Abstract
Clostridioides difficile is the main causative agent of antibiotic-associated diarrhea (AAD) in hospitals in the developed world. Both infected patients and asymptomatic colonized individuals represent important transmission sources of C. difficile. C. difficile infection (CDI) shows a large range of symptoms, from mild diarrhea to severe manifestations such as pseudomembranous colitis. Epidemiological changes in CDIs have been observed in the last two decades, with the emergence of highly virulent types and more numerous and severe CDI cases in the community. C. difficile interacts with the gut microbiota throughout its entire life cycle, and the C. difficile’s role as colonizer or invader largely depends on alterations in the gut microbiota, which C. difficile itself can promote and maintain. The restoration of the gut microbiota to a healthy state is considered potentially effective for the prevention and treatment of CDI. Besides a fecal microbiota transplantation (FMT), many other approaches to re-establishing intestinal eubiosis are currently under investigation. This review aims to explore current data on C. difficile and gut microbiota changes in colonized individuals and infected patients with a consideration of the recent emergence of highly virulent C. difficile types, with an overview of the microbial interventions used to restore the human gut microbiota.
Keywords:
Clostridioides difficile; CDI; PCR-ribotype; colonization; gut microbiota; AAD; FMT; probiotics; prebiotics; postbiotics 1. Introduction
Clostridioides difficile is an anaerobic, Gram-positive, and endospore-forming bacillus, known as the most important and common cause of nosocomial antibiotic-associated diarrhea (AAD) worldwide [1].
C. difficile spores can survive in environments for years. After the oral ingestion of spores from infected individuals or the environment, C. difficile can transitorily or permanently colonize the human intestine, especially in infants and hospitalized patients [2]. Asymptomatic individuals colonized by toxigenic C. difficile strains have received growing interest as a potential transmission source of this pathogen. In fact, C. difficile carriers have been associated with significant skin and environmental contamination, similarly to patients with a C. difficile infection (CDI) [3].
CDI presents a large range of symptoms, from mild diarrhea to severe manifestations such as pseudomembranous colitis and toxin megacolon, which can lead to the death of patients [4]. Only toxigenic C. difficile strains cause infection and these usually produce two toxins, toxin A (TcdA) and toxin B (TcdB), that are the main virulence factors of this pathogen [5]. The tcdA and the tcdB genes are located in a chromosomal element, the pathogenicity locus (PaLoc), which also harbors regulatory genes for these toxins’ expression and release into the extracellular environment [5]. Furthermore, C. difficile strains can also produce a third toxin, known as the binary toxin CDT, which enhances their virulence and adherence to intestinal cells [6]. The production of this toxin characterizes several C. difficile types, such as the PCR-ribotype (RT) 027 and the RT 078. These types are characterized by high virulence and a resistance to antibiotics, and they have been associated with severe infections, an increased frequency of CDI recurrences, and higher mortality rates [7,8].
The human gut microbiota is a complex ecosystem that has a crucial role in conferring host immunity and preventing colonization and invasion by pathogens [9]. The gut microbiota is composed of an incredible number of microorganisms that can vary between individuals and that can be altered by various factors [10]. Dysbiosis is defined as an alteration in the gut microbiota’s composition, metabolic activities, or distribution [11]. The recent advances in high-throughput sequencing, mass spectrometry, and other omics approaches have improved the volume of data available on the composition of the gut microbiota, facilitating our understanding of the causes and possible impacts of dysbiosis [12,13].
C. difficile interacts with the gut microbiota throughout its entire life cycle. C. difficile’s role as a colonizer or invader largely depends on changes in the gut microbiota’s components, which C. difficile itself can promote and maintain [14]. Antibiotic exposure is considered the primary risk for CDI. Gut microbiota perturbations due to antibiotic administration allow C. difficile spores to germinate in vegetative forms that can colonize the intestine and produce toxins, leading to the inflammation of the intestinal membrane and subsequent symptoms of CDI [15,16,17].
