Vet. Res. Commun. 2009, 33, 345–352. [Google Scholar] [CrossRef]
- Vazquez-Salgado, L.; Olveira, J.G.; Dopazo, C.P.; Bandin, I. Interspecies transmission between Solea senegalensis and Sparus aurata of reassortant nervous necrosis virus (NNV) strains and effect of stress on the outcome of the infection. Aquaculture 2022, 547, 737519. [Google Scholar]
- Garcia-Alvarez, M.A.; Arizcum, M.; Chaves-Pozo, E.; Cuesta, A. Profile of innate immunity in gilthead seabream larvae reflects mortality upon betanodavirus reassortant infection and replication. Int. J. Mol. Sci. 2022, 23, 5092. [Google Scholar] [CrossRef]
- Peruzza, L.; Pascoli, F.; Dalla Rovere, G.; Franch, R.; Ferraresso, S.; Babbucci, M.; Biasini, L.; Abbadi, M.; Panzarin, V.; Toffan, A.; et al. Transcriptome analysis reveals a complex response to the RGNNV/SJNNV reassortant nervous necrosis virus strain in sea bream larvae. Fish Shellfish Immunol. 2021, 114, 282–292. [Google Scholar] [CrossRef]
- Pereiro, P.; Figueras, A.; Novoa, B. RNA-seq analysis of juvenile sea bream (Sparus aurata) provides some clues regarding their resistance to the nodavirus RGNNV genotype. Fish Shellfish Immunol. 2023, 134, 108588. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Dios, S.; Montes, A.; Aranguren, R.; Figueras, A.; Novoa, B. Nodavirus increases the expression of Mx and inflammatory cytokines in fish brain. Mol. Immunol. 2008, 45, 218–225. [Google Scholar] [CrossRef]
- Lama, R.; Pereiro, P.; Figueras, A.; Novoa, B. Zebrafish as a vertebrate model for studying nodavirus infections. Front. Immunol. 2022, 13, 863096. [Google Scholar] [CrossRef]
- Moreno, P.; Gemez-Mata, J.; Garcia-Rosado, E.; Bejar, J.; Labella, A.M.; Souto, S.; Alonso, M.C. Differential immunogene expression profile of European sea bass (Dicentrarchus labrax, L.) in response to highly and low virulent NNV. Fish Shellfish Immunol. 2020, 106, 56–70. [Google Scholar] [CrossRef]
- Overgard, A.C.; Nerland, A.H.; Fiksdal, I.U.; Patel, S. Atlantic halibut experimentally infected with nodavirus shows increased levels of -cells markers and IFNγ transcripts. Dev. Comp. Immunol. 2012, 37, 139–150. [Google Scholar] [CrossRef]
- Aranguren, R.; Tafalla, C.; Novoa, B.; Figueras, A. Experimental transmission of encepaholaphathy and retinopathy nodavirus to sea bream, Sparus aurata L., using infection models. J. Fish Dis. 2002, 25, 317–324. [Google Scholar] [CrossRef]
- Chiang, Y.H.; Wu, Y.C.; Chi, S.C. Interleukin-1β secreted from betanodavirus-infected microglia caused the death of neurons in giant grouper brains. Dev. Comp. Immunol. 2017, 70, 19–26. [Google Scholar] [CrossRef]
- Nelson, P.T.; Soma, L.A.; Lavi, E. Microglia in diseases of the central nervous system. Ann. Med. 2002, 34, 491–500. [Google Scholar] [CrossRef]
- Williams, K.C.; Hickey, W.F. Central nervous system damage, monocytes and macrophages, and neurological disorders in AIDS. Annu. Rev. Neurosci. 2002, 25, 537–562. [Google Scholar] [CrossRef]
- Moreno, P.; Lopez-Jimena, B.; Randelli, E.; Scapigliati, G.; Buonocore, F.; Garcia-Rosado, E.; Borrego, J.J.; Alonso, M.C. Immuno-related gene transcription and antibody response in nodavirus (RGNNV and SJNNV)-infected European sea bass (Dicentrarchus labrax, L.). Fish Shellfish Immunol. 2018, 78, 270–278. [Google Scholar] [CrossRef]
- Gemez-Mata, J.; Labella, A.M.; Bandin, I.; Borrego, J.J.; Garcia-Rosado, E. Immunogene expression analysis in betanodavirus-infected Senegalese sole using an OpenArray platform. Gene 2021, 774, 145430. [Google Scholar] [CrossRef]
- Labella, A.M.; Garcia-Rosado, E.; Bandin, I.; Dopazo, C.P.; Castro, D.; Alonso, M.C.; Borrego, J.J. Transcriptomic profiles of Senegalese sole infected with nervous necrosis virus reassortants presenting different degree of virulence. Front. Immunol. 2018, 9, 1626. [Google Scholar] [CrossRef]
- Scapigliati, G.; Buonocore, F.; Randelli, E.; Casani, D.; Meloni, S.; Zarletti, G.; Tiberi, M.; Prietretti, D.; Boschi, I.; Manchado, M.; et al. Cellular and molecular immune responses of the sea bass (Dicentrarchus labrax) experimentally infected with betanodavirus. Fish Shellfish Immunol. 2010, 28, 303–311. [Google Scholar] [CrossRef]
