Clostridioides difficile and Gut Microbiota: From Colonization to Infection and Treatment
Abstract
1. Introduction
2. Healthy Gut Microbiota
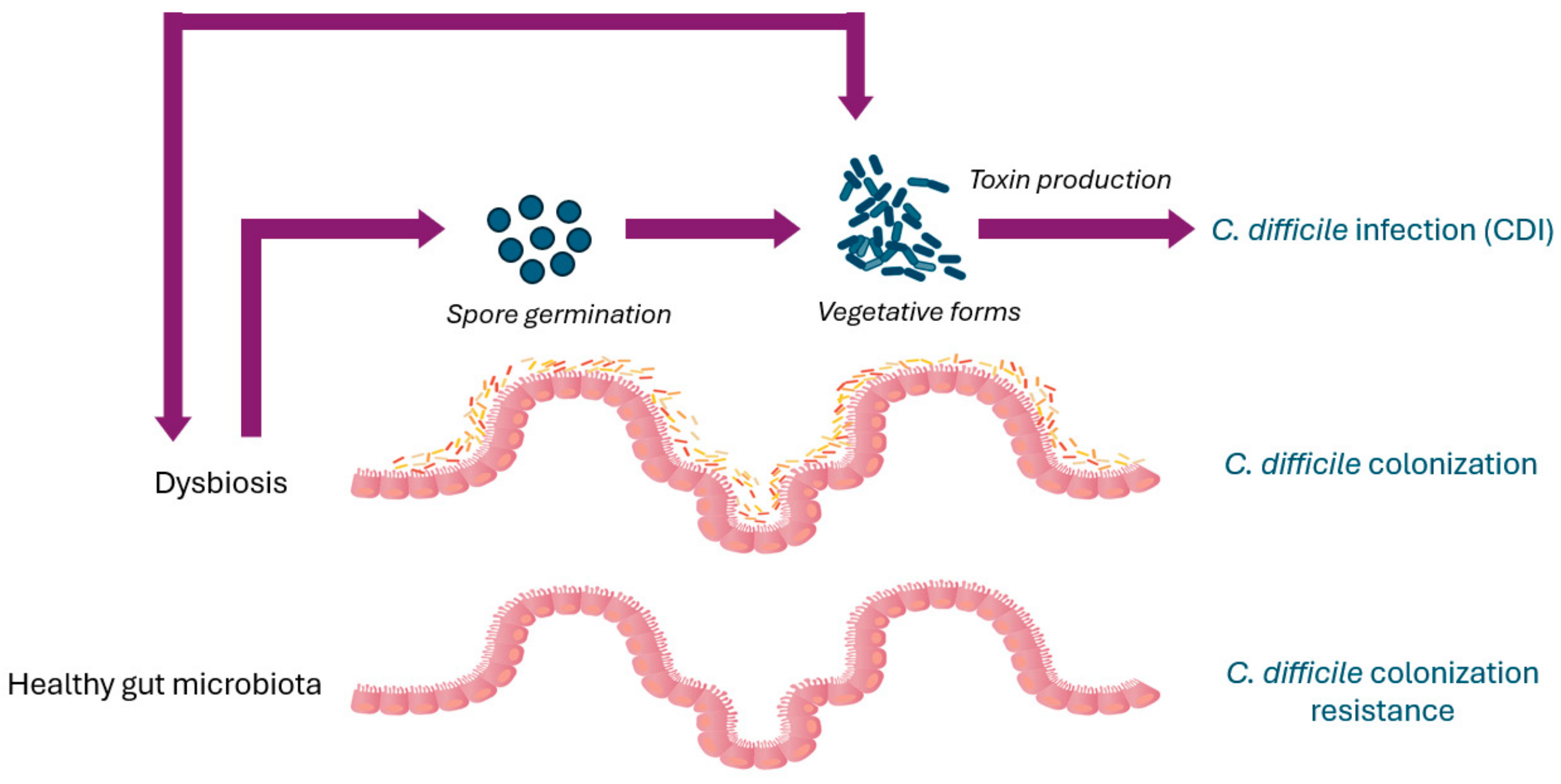
3. C. difficile Colonization Resistance
4. Asymptomatic C. difficile Colonization
5. Gut Microbiota in Asymptomatic C. difficile Carriers
5.1. Neonates and Infants
5.2. Adults
6. Gut Microbiota Dysbiosis Increases Susceptibility to C. difficile Infection
7. C. difficile Infection Pathogenesis
8. Gut Microbiota Changes in Patients with C. difficile Infection
9. Highly Virulent C. difficile Types
10. Impact of Highly Virulent C. difficile Types on Gut Microbiota
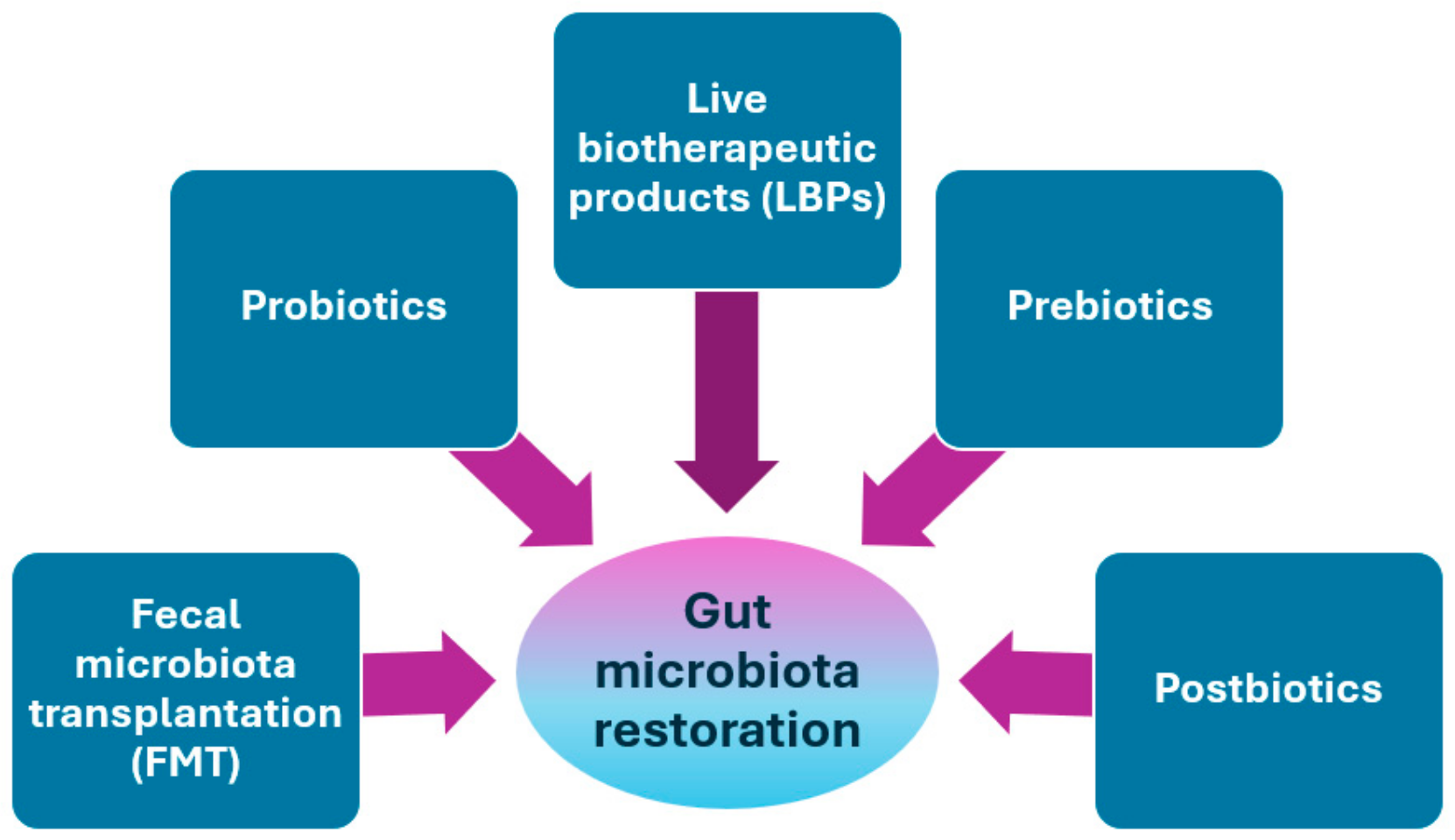
11. Microbial Interventions to Restore Gut Microbiota
11.1. Fecal Microbiota Transplantation (FMT)
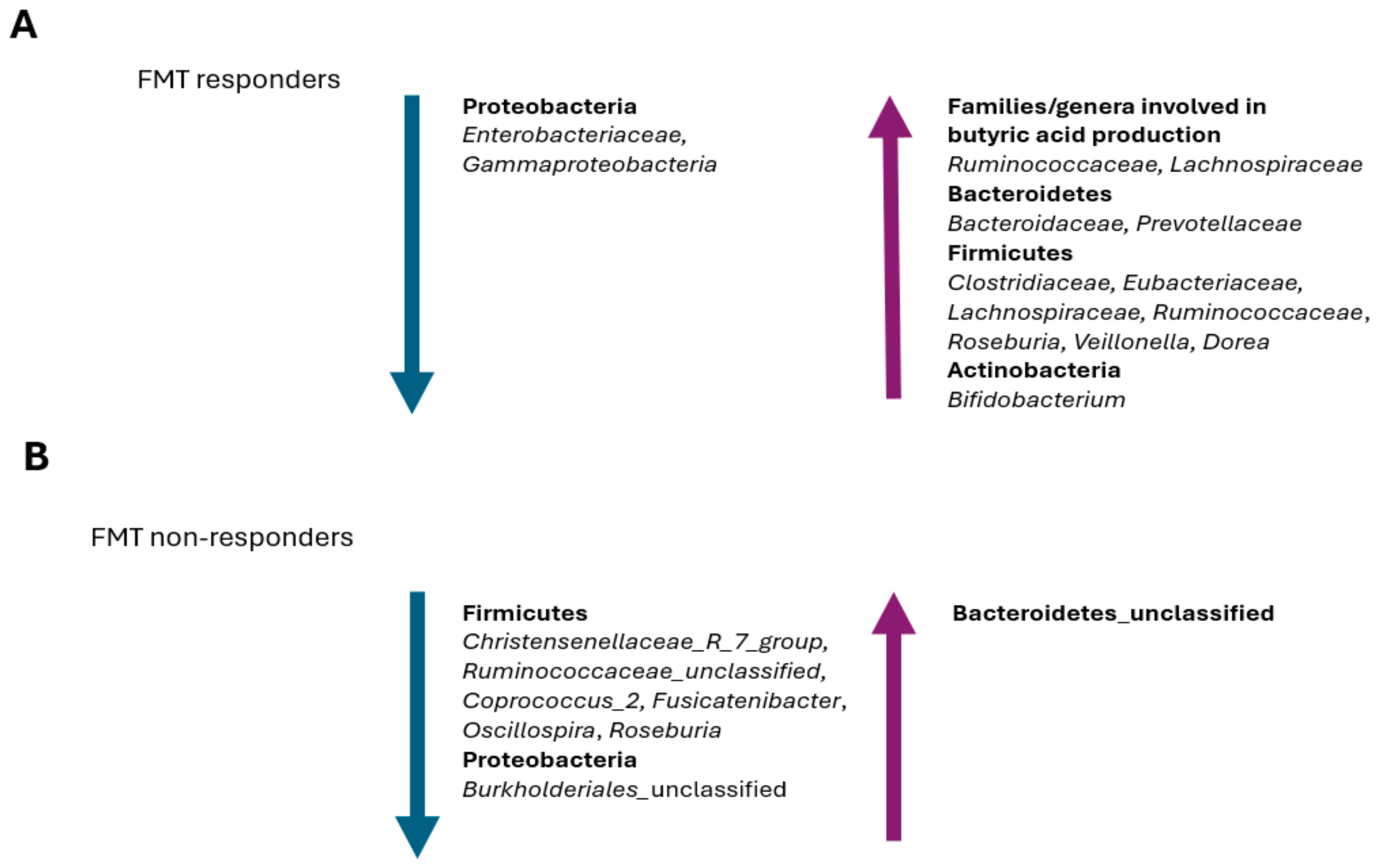
Gut Microbiota Changes in FMT Patients
11.2. Probiotics
11.3. Live Biotherapeutic Products (LBPs)
11.4. Prebiotics
11.5. Postbiotics
12. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Nasiri, M.J.; Goudarzi, M.; Hajikhani, B.; Ghazi, M.; Goudarzi, H.; Pouriran, R. Clostridioides (Clostridium) difficile infection in hospitalized patients with antibiotic-associated diarrhea: A systematic review and meta-analysis. Anaerobe 2018, 50, 32–37. [Google Scholar] [CrossRef]
- Crobach, M.J.T.; Vernon, J.J.; Loo, V.G.; Kong, L.Y.; Péchiné, S.; Wilcox, M.H.; Kuijper, E.J. Understanding Clostridium difficile colonization. Clin. Microbiol. Rev. 2018, 31, e00021-17. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.P.; Lee, J.C.; Lin, H.J.; Liu, H.C.; Wu, Y.H.; Tsai, P.J.; Ko, W.C. Clinical impact of Clostridium difficile colonization. J. Microbiol. Immunol. Infect. 2015, 48, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Leffler, D.A.; Lamont, J.T. Clostridium difficile infection. N. Engl. J. Med. 2015, 372, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Rupnik, M. Heterogeneity of large clostridial toxins: Importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 2008, 32, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Lacy, D.B. The role of toxins in Clostridium difficile infection. FEMS Microbiol. Rev. 2017, 41, 723–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Monaghan, T.; Yadegar, A.; Louie, T.; Kao, D. Insights into the evolving epidemiology of Clostridioides difficile infection and treatment: A global perspective. Antibiotics 2023, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Pedroza Matute, S.; Iyavoo, S. Exploring the gut microbiota: Lifestyle choices, disease associations, and personal genomics. Front. Nutr. 2023, 10, 1225120. [Google Scholar] [CrossRef] [PubMed]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel. Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Ufondu, A.; Lee, K.; Jayaraman, A. Emerging computational tools and models for studying gut microbiota composition and function. Curr. Opin. Biotechnol. 2020, 66, 301–331. [Google Scholar] [CrossRef] [PubMed]
- Alagiakrishnan, K.; Morgadinho, J.; Halverson, T. Approach to the diagnosis and management of dysbiosis. Front. Nutr. 2024, 11, 1330903. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.R.; Pike, C.M.; Parsons, R.J.; Rivera, A.J.; Foley, M.H.; McLaren, M.R.; Montgomery, S.A.; Theriot, C.M. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat. Commun. 2021, 12, 462. [Google Scholar] [CrossRef] [PubMed]
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: Pathogenesis and host defence. Nat. Rev. Microbiol. 2016, 14, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Loo, V.G.; Bourgault, A.M.; Poirier, L.; Lamothe, F.; Michaud, S.; Turgeon, N.; Toye, B.; Beaudoin, A.; Frost, E.H.; Gilca, R.; et al. Host and pathogen factors for Clostridium difficile infection and colonization. N. Engl. J. Med. 2011, 365, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Schäffler, H.; Breitrück, A. Clostridium difficile—From colonization to infection. Front. Microbiol. 2018, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Maida, M.; Burisch, J.; Simonelli, C.; Hold, G.; Ventimiglia, M.; Gasbarrini, A.; Cammarota, G. Efficacy of different faecal microbiota transplantation protocols for Clostridium difficile infection: A systematic review and meta-analysis. United Eur. Gastroenterol. J. 2018, 6, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, T.; Sun, J.; Liu, N. Fecal microbiota transplantation and health outcomes: An umbrella review of meta-analyses of randomized controlled trials. Front. Cell. Infect. Microbiol. 2022, 12, 899845. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Zahirović, A.; Bizjak, M.; Rupnik, M.; Štrukelj, B.; Berlec, A. New treatment approaches for Clostridioides difficile infections: Alternatives to antibiotics and fecal microbiota transplantation. Gut Microbes 2024, 16, 2337312. [Google Scholar] [CrossRef]
- Gonzales-Luna, A.J.; Carlson, T.J.; Garey, K.W. Gut microbiota changes associated with Clostridioides difficile infection and its various treatment strategies. Gut Microbes 2023, 15, 2223345. [Google Scholar] [CrossRef]
- Dixit, K.; Chaudhari, D.; Dhotre, D.; Shouche, Y.; Saroj, S. Restoration of dysbiotic human gut microbiome for homeostasis. Life Sci. 2021, 278, 119622. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Engevik, M.A.; Spinler, J.K.; Versalovic, J. Healthy human gastrointestinal microbiome: Composition and function after a decade of Exploration. Dig. Dis. Sci. 2020, 65, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The gut microbiota in the first decade of life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef]
- Schloissnig, S.; Arumugam, M.; Sunagawa, S.; Mitreva, M.; Tap, J.; Zhu, A.; Waller, A.; Mende, D.R.; Kultima, J.R.; Martin, J.; et al. Genomic variation landscape of the human gut microbiome. Nature 2013, 493, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, F. A possible perspective about the compositional models, evolution, and clinical meaning of human enterotypes. Microorganisms 2021, 9, 2341. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Ning, K. Stereotypes about enterotype: The old and new ideas. Genom. Proteom. Bioinform. 2019, 17, 4–12. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M.; Tilg, H.; van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Kachrimanidou, M.; Tsintarakis, E. Insights into the role of human gut microbiota in Clostridioides difficile infection. Microorganisms 2020, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Giel, J.L.; Sorg, J.A.; Sonenshein, A.L.; Zhu, J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE 2020, 5, e8740. [Google Scholar] [CrossRef]