The restoration of intestinal homeostasis has emerged as a promising therapeutic approach for CDI. In particular, fecal microbiota transplantation (FMT), a method used to normalize the composition of the gut microbiota of a recipient via the transplantation of a small sample of feces from a healthy donor, is widely used as a therapeutic option for recurrent CDIs [18,19]. However, alternative strategies to re-establish intestinal eubiosis for the prevention and treatment of CDI are under investigation [20,21,22].
This review aims to explore the current data on C. difficile and gut microbiota changes in colonized individuals and infected patients, with a consideration of the recent emergence of highly virulent C. difficile types, and to provide an overview of the microbial interventions used to restore the human gut microbiota in patients with CDI.
2. Healthy Gut Microbiota
The human intestinal microbiota is a dynamic functional interface between the external environment and the human body, and its composition increases in microbial diversity in the childhood, reaching stability during adulthood [23].
The components of the gut microbiota include several species of bacteria, yeasts, archaea, and viruses. The dominant bacterial phyla are Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia, with a high prevalence of Firmicutes and Bacteroidetes (90%) [24,25]. The Firmicutes phylum includes more than 200 different genera, with the Clostridium genera being prevalent (95%), while Bacteroidetes includes two predominant genera, Bacteroides and Prevotella [24].
The different regions of the gastrointestinal tract are colonized by different species, with most of the microorganisms present in the colon. In the human large intestine, the genus Bacteroides and two families belonging to the Firmicutes phylum, Lachnospiraceae and Ruminococcaceae, are prevalent (50–70%), while, in the small intestine, the Firmicutes and the Actinobacteria, the latter mainly represented by the Bifidobacterium genus, are the prevalent phyla [26].
The gut microbiota’s composition changes with the age. For example, while the intestinal community of infants is rich in Bifidobacterium spp., in elderly individuals a decreased proportion of Bifidobacterium spp., Lactobacillus, and the Faecalibacterium and Ruminococcaceae families have been observed, with an increased abundance of Proteobacteria, Bacteroidetes, and Clostridium spp. [24,27,28].
Although the composition of the gut microbiota is different in different individuals, characterized by specific bacterial strains [29], a classification based on its taxonomic composition proposes three different enterotypes: enterotype I, more frequently enriched in Bacteroides; enterotype II, more frequently enriched in Prevotella; and enterotype III, more frequently enriched in Ruminococcus [30]. However, there are controversial opinions on this classification, which is not accepted by the entire scientific community [31,32].
3. C. difficile Colonization Resistance
The components of a healthy gut microbiota form a stable community and their biochemical activities have a beneficial impact on the host, conferring resistance to non-native bacteria, developing the host immune system, and exerting important metabolic functions [33,34]. The capability of the gut microbiota to prevent colonization by pathogens, including C. difficile, is known as colonization resistance (Figure 1). Different mechanisms are involved in the colonization resistance against C. difficile, such as the production of bile acids and metabolites, competition for nutrients, and the stimulation of the immune response of the host [35]. The administration of antibiotics has been demonstrated to cause a shift of the bile acid pool, with a consequent increase in C. difficile spore germination [36,37]. Furthermore, the reduction in bacteria with a bile-metabolizing capacity results in the enrichment of primary bile acids, which promotes the germination of C. difficile spores, and in the loss of secondary bile acids, which inhibit C. difficile growth [38,39,40,41]. Among the bacteria with a bile-metabolizing capacity, Clostridium scindens has been associated with C. difficile colonization resistance in a gnotobiotic mouse model [38,42], while Clostridium butyricum and Roseburia intestinalis, both butyrate producers, are strongly associated with CDI-negative samples [43].
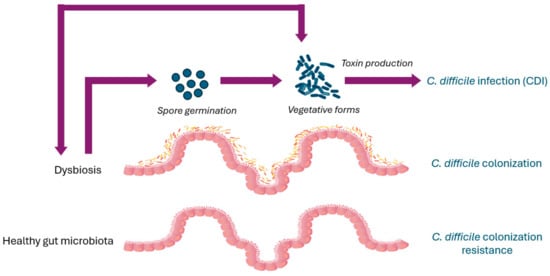
Figure 1.
Clostridioides difficile pathogenesis and gut microbiota.