- Moreno, M.; Gemez-Mata, J.; Alvarez-Torres, D.; Garcia-Rosado, E.; Bejar, J.; Alonso, M.C. Genomic characterization and transcription analysis of European sea bass (Dicentrarchus labrax) rtp3 genes. Mol. Immunol. 2023, 163, 243–248. [Google Scholar] [CrossRef]
- Liu, P.; Wang, L.; Ye, B.Q.; Huang, S.; Wong, S.M.; Yue, G.H. Characterization of a novel disease resistance gene rtp3 and its association with VNN disease resistance in Asian seabass. Fish Shellfish Immunol. 2017, 61, 61–67. [Google Scholar] [CrossRef]
- Lama, R.; Pereiro, P.; Valenzuela-Muñoz, V.; Gallardo-Escarate, C.; Tort, L.; Figueras, A.; Novoa, B. RNA-seq analysis of European sea bass (Dicentrarchus labrax L.) infected with nodavirus reveals powerful modulation of the stress response. Vet. Res. 2020, 51, 64. [Google Scholar] [CrossRef]
- Dai, Z.; Li, J.; Hu, C.; Wang, F.; Shi, X.; Hou, Q.; Huang, W.; Lin, G. Transcriptome data analysis of grass carp (Ctenopharyngodon idella) infected by reovirus provides insights into two immune-related genes. Fish Shellfish Immunol. 2017, 64, 68–77. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Sen, G.C. IRF-3 and Bax: A deadly affair. Cell Cycle 2010, 9, 2479–2480. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Yamashita, M.; Peters, K.L.; Smith, K.; Desai, A.; Williams, R.G.; Sen, G.C. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010, 29, 10. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Yamashita, M.; Zhang, Y.; Sen, G.C. The IRF3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA or DNA, inhibits virus replication. J. Virol. 2011, 85, 3708–3716. [Google Scholar] [CrossRef]
- Heylbroeck, C.; Balachandran, S.; Servant, M.J.; DeLuca, C.; Barber, G.N.; Lin, R.; Hiscott, J. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 2000, 74, 3781–3792. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Valero, Y.; Lozano, M.T.; Rodriguez-Cerezo, P.; Miao, L.; Campo, V.; Esteban, M.A.; Cuesta, A. Fish Granzyme B in fish innate cell-mediated cytotoxicity. Front. Immunol. 2019, 10, 2579. [Google Scholar] [CrossRef]
- Angosto, D.; Lopez-Castejon, G.; Lopez-Muñoz, A.; Sepulcre, M.P.; Arizcun, M.; Meseguer, J.; Mulero, V. Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of Il-1β. Innate Immun. 2012, 18, 815–824. [Google Scholar] [CrossRef]
- Lanneau, D.; Brunet, M.; Frisan, E.; Solary, E.; Fontenay, M.; Garrido, C. Heat shock proteins: Essential proteins for apoptosis regulation. J. Cell. Mol. Med. 2008, 12, 743–761. [Google Scholar] [CrossRef]
- Chang, J.-S.; Chi, S.-C. GHSC70 is involved in the cellular entry of nervous necrosis virus. J. Virol. 2015, 89, 61–70. [Google Scholar] [CrossRef]
- Zenke, K.; Okinaka, Y. Multiple isoforms of HSP70 and HSP90 required for betanodavirus multiplication in medaka cells. Arch. Virol. 2022, 167, 1961–1975. [Google Scholar] [CrossRef]
- Sung, Y.Y.; MacRae, T.H. Heat shock proteins and disease control in aquatic organisms. J. Aquac. Res. Dev. 2011, S2, 006. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gemez-Mata, J.; Moreno, P.; Alvarez-Torres, D.; Garcia-Rosado, E.; Bejar, J.; Alonso, M.C. Comparative Analysis of Immune Gene Transcription in Sea Bream (Sparus aurata) Challenged with RGNNV or RGNNV/SJNNV Betanodaviruses. Pathogens 2024, 13, 478. https://doi.org/10.3390/pathogens13060478
Gemez-Mata J, Moreno P, Alvarez-Torres D, Garcia-Rosado E, Bejar J, Alonso MC. Comparative Analysis of Immune Gene Transcription in Sea Bream (Sparus aurata) Challenged with RGNNV or RGNNV/SJNNV Betanodaviruses. Pathogens. 2024; 13(6):478. https://doi.org/10.3390/pathogens13060478
Chicago/Turabian StyleGemez-Mata, Juan, Patricia Moreno, Daniel Alvarez-Torres, Esther Garcia-Rosado, Julia Bejar, and M. Carmen Alonso. 2024. "Comparative Analysis of Immune Gene Transcription in Sea Bream (Sparus aurata) Challenged with RGNNV or RGNNV/SJNNV Betanodaviruses" Pathogens 13, no. 6: 478. https://doi.org/10.3390/pathogens13060478