- Antunes, L.C.M.; Finlay, B.B. A comparative analysis of the effect of antibiotic treatment and enteric infection on intestinal homeostasis. Gut Microbes 2011, 2, 105–108. [Google Scholar] [CrossRef]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.M.; Yalcinkaya, N.; Wu, Q.; Swennes, A.; Tessier, M.E.; Roberts, P.; Miyajima, F.; Savidge, T.; Sorg, J.A. Bile acid-independent protection against Clostridioides difficile infection. PLoS Pathog. 2021, 17, e1010015. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: Role of secondary bile acids. Cell Chem. Biol. 2019, 26, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Thanissery, R.; Winston, J.A.; Theriot, C.M. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 2017, 45, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Studer, N.; Desharnais, L.; Beutler, M.; Brugiroux, S.; Terrazos, M.A.; Menin, L.; Schürch, C.M.; McCoy, K.D.; Kuehne, S.A.; Minton, N.P.; et al. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell Infect. Microbiol. 2016, 6, 191. [Google Scholar] [CrossRef] [PubMed]
- Chen See, J.R.; Leister, J.; Wright, J.R.; Kruse, P.I.; Khedekar, M.V.; Besch, C.E.; Kumamoto, C.A.; Madden, G.R.; Stewart, D.B.; Lamendella, R. Clostridioides difficile infection is associated with differences in transcriptionally active microbial communities. Front. Microbiol. 2024, 15, 1398018. [Google Scholar] [CrossRef] [PubMed]
- Revolinski, S.L.; Munoz-Price, L.S. Clostridium difficile exposures, colonization, and the microbiome: Implications for prevention. Infect. Control. Hosp. Epidemiol. 2018, 39, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, R.D. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun. 1984, 45, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.M.; Sorg, J.A. Gut associated metabolites and their roles in Clostridioides difficile pathogenesis. Gut Microbes 2022, 14, 2094672. [Google Scholar] [CrossRef]
- O’Reilly, C.; O’Connor, P.M.; O’Sullivan, Ó.; Rea, M.C.; Hill, C.; Ross, R.P. Impact of nisin on Clostridioides difficile and microbiota composition in a faecal fermentation model of the human colon. J. Appl. Microbiol. 2022, 132, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. Clostridioides difficile infection: Microbe-microbe interactions and live biotherapeutics. Front. Microbiol. 2023, 14, 1182612. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.D.; Theriot, C.M. Contribution of inhibitory metabolites and competition for nutrients to colonization resistance against Clostridioides difficile by commensal Clostridium. Microorganisms 2021, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Moya, A.; Gosalbes, M.J.; Latorre, A. Colonization resistance of the gut microbiota against Clostridium difficile. Antibiotics 2015, 4, 337–357. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, e23. [Google Scholar] [CrossRef] [PubMed]
- Umesaki, Y.; Setoyama, H.; Matsumoto, S.; Imaoka, A.; Itoh, K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 1999, 67, 3504–3511. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Ariyoshi, T.; Kuroki, Y.; Eguchi, S.; Higashi, S.; Mori, T.; Nonogaki, T.; Iwasaki, K.; Yamashita, M.; Asai, N.; et al. Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci. Rep. 2021, 11, 15007. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Nagao-Kitamoto, H.; Kitamoto, S.; Kim, C.H.; Kamada, N. The butyrate-producing bacterium Clostridium butyricum suppresses Clostridioides difficile infection via neutrophil- and antimicrobial cytokine–dependent but GPR43/109a-independent mechanisms. J. Immunol. 2021, 206, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Osaki, T.; Hanawa, T.; Kurata, S.; Sugiyama, E.; Takahashi, M.; Tanaka, M.; Taguchi, H.; Kamiya, S. Establishment of an endogenous Clostridium difficile rat infection model and evaluation of the effects of Clostridium butyricum MIYAIRI 588 probiotic strain. Front. Microbiol. 2018, 9, 1264. [Google Scholar] [CrossRef]
- Lee, J.C.; Chiu, C.W.; Tsai, P.J.; Lee, C.C.; Huang, I.H.; Ko, W.C.; Hung, Y.P. Clostridium butyricum therapy for mild-moderate Clostridioides difficile infection and the impact of diabetes mellitus. Biosci. Microbiota Food Health 2022, 41, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Artacho, A.; Ott, S.J.; Moya, A.; Gosalbes, M.J.; Latorre, A. Structural and functional changes in the gut microbiota associated to Clostridium difficile infection. Front. Microbiol. 2014, 5, 335. [Google Scholar]
- Dai, Z.L.; Wu, G.; Zhu, W.Y. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768–1786. [Google Scholar] [CrossRef] [PubMed]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, e4548. [Google Scholar] [CrossRef] [PubMed]
- Rojo, D.; Gosalbes, M.J.; Ferrari, R.; Pérez-Cobas, A.E.; Hernández, E.; Oltra, R.; Buesa, J.; Latorre, A.; Barbas, C.; Ferrer, M.; et al. Clostridium difficile heterogeneously impacts intestinal community architecture but drives stable metabolome responses. ISME J. 2015, 9, 2206–2220. [Google Scholar] [CrossRef] [PubMed]
- Furuya-Kanamori, L.; Marquess, J.; Yakob, L.; Riley, T.V.; Paterson, D.L.; Foster, N.F.; Huber, C.A.; Clements, A.C. Asymptomatic Clostridium difficile colonization: Epidemiology and clinical implications. BMC Infect. Dis. 2015, 15, 516. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Soto Ocana, J.; Zackular, J.P. From nursery to nursing home: Emerging concepts in Clostridioides difficile pathogenesis. Infect. Immun. 2020, 88, e00934-19. [Google Scholar] [CrossRef] [PubMed]
- Shirley, D.A.; Tornel, W.; Warren, C.A.; Moonah, S. Clostridioides difficile infection in children: Recent updates on epidemiology, diagnosis, therapy. Pediatrics 2023, 52, e2023062307. [Google Scholar] [CrossRef] [PubMed]
- Semon, A.K.; Keenan, O.; Zackular, J.P. Clostridioides difficile and the microbiota early in life. J. Pediatr. Infect. Dis. Soc. 2021, 10 (Suppl. 3), S3–S7. [Google Scholar] [CrossRef]
- Tonooka, T.; Sakata, S.; Kitahara, M.; Hanai, M.; Ishizeki, S.; Takada, M.; Sakamoto, M.; Benno, Y. Detection and quantification of four species of the genus Clostridium in infant feces. Microbiol. Immunol. 2005, 49, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, M.; Baharav, N.; Melzer, E.; Regev-Yochay, G.; Yahav, D. Screening for asymptomatic Clostridioides difficile carriage among hospitalized patients: A narrative review. Infect. Dis. Ther. 2023, 12, 2223–2240. [Google Scholar] [CrossRef]