Metabolites produced by the gut microbiota (or metabolome), such as sugars, amino acids, and lipids, may also have an impact on C. difficile colonization resistance [44,45]. For example, short-chain fatty acids (SCFAs), such as butyrate, have been associated with colonization resistance against C. difficile, since they are able to inhibit the growth of this pathogen and to affect the immunological response of the intestinal mucosa in vitro [44,46]. Intestinal microbiota components also produce bacteriocins, which are antimicrobial peptides with a narrow or broad spectrum of activity against other bacteria and pathogens [47]. Bacteriocins produced by strains of Bacillus, Lactococcus, and Enterococcus have been demonstrated to exert an antimicrobial effect against C. difficile in vitro [48,49,50].
The intestinal microbiota’s components can also directly compete with C. difficile through the consumption of nutrients; in particular, bacteria with similar nutritional requirements can protect the host. Interestingly, it has been observed that colonization by a nontoxigenic strain of C. difficile protects against colonization by toxigenic C. difficile in murine models [51].
Interestingly, the gut microbiota interacts with the innate and adaptive immune system of the host and modulates its activity to maintain homeostasis and inhibit inflammation via metabolic products that interact with and regulate immune cells [52,53]. The stimulation of local innate lymphoid cells ILC1s by microbiota components has been found to be implicated in the protection against C. difficile through interferon (IFN)-γ production [34]. Among the gut microbiota’s components, the commensal Clostridia can promote the development of intraepithelial lymphocytes (IELs), particularly alpha beta T-cell receptor (TcR)-bearing T cells and immunoglobulin A (IgA)-producing cells, through a gradient of SCFAs and secondary bile acids, contributing to the maintenance of overall gut function [54,55]. Several authors have reported that C. butryicum promotes resistance to CDI through metabolic and immune modulation, suggesting that this bacterium might be potentially used as a treatment for CDI [56,57,58,59]. Finally, the products of some metabolic pathways related to tryptophan or polyamines biosynthesis metabolism, which have an anti-inflammatory effect, can contribute to the protection against CDI and to the restoration of gut homeostasis [60,61,62,63].
4. Asymptomatic C. difficile Colonization
C. difficile colonization is usually defined as the detection of C. difficile or its toxins in the absence of diarrhea or without findings consistent with pseudomembranous colitis [2,64].
Unusually high levels of C. difficile asymptomatic colonization have been reported in neonates and infants, with rates up to 90% and levels of bacterial burdens similar to those found in adults with CDI [65]. C. difficile rates of colonization remain high (22%) up to 2 years of age, declining to 1–3% with the maturation of both the gut microbiota and the immune system of the host [66].
Among the possible factors protecting infants from CDI, it has been hypothesized that the lack of the receptors for C. difficile toxins in the intestine cells and the acquisition of maternal antibodies via breastmilk could have a role [67]. Also, it has been hypothesized that breastfed babies may be more protected from colonization by C. difficile due to a reduced mean fecal pH (5.29 vs. pH 6.48 of bottle-fed infants), possibly related to the reduced buffering capacity of breastmilk [68].
Relatively low percentages of asymptomatic C. difficile colonization have been reported in adults, although these percentages can dramatically increase in individuals who have contact with healthcare structures, particularly the elderly, with rates up to 51% [17,69]. It has been hypothesized that the development of antibodies may be one of the mechanisms involved in C. difficile’s asymptomatic colonization due to the very high levels of antibodies against the TcdA observed in C. difficile carriers [70]. Antibodies against C. difficile toxins may contribute to CDI protection directly, by a toxin neutralizing effect, and indirectly, by limiting epithelial damage and restoring colonization resistance [71,72,73].
Asymptomatic C. difficile carriers are usually undetected due to a lack of routine screening, but there has been increased attention given to them as a reservoir of this pathogen in both healthcare facilities and in the community. The importance of asymptomatic C. difficile carriers in hospital wards has been highlighted in a recent study, showing that 2.6% of patients not exposed to asymptomatic carriers develop CDI, while this percentage rises to 4.6% when the patients are exposed to carriers [74]. Another study using a mathematical model to analyze the transmission of C. difficile in hospital wards has demonstrated that the contribution of colonized patients at admission and patients with CDI to new colonizations are similar [75]. C. difficile carriers also have an important role in the community; in fact, a recent study estimating the risks of community-associated CDI (CA-CDI) has reported increased CDI rates among residents ≥ 65 years old in comparison to controls residing at home [76]. Furthermore, close contact with infants ≤ 2 years, frequently colonized by C. difficile, has been identified as a potential risk factor for CA-CDI in a prospective case–control study in the UK [77].