- De Roo, A.C.; Regenbogen, S.E. Clostridium difficile infection: An epidemiology update. Clin. Colon. Rectal. Surg. 2020, 33, 49–57. [Google Scholar] [CrossRef]
- Kyne, L.; Warny, M.; Qamar, A.; Kelly, C.P. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 2000, 342, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, M.H.; Gerding, D.N.; Poxton, I.R.; Kelly, C.; Nathan, R.; Birch, T.; Cornely, O.A.; Rahav, G.; Bouza, E.; Lee, C.; et al. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N. Engl. J. Med. 2017, 376, 305–317. [Google Scholar] [CrossRef]
- Dieterle, M.G.; Young, V.B. Reducing recurrence of C. difficile infection. Cell 2017, 169, 375. [Google Scholar] [CrossRef] [PubMed]
- Blixt, T.; Gradel, K.O.; Homann, C.; Seidelin, J.B.; Schonning, K.; Lester, A.; Houlind, J.; Stangerup, M.; Gottlieb, M.; Knudsen, J.D. Asymptomatic carriers contribute to nosocomial Clostridium difficile infection: A cohort study of 4508 patients. Gastroenterology 2017, 152, 1031–1041.e2. [Google Scholar] [CrossRef] [PubMed]
- Lanzas, C.; Dubberke, E.R.; Lu, Z.; Reske, K.A.; Gröhn, Y.T. Epidemiological model for Clostridium difficile transmission in healthcare settings. Infect. Control. Hosp. Epidemiol. 2011, 32, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Charis, A.; Ning Yu, M.; Lockhart, M.C.; McGuigan, C.C.; Wiuff, C.; Davey, P.G.; Donnan, P.T. Community-associated Clostridium difficile infection among older people in Tayside, Scotland, is associated with antibiotic exposure and care home residence: Cohort study with nested case–control. J. Antimicrob. Chemother. 2013, 68, 2927–2933. [Google Scholar]
- Wilcox, M.H.; Mooney, L.; Bendall, R.; Settle, C.D.; Fawley, W.N. A case-control study of community-associated Clostridium difficile infection. J. Antimicrob. Chemother. 2008, 62, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Zacharioudakis, I.M.; Zervou, F.N.; Pliakos, E.E.; Ziakas, P.D.; Mylonakis, E. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: A systematic review and meta-analysis. Am. J. Gastroenterol. 2015, 110, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Nissle, K.; Kopf, D.; Rösler, A. Asymptomatic and yet C. difficile-toxin positive? Prevalence and risk factors of carriers of toxigenic Clostridium difficile among geriatric in-patients. BMC Geriatr. 2016, 16, 185. [Google Scholar] [CrossRef]
- Rousseau, C.; Levenez, F.; Fouqueray, C.; Doré, J.; Collignon, A.; Lepage, P. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J. Clin. Microbiol. 2011, 49, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Sakata, S.; Tonooka, T.; Ishizeki, S.; Takada, M.; Sakamoto, M.; Fukuyama, M.; Benno, Y. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol. Lett. 2005, 243, 417–423. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere 2019, 3, e00041-18. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef]
- Tun, H.M.; Peng, Y.; Chen, B.; Konya, T.B.; Morales-Lizcano, N.P.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; et al. Ethnicity associations with Food sensitization are mediated by gut microbiota development in the first year of life. Gastroenterology 2021, 161, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Liu, X.; Jia, X.; Cheng, Y.; Luo, Y.; Yuan, L.; Wang, Y.; Zhao, C.; Guo, S.; Li, L.; et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci. Rep. 2014, 4, 7485. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Lepage, P.; Jolivet, S.; Delannoy, J.; Mesa, V.; Ancel, P.Y.; Rozé, J.C.; Butel, M.J.; Barbut, F.; Aires, J. Gut microbiota diversity of preterm neonates is associated with Clostridioides difficile colonization. Front. Cell Infect. Microbiol. 2022, 12, 907323. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.Y.; Zhang, H.; Brannan, L.E.; Carman, R.J.; Boone, J.H. Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Microbiome 2016, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Th, L.; Dong, D.; Jiang, C.; Li, Z.; Wang, X.; Peng, Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 2015, 34, 1–7. [Google Scholar]
- Vázquez-Cuesta, S.; Villar, L.; García, N.L.; Fernández, A.I.; Olmedo, M.; Alcalá, L.; Marín, M.; Muñoz, P.; Bouza, E.; Reigadas, E. Characterization of the gut microbiome of patients with Clostridioides difficile infection, patients with non-C. difficile diarrhea, and C. difficile-colonized patients. Front. Cell Infect. Microbiol. 2023, 13, 1130701. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.T.; Ducarmon, Q.R.; Terveer, E.M.; Harmanus, C.; Sanders, I.M.J.G.; Verduin, K.M.; Kuijper, E.J.; Zwittink, R.D. The bacterial gut microbiota of adult patients infected, colonized or noncolonized by Clostridioides difficile. Microorganisms 2020, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.; Miller, M.A.; Edens, T.J.; Mehrotra, S.; Dewar, K.; Manges, A.R. Bloom and bust: Intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Henderickx, J.G.E.; Crobach, M.J.T.; Terveer, E.M.; Smits, W.K.; Kuijper, E.J.; Zwittink, R.D. Fungal and bacterial gut microbiota differ between Clostridioides difficile colonization and infection. Microbiome Res. Rep. 2023, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, M.; Meng, J.; Wang, L.; Chen, M. A review of the interaction between diet composition and gut microbiota and its impact on associated disease. J. Future Foods 2024, 4, 221–232. [Google Scholar] [CrossRef]
- Britton, R.A.; Young, V.B. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 2014, 146, 1547–1553. [Google Scholar] [CrossRef] [PubMed]
- Vila, A.V.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.M.A.E.; Masclee, A.A.M.; Fu, J.; et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Hryckowian, A.J.; Treuren, W.V.; Smits, S.A.; Davis, N.M.; Gardner, J.O.; Bouley, D.M.; Sonnenburg, J.L. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat. Microbiol. 2018, 3, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Lange, K.; Buerger, M.; Stallmach, A.; Bruns, T. Effects of antibiotics on gut microbiota. Dig. Dis. 2016, 34, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, D.; Nigam, P.S. Antibiotic-therapy-induced gut dysbiosis affecting gut microbiota-brain axis and cognition: Restoration by intake of probiotics and synbiotics. Int. J. Mol Sci. 2023, 24, 3074. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Neulinger, S.C.; Heinsen, F.A.; Knecht, C.; Schilhabel, A.; Schmitz, R.A.; Zimmermann, A.; dos Santos, V.M.; Ferrer, M.; Rosenstiel, P.C.; et al. Effects of beta-lactam antibiotics and fluoroquinolones on human gut microbiota in relation to Clostridium difficile associated diarrhea. PLoS ONE 2014, 9, e89417. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; de Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota—A systematic review. J. Infect. 2019, 79, 471–489. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, K.; Khanna, S. Gut microbiome and Clostridioides difficile infection: A closer look at the microscopic interface. Ther. Adv. Gastroenterol. 2021, 14, 1756284821994736. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.; O’Neill, F.J.; Wilcox, M.H. Effects of cefotaxime and desacetylcefotaxime upon Clostridium difficile proliferation and toxin production in a triple-stage chemostat model of the human gut. J. Antimicrob. Chemother. 2003, 52, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Altman, K.W.; Chhaya, V.; Hammer, N.D.; Pavlova, S.; Vesper, B.J.; Tao, L.; Radosevich, J.A. Effect of proton pump inhibitor pantoprazole on growth and morphology of oral Lactobacillus strains. Laryngoscope 2008, 118, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Clooney, A.G.; Bernstein, C.N.; Leslie, W.D.; Vagianos, K.; Sargent, M.; Laserna-Mendieta, E.J.; Claesson, M.J.; Targownik, L.E. A comparison of the gut microbiome between long-term users and non-users of proton pump inhibitors. Aliment. Pharmacol. Ther. 2016, 43, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Indiani, C.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.F.C.; Darrieux, M.; Parisotto, T.M. Childhood obesity and Firmicutes/Bacteroidetes ratio in the gut microbiota: A systematic review. Child Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Buddle, J.E.; Fagan, R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence 2023, 14, 2150452. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Johanesen, P.A.; Carter, G.P.; Rose, E.; Lyras, D. Clostridium difficile virulence factors: Insights into an anaerobic spore-forming pathogen. Gut Microbes 2014, 5, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Hundsberger, T.; Leukel, P.; Sauerborn, M.; Eichel-Streiber, C.V. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 1996, 181, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.H.; Tang, Y.J.; Silva, J., Jr. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 2000, 18, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Kordus, S.L.; Thomas, A.K.; Lacy, D.B. Clostridioides difficile toxins: Mechanisms of action and antitoxin therapeutics. Nat. Rev. Microbiol. 2022, 20, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Skinner, A.M.; Phillips, S.T.; Merrigan, M.M.; O’Leary, K.J.; Sambol, S.P.; Siddiqui, F.; Peterson, L.R.; Gerding, D.N.; Johnson, S. The relative role of toxins A and B in the virulence of Clostridioides difficile. J. Clin. Med. 2020, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Gerding, D.N.; Johnson, S.; Rupnik, M.; Aktories, K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes 2014, 5, 15–27. [Google Scholar] [CrossRef]
- Papatheodorou, P.; Minton, N.P.; Aktories, K.; Barth, H. An updated view on the cellular uptake and mode-of-action of Clostridioides difficile toxins. Adv. Exp. Med. Biol. 2024, 1435, 219–247. [Google Scholar] [PubMed]
- Aktories, K.; Wegner, A. Mechanisms of the cytopathic action of actin-ADP-ribosylating toxins. Mol Microbiol. 1992, 6, 2905–2908. [Google Scholar] [CrossRef] [PubMed]
- Schwan, C.; Stecher, B.; Tzivelekidis, T.; van Ham, M.; Rohde, M.; Hardt, W.D.; Wehland, J.; Aktories, K. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009, 5, e1000626. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.J.; Ballard, J.D. Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol. Mol. Biol. Rev. 2013, 77, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Lanis, J.M.; Barua, S.; Ballard, J.D. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 2010, 6, e1001061. [Google Scholar] [CrossRef]
- Theriot, C.M.; Young, V.B. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu. Rev. Microbiol. 2015, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Young, V.B. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes 2014, 5, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.M.; Rogers, M.A.; Ring, C.; Mogle, J.; Petrosino, J.P.; Young, V.B.; Aronoff, D.M.; Schloss, P.D. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 2014, 5, e01021-14. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.D.; Ann, H.W.; Lee, W.J.; Kim, J.H.; Seong, H.; Kim, J.H.; Ahn, J.Y.; Jeong, S.J.; Ku, N.S.; Yeom, J.S.; et al. Characteristics of faecal microbiota in Korean patients with Clostridioides difficile-associated diarrhea. Infect. Chemother. 2019, 51, 365–375. [Google Scholar] [CrossRef]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Maier, L.; Hardt, W.D. ’Blooming’ in the gut: How dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 2013, 11, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Yi, J.; Kim, J.H.; Lee, S.; Moon, H.W. Composition of gut microbiota in patients with toxigenic Clostridioides (Clostridium) difficile: Comparison between subgroups according to clinical criteria and toxin gene load. PLoS ONE 2019, 14, e0212626. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Taminiau, B.; Rodriguez, C.; Daube, G. Gut microbiota composition associated with Clostridioides difficile colonization and infection. Pathogens 2022, 11, 781. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Arguello, E.S.; Jenq, R.R.; Littmann, E.; Kim, G.J.; Miller, L.C.; Ling, L.; Figueroa, C.; Robilotti, E.; Perales, M.A.; et al. Protective factors in the intestinal microbiome against Clostridium difficile infection in recipients of allogeneic hematopoietic stem cell transplantation. J. Infect. Dis. 2017, 215, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Fishbein, S.R.; Robinson, J.I.; Hink, T.; Reske, K.A.; Newcomer, E.P.; Burnham, C.D.; Henderson, J.P.; Dubberke, E.R.; Dantas, G. Multi-omics investigation of Clostridioides difficile-colonized patients reveals pathogen and commensal correlates of C. difficile pathogenesis. eLife 2022, 11, e72801. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Labbe, A.; Loo, V.G.; Atherton, J.K.; Behr, M.A.; Masson, L.; Tellis, P.A.; Brousseau, R. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridium difficile-associated disease. J. Infect. Dis. 2010, 202, 1877–1884. [Google Scholar] [CrossRef]