Finally, individuals colonized by toxigenic strains have a higher risk of developing CDI compared to non-colonized individuals; therefore, colonization with C. difficile is considered a crucial factor in the progression to CDI [78,79].
5. Gut Microbiota in Asymptomatic C. difficile Carriers
Age, as well as other factors, such as the use of antibiotics, proton pump inhibitors (PPIs), and hospitalization, may affect the gut microbiota and predispose a person to C. difficile colonization (Figure 1). For this reason, several studies have investigated the alterations in the gut community structure of neonates/infants and adults colonized by C. difficile, compared to healthy individuals and patients with CDI, providing important information on the intestinal microbiota’s components in relation to the age.
5.1. Neonates and Infants
The high rates of C. difficile colonization observed in infants suggest that the microbiota in the early life is a favorable environment for this pathogen. Interestingly, many bacteria that antagonize C. difficile, such as Lactobacillus species, have been negatively associated with C. difficile colonization in neonates (Figure 2A). In fact, it is known that Lactobacillus gasseri and Lactobacillus acidophilus reduce the C. difficile burden and inhibit the expression of the TcdA, respectively [67]. In infants, C. difficile colonization has also been negatively associated with Bifidobacterium longum, which is able to inhibit C. difficile growth and toxin production by decreasing the intestinal pH, while Ruminococcus gnavus and Klebsiella pneumoniae are more frequently detected in the microbiota of colonized infants [67,80,81,82] (Figure 2A). Antibiotic treatment has been associated with a decrease in microbial diversity and often with an outgrowth of Enterobacteriaceae [83]. Interestingly, infants born via cesarean section have been found to be enriched in Enterococcus, which is associated with increased rates of C. difficile colonization [67].

Figure 2.
Prevalent changes observed in the gut microbiota of (A) infants and adults asymptomatic C. difficile carriers and (B) patients with CDI.
Different authors have observed a correlation between an increase in specific gut anaerobes and a reduction in C. difficile colonization in infants and children [67,80,84,85]. Although a Bacteroides enrichment has been found in neonates with vaginal delivery and an exclusively breastmilk diet, which are negatively associated with C. difficile colonization, it is still unclear whether Bacteroides may have a direct or indirect effect on CDI protection in early infancy [67,86].
The analysis of the microbiota composition of an infant colonized by C. difficile has shown important changes during weaning, with an increase in the relative abundance of Bacteroides spp., Blautia spp., Parabacteroides spp., Coprococcus spp., Ruminococcus spp., and Oscillospira spp., suggesting that the increased level of some bacterial groups during weaning can be associated with a reduction in C. difficile colonization [87].
Although further studies are necessary to better understand C. difficile colonization as a potential protection from developing CDI, some data support the fact that neonates and infants might potentially benefit from asymptomatic C. difficile colonization. In particular, the IgA and IgG responses to C. difficile toxins might be protective against CDI in infants colonized by toxigenic C. difficile strains and, also, in breastfed infants [4,65].
5.2. Adults
A lower microbial diversity and species richness has been reported in the gut microbiota of adults colonized by C. difficile compared to healthy subjects [88]. A depletion or lack of butyrate-producing bacteria, such as Ruminococcaceae (Faecalibacterium and Ruminococcus), Lachnospiraceae (Blautia and Coprococcus), and Bifidobacterium, has been reported by Vázquez-Cuestain et al. in C. difficile carriers [89] (Figure 2A). The authors further identified the genera Parabacteroides, Faecalicoccus, Flavonifractor, and Clostridium_XVIII as biomarkers for patients colonized by C. difficile (Figure 2A).
Other authors have also found that the gut microbiota of asymptomatic carriers exhibits a relative abundance of Clostridium and Eubacterium compared to patients with CDI, and that Veillonella, Fusicatenibacter, and Eubacterium species may play a role in preventing CDI in asymptomatic C. difficile carriers [90,91].