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The gut bacterial community potentiates Clostridioides difficile infection severity. mBio 2022, 13, e0118322. [Google Scholar] [CrossRef] [PubMed]
- Magdy Wasfy, R.; Mbaye, B.; Borentain, P.; Tidjani Alou, M.; Murillo Ruiz, M.L.; Caputo, A.; Andrieu, C.; Armstrong, N.; Million, M.; Gerolami, R. Ethanol-producing Enterocloster bolteae is enriched in chronic hepatitis B-associated gut dysbiosis: A case-control culturomics Study. Microorganisms 2023, 11, 2437. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Berkell, M.; Mysara, M.; Xavier, B.B.; van Werkhoven, C.H.; Monsieurs, P.; Lammens, C.; Ducher, A.; Vehreschild, M.J.G.T.; Goossens, H.; de Gunzburg, J.; et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 2021, 12, 2241. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Montassier, E.; Schmidt, B.; Patel, R.; Knights, D.; Pardi, D.S.; Kashyap, P. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment. Pharmacol. Ther. 2016, 44, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Gazzola, A.; Panelli, S.; Corbella, M.; Merla, C.; Comandatore, F.; De Silvestri, A.; Piralla, A.; Zuccaro, V.; Bandi, C.; Marone, P.; et al. Microbiota in Clostridioides difficile-associated diarrhea: Comparison in recurrent and non-recurrent infections. Biomedicines 2020, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Chen, C.; Bobr, A.; Yao, D.; Lu, Y.; Nelson, V.M.; Sadowsky, M.J.; Khoruts, A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G310–G319. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, J.R.; Kearney, S.; Li, N.; Bogart, E.; Bullock, K.; Gerber, G.K.; Bry, L.; Clish, C.B.; Alm, E.; Korzenik, J.R. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 2016, 43, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.K.; Brensinger, C.M.; Wu, Q.; Lewis, J.D. Increasing incidence of multiply recurrent Clostridium difficile infection in the United States: A cohort study. Ann. Intern. Med. 2017, 167, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Davies, K.; Barbut, F. Ribotypes and new virulent strains across Europe. Adv. Exp. Med. Biol. 2024, 1435, 151–168. [Google Scholar] [PubMed]
- McDonald, L.C.; Killgore, G.E.; Thompson, A.; Owens, R.C.; Kazakova, S.V.; Sambol, S.P.; Johnson, S.; Gerding, D.N. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 2005, 353, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Beldavs, Z.G.; Winston, L.G.; Dumyati, G.; Holzbauer, S.; Dunn, J.; Farley, M.M.; Lyons, C.; Johnston, H.; Phipps, E.; et al. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin. Infect. Dis. 2014, 58, 1394–1400. [Google Scholar]
- Bauer, M.P.; Notermans, D.W.; van Benthem, B.H.; Brazier, J.S.; Wilcox, M.H.; Rupnik, M.; Monnet, D.L.; van Dissel, J.T.; Kuijper, E.J.; Group, E.S. Clostridium difficile infection in Europe: A hospital-based survey. Lancet 2011, 377, 63–73. [Google Scholar] [CrossRef]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar]
- Curry, S.R.; Marsh, J.W.; Muto, C.A.; O’Leary, M.M.; Pasculle, A.W.; Harrison, L.H. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 2007, 45, 215–221. [Google Scholar] [CrossRef]
- Warny, M.; Pepin, J.; Fang, A.; Killgore, G.; Thompson, A.; Brazier, J.; Frost, E.; McDonald, L.C. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 2005, 366, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Meléndez, A.; Morfin-Otero, R.; Villarreal-Treviño, L.; Baines, S.D.; Camacho-Ortíz, A.; Garza-González, E. Molecular epidemiology of predominant and emerging Clostridioides difficile ribotypes. J. Microbiol. Methods 2020, 175, 105974. [Google Scholar] [CrossRef]
- Spigaglia, P.; Mastrantonio, P.; Barbanti, F. Antibiotic resistances of Clostridioides difficile. Adv. Exp. Med. Biol. 2024, 1435, 169–198. [Google Scholar]
- Goorhuis, A.; Bakker, D.; Corver, J.; Debast, S.B.; Harmanus, C.; Notermans, D.W.; Bergwerff, A.A.; Dekker, F.W.; Kuijper, E.J. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 2008, 47, 1162–1170. [Google Scholar] [CrossRef]
- Knight, D.R.; Kullin, B.; Androga, G.O.; Barbut, F.; Eckert, C.; Johnson, S.; Spigaglia, P.; Tateda, K.; Tsai, P.J.; Riley, T.V. Evolutionary and genomic insights into Clostridioides difficile sequence type 11: A diverse zoonotic and antimicrobial-resistant lineage of global One Health importance. mBio 2019, 10, e00446-19. [Google Scholar] [CrossRef] [PubMed]
- Stabler, R.A.; Dawson, L.F.; Valiente, E.; Cairns, M.D.; Martin, M.J.; Donahue, E.H.; Riley, T.V.; Songer, J.G.; Kuijper, E.J.; Dingle, K.E.; et al. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS ONE 2012, 7, e31559. [Google Scholar] [CrossRef]
- Nyc, O.; Pituch, H.; Matejkova, J.; Obuch-Woszczatynski, P.; Kuijper, E.J. Clostridium difficile PCR ribotype 176 in the Czech Republic and Poland. Lancet 2011, 377, 1407. [Google Scholar] [CrossRef] [PubMed]
- Valiente, E.; Dawson, L.F.; Cairns, M.D.; Stabler, R.A.; Wren, B.W. Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J. Med. Microbiol. 2012, 61, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Khanna, S. Community-acquired Clostridium difficile infection: An increasing public health threat. Infect. Drug Resist. 2014, 7, 63–72. [Google Scholar] [PubMed]
- Jenior, M.L.; Leslie, J.L.; Young, V.B.; Schloss, P.D. Clostridium difficile alters the structure and metabolism of distinct cecal microbiomes during initial infection to promote sustained colonization. mSphere 2018, 3, e00261-18. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.; Auchtung, J.; Collins, J.; Britton, R. Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates. Infect. Immun. 2014, 82, 2815–2825. [Google Scholar] [CrossRef] [PubMed]
- Skraban, J.; Dzeroski, S.; Zenko, B.; Mongus, D.; Gangl, S.; Rupnik, M. Gut microbiota patterns associated with colonization of different Clostridium difficile ribotypes. PLoS ONE 2013, 8, e58005. [Google Scholar] [CrossRef] [PubMed]
- Horvat, S.; Mahnic, A.; Breskvar, M.; Dzeroski, S.; Rupnik, M. Evaluating the effect of Clostridium difficile conditioned medium on fecal microbiota community structure. Sci. Rep. 2017, 27, 16448. [Google Scholar] [CrossRef]
- Horvat, S.; Rupnik, M. Interactions between Clostridioides difficile and fecal microbiota in vitro batch model: Growth, sporulation, and microbiota changes. Front. Microbiol. 2018, 9, 1633. [Google Scholar] [CrossRef] [PubMed]
- Horvat, S.; Mahnic, A.; Makuc, D.; Pečnik, K.; Plavec, J.; Rupnik, M. Children gut microbiota exhibits a different composition and metabolic profile after in vitro exposure to Clostridioides difficile and increases its sporulation. Front. Microbiol. 2022, 13, 1042526. [Google Scholar] [CrossRef]
- Vincent, C.; Manges, A.R. Antimicrobial use, human gut microbiota and Clostridium difficile colonization and infection. Antibiotics 2015, 4, 230–253. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, I.M.; Chifiriuc, M.C.; Pircalabioru, G.G.; Filip, R.; Bolocan, A.; Lazăr, V.; Diţu, L.M.; Bleotu, C. Gut dysbiosis and Clostridioides difficile infection in neonates and adults. Front. Microbiol. 2022, 12, 651081. [Google Scholar] [CrossRef] [PubMed]
- Stallhofer, J.; Steube, A.; Katzer, K.; Stallmach, A. Microbiota-based therapeutics as new standard-of-care treatment for recurrent Clostridioides difficile infection. Visc. Med. 2024, 40, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Wortelboer, K.; Nieuwdorp, M.; Herrema, H. Fecal microbiota transplantation beyond Clostridioides difficile infections. EBioMedicine 2019, 44, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Masucci, L.; Ianiro, G.; Bibbo, S.; Dinoi, G.; Costamagna, G.; Sanguinetti, M.; Gasbarrini, A. Randomised clinical trial: Faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 2015, 41, 835–843. [Google Scholar] [CrossRef]
- Du, C.; Luo, Y.; Walsh, S.; Grinspan, A. Oral fecal microbiota transplant capsules are safe and effective for recurrent Clostridioides difficile infection: A systematic review and meta-analysis. J. Clin. Gastroenterol. 2021, 55, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Gangwani, M.K.; Aziz, M.; Aziz, A.; Priyanka, F.; Weissman, S.; Phan, K.; Dahiya, D.S.; Ahmed, Z.; Sohail, A.H.; Lee-Smith, W.; et al. Fresh versus frozen versus lyophilized fecal microbiota transplant for recurrent Clostridium difficile infection: A systematic review and network meta-analysis. J. Clin. Gastroenterol. 2023, 57, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Ramai, D.; Zakhia, K.; Fields, P.J.; Ofosu, A.; Patel, G.; Shahnazarian, V.; Lai, J.K.; Dhaliwal, A.; Reddy, M.; Chang, S. Fecal microbiota transplantation (FMT) with colonoscopy is superior to enema and nasogastric tube while comparable to capsule for the treatment of recurrent Clostridioides difficile infection: A systematic review and meta-analysis. Dig. Dis. Sci. 2021, 66, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Hvas, C.L.; Dahl Jørgensen, S.M.; Jørgensen, S.P.; Storgaard, M.; Lemming, L.; Hansen, M.M.; Erikstrup, C.; Dahlerup, J.F. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent Clostridium difficile infection. Gastroenterology 2019, 156, 1324–1332.e3. [Google Scholar] [CrossRef] [PubMed]
- Rokkas, T.; Gisbert, J.P.; Gasbarrini, A.; Hold, G.L.; Tilg, H.; Malfertheiner, P.; Megraud, F.; O’Morain, C.A. A network meta-analysis of randomized controlled trials exploring the role of fecal microbiota transplantation in recurrent Clostridium difficile infection. United Eur. Gastroenterol. J. 2019, 7, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, W.; Li, P.; Wen, Q.; Cui, B.; Zhang, F. Washed preparation of faecal microbiota changes the transplantation related safety, quantitative method and delivery. Microb. Biotechnol. 2022, 15, 2439–2449. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ai, R.J.; Xu, J.; Wen, Q.; Pan, H.Q.; Zhang, Z.H.; Ning, W.; Fang, Y.; Ding, D.F.; Wang, Q.; et al. Washed microbiota transplantation for Clostridioides difficile infection: A national multicenter real-world study. J. Dig. Dis. 2023, 24, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Kelly, C.R.; Mullish, B.H.; Allegretti, J.R.; Kassam, Z.; Putignani, L.; Fischer, M.; Keller, J.J.; Costello, S.P.; et al. International consensus conference on stool banking for faecal microbiota transplantation in clinical practice. Gut 2019, 68, 2111–2121. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.J.; Ooijevaar, R.E.; Hvas, C.L.; Terveer, E.M.; Lieberknecht, S.C.; Högenauer, C.; Arkkila, P.; Sokol, H.; Gridnyev, O.; Mégraud, F.; et al. A standardised model for stool banking for faecal microbiota transplantation: A consensus report from a multidisciplinary UEG working group. United Eur. Gastroenterol. J. 2021, 9, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.A.; Babakhani, F.; Sears, P.; Nguyen, L.; Sorg, J.A. Both fidaxomicin and vancomycin inhibit outgrowth of Clostridium difficile spores. Antimicrob. Agents Chemother. 2013, 57, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Khoruts, A.; Sadowsky, M.J. Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Seekatz, A.M.; Aas, J.; Gessert, C.E.; Rubin, T.A.; Saman, D.M.; Bakken, J.S.; Young, V.B. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 2014, 5, e00893-14. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Chang, T.E.; Wang, Y.P.; Lee, K.C.; Lin, Y.T.; Chiou, J.J.; Huang, C.W.; Yang, U.C.; Li, F.Y.; Huang, H.C.; et al. Alteration of gut microbial composition associated with the therapeutic efficacy of fecal microbiota transplantation in Clostridium difficile infection. J. Formos. Med. Assoc. 2022, 121, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Shankar, V.; Hamilton, M.J.; Khoruts, A.; Kilburn, A.; Unno, T.; Paliy, O.; Sadowsky, M.J. Species and genus level resolution analysis of gut microbiota in Clostridium difficile patients following fecal microbiota transplantation. Microbiome 2014, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Vazquez-Baeza, Y.; Gonzalez, A.; Weiss, S.; Schmidt, B.; Muniz-Pedrogo, D.A.; Rainey, J.F.; Kammer, P.; Nelson, H.; Sadowsky, M.; et al. Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 2017, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Aggarwala, V.; Mogno, I.; Li, Z.; Yang, C.; Britton, G.J.; Chen-Liaw, A.; Mitcham, J.; Bongers, G.; Gevers, D.; Clemente, J.C.; et al. Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes. Nat. Microbiol. 2021, 6, 1309–1318. [Google Scholar] [CrossRef]