Finally, a study has recently demonstrated that the gut microbiota of C. difficile-colonized patients shows a higher relative abundance of the genera Aspergillus and Penicillium compared to patients with CDI and healthy controls, suggesting that indirect interactions between fungi and bacteria may have some effects on C. difficile and on treatment effectiveness [92] (Figure 2A).
All these data indicate a decrease in species richness/diversity in the gut microbiota of asymptomatic C. difficile carriers and that the presence of certain bacterial groups might be protective, preventing C. difficile overgrowth or the transition from asymptomatic colonization to infection.
6. Gut Microbiota Dysbiosis Increases Susceptibility to C. difficile Infection
The composition and function of the gut microbiota are influenced by exposure to a changing environment and factors such as antibiotics, age, medical treatments, diet, etc. [24,93]. The imbalance of the gut microbiota ecosystem, or dysbiosis, determines a shift in the composition and/or function of bacterial components, which can lead to increased CDI susceptibility [38,94,95,96] (Figure 1).
The excessive/wrong use of antibiotics, particularly those with a broad spectrum of action, represents the main risk for CDI, reducing the gut microbiota’s diversity and increasing gut colonization [97,98,99]. Gut microbiota changes due to an exposure to antibiotics can persist for a long time. In fact, treatment with ciprofloxacin, clindamycin, clarithromycin, or metronidazole has been demonstrated to cause alterations in the gut microbiome lasting up to four years [100,101,102]. Furthermore, antibiotics can affect the metabolic function of the endogenous microbiota. For example, Bacteroides spp. produce succinate and liberate sialic acid and fucose, promoting C. difficile colonization, only when the host glycans became available in the gut after antibiotic exposure [103,104]. Changes in the components of the gut microbiota also depend on the different classes of antibiotics used [105]. A study on the effects of antibiotics associated with CDIs in a mouse model has demonstrated that the usage of clindamycin determines a decrease in obligate anaerobic bacteria and a prevalence of Enterobacteriaceae, while a prevalence of Pseudomonadaceae and Lactobacillaceae has been observed after cephalosporin exposure, and a lower prevalence of Bacteroidetes and a higher prevalence of Proteobacteria when tigecycline is used [106]. Furthermore, a decrease in Bacteroidetes and Bifidobacteria has been observed using a cefotaxime instillation in a human gut model, with subsequent C. difficile germination and the production of high levels of toxins [107].
Interestingly, the target population of CDIs is represented by patients ≥ 65 years old, and increasing age may be implicated in intestinal microbiota changes. In fact, lower microbial diversity and an altered microbiota composition have been observed in elderly people, with a decrease in protective bacteria such as Bifidobacteria, Firmicutes, and Bacteroidetes and an increment in those associated with disease, such as Proteobacteria [108]. In addition, the more frequent use of antibiotics, frequent hospitalizations, and the comorbid conditions that commonly occur in the elderly contribute to their decreased resistance to C. difficile’s colonization and infection [4].
The use of PPIs may also have an impact on the gut microbiota. Studies in vitro have demonstrated that PPIs cause a decrease in the relative abundance of Bacteroidetes and an increase in Firmicutes, particularly Lactobacillus, which may increase susceptibility to C. difficile [109,110].
Furthermore, obesity, autoimmune and allergic diseases, diabetes, and inflammatory bowel disease may have a role in gut microbiota perturbations [111]. Interestingly, an interaction between the gut microbiota and obesity has been reported in humans, resulting in a decrease in Bacteroidetes and an increase in Firmicutes, although a clear trend between this ratio and obesity status has not been demonstrated [112,113].
7. C. difficile Infection Pathogenesis
C. difficile transmission occurs via the fecal–oral route, with the ingestion of either vegetative forms or spores. C. difficile spores are metabolically inert, extremely resilient, and resistant to oxygen, so they can survive for long periods in the environment [114]. C. difficile germination initiates in response to chemical signals, such as bile acid taurocholate, that indicate that environmental conditions may be conducive to the survival of the vegetative cells and their growth [115]. After the induction of the spores’ germination and the replication of vegetative cells, the production of toxins leads to the symptomatic disease (Figure 1).