- Wei, S.; Bahl, M.I.; Baunwall, S.M.D.; Dahlerup, J.F.; Hvas, C.L.; Licht, T.R. Gut microbiota differs between treatment outcomes early after fecal microbiota transplantation against recurrent Clostridioides difficile infection. Gut Microbes 2022, 14, 2084306. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Cui, J.; Ye, C.; Zhao, J.; Yang, B.; Xu, Y.; Ji, S.; Wang, L.; Lv, X.; Ma, C.; et al. Depletion of butyrate-producing microbes of the Firmicutes predicts nonresponse to FMT therapy in patients with recurrent Clostridium difficile infection. Gut Microbes 2023, 15, 2236362. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Kaiser, T.; Vaughn, B.P.; Graiziger, C.T.; Hamilton, M.J.; Rehman, T.U.; Song, K.; Khoruts, A.; Sadowsky, M.J. Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation. Microbiome 2018, 6, 166. [Google Scholar] [CrossRef] [PubMed]
- Fachi, J.L.; Felipe, J.S.; Pral, L.P.; da Silva, B.K.; Corrêa, R.O.; de Andrade, M.C.P.; da Fonseca, D.M.; Basso, P.J.; Câmara, N.O.S.; de Sales, E.S.É.; et al. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep. 2019, 27, 750–761.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, J.; Gao, W.; Li, L.; Li, P.; Zhang, L.; Gong, Y.N.; Peng, X.; Xi, J.J.; Chen, S.; et al. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature 2014, 513, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, J.J.; Allegretti, J.R.; Gibson, T.E.; McClure, E.; Delaney, M.; Bry, L.; Gerber, G.K. Gut metabolites predict Clostridioides difficile recurrence. Microbiome 2022, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Pakpour, S.; Bhanvadia, A.; Zhu, R.; Amarnani, A.; Gibbons, S.M.; Gurry, T.; Alm, E.J.; Martello, L.A. Identifying predictive features of Clostridium difficile infection recurrence before, during, and after primary antibiotic treatment. Microbiome 2017, 5, 148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhong, H.; Li, Y.; Shi, Z.; Ren, H.; Zhang, Z.; Zhou, X.; Tang, S.; Han, X.; Lin, Y.; et al. Sex- and age-related trajectories of the adult human gut microbiota shared across populations of different ethnicities. Nat. Aging 2021, 1, 87–100. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.C.; Vatanen, T.; Cutfield, W.S.; O’Sullivan, J.M. The super-donor phenomenon in fecal microbiota transplantation. Front. Cell. Infect. Microbiol. 2019, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Valles-Colomer, M.; Bacigalupe, R.; Vieira-Silva, S.; Suzuki, S.; Darzi, Y.; Tito, R.Y.; Yamada, T.; Segata, N.; Raes, J.; Falony, G. Variation and transmission of the human gut microbiota across multiple familial generations. Nat. Microbiol. 2022, 7, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Almeida, A.; Kumar, N.; Vervier, K.; Adoum, A.T.; Viciani, E.; Dawson, N.J.R.; Forster, S.C.; Cormie, C.; Goulding, D.; et al. Host adaptation in gut Firmicutes is associated with sporulation loss and altered transmission cycle. Genome Biol. 2021, 22, 204. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.Y.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73 (Suppl. 2), 386S–392S. [Google Scholar] [CrossRef] [PubMed]
- Liévin-Le Moal, V.; Servin, A.L. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: From probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin. Microbiol. Rev. 2014, 27, 167–199. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Surawicz, C.M.; Greenberg, R.N.; Fekety, R.; Elmer, G.W.; Moyer, K.A.; Melcher, S.A.; Bowen, K.E.; Cox, J.L.; Noorani, Z.; et al. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA 1994, 271, 913–918. [Google Scholar] [CrossRef]
- Surawicz, C.M.; McFarland, L.V.; Greenberg, R.N.; Rubin, M.; Fekety, R.; Mulligan, M.E.; Garcia, R.J.; Brandmarker, S.; Bowen, K.; Borjal, D.; et al. The search for a better treatment for recurrent Clostridium difficile disease: Use of high-dose vancomycin combined with Saccharomyces boulardii. Clin. Infect. Dis. 2000, 31, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Wullt, M.; Hagslatt, M.L.; Odenholt, I. Lactobacillus plantarum 299v for the treatment of recurrent Clostridium difficile-associated diarrhoea: A double-blind, placebo-controlled trial. Scand. J. Infect. Dis. 2003, 35, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Klarin, B.; Wullt, M.; Palmquist, I.; Molin, G.; Larsson, A.; Jeppsson, B. Lactobacillus plantarum 299v reduces colonisation of Clostridium difficile in critically ill patients treated with antibiotics. Acta Anaesthesiol. Scand. 2008, 52, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Kujawa-Szewieczek, A.; Adamczak, M.; Kwiecień, K.; Dudzicz, S.; Gazda, M.; Więcek, A. The effect of Lactobacillus plantarum 299v on the incidence of Clostridium difficile infection in high-risk patients treated with antibiotics. Nutrients 2015, 7, 10179–10188. [Google Scholar] [CrossRef] [PubMed]
- Barker, A.K.; Duster, M.; Valentine, S.; Hess, T.; Archbald-Pannone, L.; Guerrant, R.; Safdar, N. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J. Antimicrob. Chemother. 2017, 72, 3177–3180. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Sun, Y.; Mu, P.; Zheng, T.; Mu, H.; Deng, F.; Deng, Y.; Wen, J. Lactobacillus rhamnosus GG supplementation modulates the gut microbiota to promote butyrate production, protecting against deoxynivalenol exposure in nude mice. Biochem. Pharmacol. 2020, 175, 113868. [Google Scholar] [CrossRef] [PubMed]
- Carucci, L.; Nocerino, R.; Paparo, L.; De Filippis, F.; Coppola, S.; Giglio, V.; Cozzolino, T.; Valentino, V.; Sequino, G.; Bedogni, G.; et al. Therapeutic effects elicited by the probiotic Lacticaseibacillus rhamnosus GG in children with atopic dermatitis. The results of the ProPAD trial. Pediatr. Allergy Immunol. 2022, 33, e13836. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.W.; Tsai, P.J.; Lee, C.C.; Ko, W.C.; Hung, Y.P. Application of microbiome management in therapy for Clostridioides difficile Infections: From fecal microbiota transplantation to probiotics to microbiota-preserving antimicrobial agents. Pathogens 2021, 10, 649. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, J.Z.; Yap, C.; Lytvyn, L.; Lo, C.K.; Beardsley, J.; Mertz, D.; Johnston, B.C. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst. Rev. 2017, 12, CD006095. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zeng, L.; Yan, Z.; Jia, J.; Gao, J.; Wei, Y. The mechanisms and safety of probiotics against toxigenic Clostridium difficile. Expert. Rev. Anti. Infect. Ther. 2020, 18, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, J.Y.; Peng, X.; Xiao, K.Y.; Xu, Q.; Wang, C. Which probiotic has the best effect on preventing Clostridium difficile-associated diarrhea? A systematic review and network meta-analysis. J. Dig. Dis. 2020, 21, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Varela, L.; Gueimonde, M.; Ruas-Madiedo, P. Probiotics for prevention and treatment of Clostridium difficile infection. Adv. Exp. Med. Biol. 2018, 1050, 161–176. [Google Scholar] [PubMed]