The major virulence factors produced by C. difficile are the enterotoxin TcdA and the cytotoxin TcdB [116]. The genes encoding for these toxins and for the accessory proteins TcdR, TcdC, and TcdE, involved in the regulation of toxin production and in their extracellular extrusion, are located on a chromosomal element of 19.6 kb, known as the pathogenicity locus (PaLoc) [117,118]. TcdA and TcdB show approximately 66% amino acid sequence similarity, and both present two conserved domains, a catalytic glucosyltransferase domain and a cysteine protease domain, necessary for the auto-processing and delivery of the catalytic domain to the cytosol of the intoxicated cells [6]. In the cytosol, the catalytic domain inactivates several cellular proteins belonging to the RAS homolog (Rho) family that catalyze the hydrolysis of guanosine triphosphate (GTPases), which are crucial in the regulation of the actin cytoskeleton, leading to alterations in cellular integrity and morphology, a loss of the tight junctions, and the increased permeability of the intestine epithelial layer [118,119]. Although TcdA and TcdB act synergistically, strains with a TcdA−/TcdB+ phenotype may cause severe infections and even mortality [120].
In addition to TcdA and TcdB, several C. difficile types, prevalently those which are highly virulent, also produce an additional toxin, the binary toxin CDT, which has been associated with more severe diseases, an elevated risk of recurrence, and high mortality rates [121,122]. CDT belongs to the family of Clostridial iota-like toxins and shows two components, the enzymatic component CdtA and the binding component CdtB, encoded by genes located on a 6.2 kb region designated as the Cdtlocus (CdtLoc) [123]. The internalization of the enzymatic component CdtA into the cytosol results in the adenosine diphosphate (ADP)-ribosylation of monomeric actin and in the disorganization of the cytoskeleton, with the formation of microtubule-based cell protrusions and an increased adhesion of bacteria to intestinal cells [123,124]. Since CDT targets a broader array of cells in vivo, higher rates of CDI recurrences and lower clinical cure rates are reported for patients infected with strains producing CDT compared to those infected by historical strains [125]. In addition to CDT production, it has been demonstrated that highly virulent C. difficile strains produce a TcdB exhibiting broader tropism and cytotoxicity activity compared to the TcdB of historical strains, leading to more rapid cell entry and the increased activity of the toxin [126].
8. Gut Microbiota Changes in Patients with C. difficile Infection
In general, a decrease in species richness and microbial diversity has been observed in patients with CDI [35]. In addition, a decrease in butyrate-producing Ruminococcaceae, Lachnospiraceae, and Clostridia clusters IV and XIVa bacteria has been observed [127,128,129] (Figure 2B).
Ruminococcaceae and Lachnospiraceae from the Firmicutes phylum metabolize primary bile acids into secondary bile acids; therefore, if the abundances of these bacterial families decrease, there is also a decrease in secondary bile acids and a consequent decrease in the inhibition of C. difficile germination, facilitating the development of a CDI. In particular, a significant reduction in Lachnospira, Odoribacter, Coprococcus, and Anaerostipes, as well as in Prevotella and Faecalibacterium, has been reported as an important finding in patients with CDIs [130,131,132,133]. Interestingly, the results obtained in different studies support a depletion of the genera producing butyric acid not only in the case of severe CDIs, but also with mild disease or C. difficile colonization [131,133]. Furthermore, Antharam et al. have found a decrease in the bacteria producing butyric acid, compared to healthy individuals, not only in the intestinal microbial community of patients with CDI but also in those of patients with non-C. difficile AAD [134].