- Plummer, S.; Weaver, M.A.; Harris, J.C.; Dee, P.; Hunter, J. Clostridium difficile pilot study: Effects of probiotic supplementation on the incidence of C. difficile diarrhoea. Int. Microbiol. 2004, 7, 59–62. [Google Scholar] [PubMed]
- Van Prehn, J.; Reigadas, E.; Vogelzang, E.H.; Bouza, E.; Hristea, A.; Guery, B.; Krutova, M.; Norén, T.; Allerberger, F.; Coia, J.E.; et al. European society of clinical microbiology and infectious diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin. Microbiol. Infect. 2021, 27 (Suppl. 2), S1–S21. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.R.; Fischer, M.; Allegretti, J.R.; LaPlante, K.; Stewart, D.B.; Limketkai, B.N.; Stollman, N.H. ACG clinical guidelines: Prevention, diagnosis, and treatment of Clostridioides difficile Infections. Am. J. Gastroenterol. 2021, 116, 1124–1147. [Google Scholar] [CrossRef] [PubMed]
- Besselink, M.G.; van Santvoort, H.C.; Buskens, E.; Boermeester, M.A.; van Goor, H.; Timmerman, H.M.; Nieuwenhuijs, V.B.; Bollen, T.L.; van Ramshorst, B.; Witteman, B.J.; et al. Probiotic prophylaxis in predicted severe acute pancreatitis: A randomised, double-blind, placebo-controlled trial. Lancet 2008, 371, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Assi, M.; Lee, C.; Yoho, D.; Louie, T.; Knapple, W.; Aguilar, H.; Garcia-Diaz, J.; Wang, G.P.; Berry, S.M.; et al. Efficacy and safety of RBX2660 in PUNCH CD3, a phase III, randomized, double-blind, placebo-controlled trial with a bayesian primary analysis for the prevention of recurrent Clostridioides difficile infection. Drugs 2022, 82, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Papazyan, R.; Ferdyan, N.; Srinivasan, K.; Gonzalez, C.; Shannon, W.D.; Blount, K.; Fuchs, B.C. Human fecal bile acid analysis after investigational microbiota-based live biotherapeutic delivery for recurrent Clostridioides difficile infection. Microorganisms 2023, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Doosetty, S.; Umeh, C.; Eastwood, W.; Samreen, I.; Penchala, A.; Kaur, H.; Chilinga, C.; Kaur, G.; Mohta, T.; Nakka, S.; et al. Efficacy of Fecal Microbiota (REBYOTA) in recurrent Clostridium difficile infections: A systematic review and meta-analysis. Cureus 2024, 16, e58862. [Google Scholar] [CrossRef] [PubMed]
- Feuerstadt, P.; Louie, T.J.; Lashner, B.; Wang, E.E.L.; Diao, L.; Bryant, J.A.; Sims, M.; Kraft, C.S.; Cohen, S.H.; Berenson, C.S.; et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 2022, 386, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Louie, T.; Golan, Y.; Khanna, S.; Bobilev, D.; Erpelding, N.; Fratazzi, C.; Carini, M.; Menon, R.; Ruisi, M.; Norman, J.M.; et al. VE303, a defined bacterial consortium, for prevention of recurrent Clostridioides difficile infection: A randomized clinical trial. JAMA 2023, 329, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Gerding, D.N.; Meyer, T.; Lee, C.; Cohen, S.H.; Murthy, U.K.; Poirier, A.; Van Schooneveld, T.C.; Pardi, D.S.; Ramos, A.; Barron, M.A.; et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: A randomized clinical trial. JAMA 2015, 313, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Delzenne, N.M.; Cani, P.D.; Walter, J. Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.; Montero, I.; Fernandez-Gosende, M.; Martinez, N.; Cantabrana, C.H.; Rios-Covian, D. Past, present, and future of microbiome-based therapies. Microbiome Res. Rep. 2024, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Rätsep, M.; Kõljalg, S.; Sepp, E.; Smidt, I.; Truusalu, K.; Songisepp, E.; Stsepetova, J.; Naaber, P.; Mikelsaar, R.H.; Mikelsaar, M. A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe 2017, 47, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Varela, L.; Hernández-Barranco, A.M.; Ruas-Madiedo, P.; Gueimonde, M. Effect of Bifidobacterium upon Clostridium difficile Growth and toxicity when co-cultured in different prebiotic Substrates. Front. Microbiol. 2016, 7, 738. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, N.; Feng, Y.; Zhou, M.; Li, H.; Zhang, X.; Ma, X. From probiotics to postbiotics: Concepts and applications. Anim. Res. One Health 2023, 1, 92–114. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, S.; Li, S.; Jiang, W.; Wang, J.; Xiao, J.; Chen, T.; Ma, J.; Khan, M.Z.; Wang, W.; et al. Unlocking the power of postbiotics: A revolutionary approach to nutrition for humans and animals. Cell Metab. 2024, 36, 725–744. [Google Scholar] [CrossRef] [PubMed]
- Mantziari, A.; Salminen, S.; Szajewska, H.; Malagón-Rojas, J.N. Postbiotics against pathogens commonly involved in pediatric infectious diseases. Microorganisms 2020, 8, 1510. [Google Scholar] [CrossRef] [PubMed]
- Luenglusontigit, P.; Sathapondecha, P.; Saengsuwan, P.; Surachat, K.; Boonserm, P.; Singkhamanan, K. Effects of postbiotic from bacteriocin-like inhibitory substance producing Enterococcus faecalis on toxigenic Clostridioides difficile. J. Health Sci. Med. Res. 2023, 41, e2023918. [Google Scholar] [CrossRef]
- Piqué, N.; Berlanga, M.; Miñana-Galbis, D. Health benefits of heat-killed (Tyndallized) probiotics: An overview. Int. J. Mol. Sci. 2019, 20, 2534. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.; Oliveira, A.L.; Oliveira, C.; Pintado, M.; Amaro, A.; Madureira, A.R. Current postbiotics in the cosmetic market—An update and development opportunities. Appl. Microbiol. Biotechnol. 2022, 106, 5879–5891. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spigaglia, P. Clostridioides difficile and Gut Microbiota: From Colonization to Infection and Treatment. Pathogens 2024, 13, 646. https://doi.org/10.3390/pathogens13080646
Spigaglia P. Clostridioides difficile and Gut Microbiota: From Colonization to Infection and Treatment. Pathogens. 2024; 13(8):646. https://doi.org/10.3390/pathogens13080646
Chicago/Turabian StyleSpigaglia, Patrizia. 2024. "Clostridioides difficile and Gut Microbiota: From Colonization to Infection and Treatment" Pathogens 13, no. 8: 646. https://doi.org/10.3390/pathogens13080646
APA StyleSpigaglia, P. (2024). Clostridioides difficile and Gut Microbiota: From Colonization to Infection and Treatment. Pathogens, 13(8), 646. https://doi.org/10.3390/pathogens13080646