An increase in the phyla Firmicutes, Proteobacteria, and Actinobacteria and members of the Gammaproteobacteria, Lactobacillaceae, and Enterococcaceae families has been reported in the gut microbiota of people suffering from a CDI compared to healthy people, in addition to a decrease in the relative abundance of Bacteroidetes [134,135,136,137,138] (Figure 2B). However, regarding the phylum Bacteroidetes, data from the literature are not concordant, since some studies show a general decrease in abundance, while others show an increase [135]. Several studies have also reported an increase in the genera Parabacteroides, Enterococcus, Klebsiella, and Akkermansia in the microbiota of patients with CDIs, a phenomenon resulting from the competition for a reduced ecological niche [88,127,133]. In addition, Han et al. suggest the depletion of some species, such as Phascolarctobacterium spp., and genera, such as Lachnospira, Butyricimonas, Catenibacterium, Paraprevotella, Odoribacter and Anaerostipes, as a signature change in CDIs [133] (Figure 2B).
Interestingly, there is evidence that the gut microbiota can modulate a CDI’s severity, and it has been demonstrated that the relative abundance of Akkermansia, Anaerotignum, Blautia, Enterocloster, Lactonifactor, and Monoglobus is associated with a decrease in C. difficile toxin production, a low histopathologic scoring of the cecal tissue, and low mortality in a mouse model [139].
The study of Chen et al. showed that some pathogens are strongly associated with CDIs, such as Enterocloster bolteae and Ruminoccocus gnavus [43]. E. bolteae is an opportunistic pathogen linked to the expression of inflammatory genes; similarly, R. gnavus produces glucorhamnan, a potent inflammatory polysaccharide [140,141]. Opportunistic pathogens, such as Bacteroides, Enterococcus, and Klebsiella, as well as Prevotellaceae, Eggerthella, and Helicobacter, have been commonly detected in CDI cases with a higher histopathologic score and worse outcomes [139,142].
Profound alterations were found in patients with recurrent CDI who had received multiple antibiotic treatments [143]. The available literature indicates that the gut microbiota of patients with recurrent CDI shows a reduction in the diversity and richness of its components, as observed in patients with a primary CDI, although some taxa can display different trends in patients with recurrent CDI compared to healthy individuals and patients with a primary CDI. In recent studies analyzing the fecal samples of patients with recurrent CDI, a relative reduction in Bacteroidetes and Firmicutes and an increase in Proteobacteria’s abundance compared to healthy individuals have been observed [143,144]. Khanna et al. have reported that patients with recurrent CDI had a statistically significant increase in Enterobacteriaceae, Lachnospiraceae, Streptococci, Parabacteroides, and Veillonella levels compared to patients with CDI [144]. In addition, Gazzola et al. have reported a strong reduction in gut commensal Parabacteroides in patients with recurrences compared to patients with primary CDI [145]; however, this result disagrees with that of Khanna et al., who suggest that the increase in Parabacteroides is a predictor of recurrence [144].
Weingarden et al. have reported that two primary bile acids, cholic acid and chenodoxycholic acid, are significantly elevated in patients with a recurrent CDI [146]. A more recent investigation confirmed these results, reporting an increase in primary bile acids in the stools of patients with recurrences compared to healthy controls and patients with a primary CDI, which are associated with a reduced abundance of species capable of metabolizing bile acids, such as Collinsella, and an increase in the relative abundance of Enterobacteriaceae and Lactobacillus [147].
The data available indicate that there are significant differences in the gut microbiota composition of CDI patients compared to those of carriers and healthy individuals, suggesting that the presence or absence of certain bacteria taxa are often more impactful in determining the development of a CDI than diversity or species richness alone.
9. Highly Virulent C. difficile Types
The epidemiology of CDIs has been changing in recent decades. Until the end of the 20th century, CDI was considered a complication of antimicrobial exposure and occurred mainly in the hospital environment [148]. Since then, CDIs have been more frequently detected in the community and in populations usually not affected by CDI (antibiotic-naïve people, peripartum women, and children) [8]. Also, the incidence of CDI recurrence has risen, sometimes disproportionately compared to CDIs. For example, between 2001 and 2012, the increase in CDI recurrences was four times higher compared to that of primary CDIs diagnosed in the USA [149].
An increase in C. difficile diversity has been observed worldwide, with the emergence of more virulent and antibiotic-resistant C. difficile types, which are considered the main driver of recent CDI epidemiological changes [7,150].
Starting in the early 2000s, the PCR-ribotype (RT) 027 has been reported as cause of many outbreaks in the hospitals of North America and Europe [151,152,153]. Its acquisition of a resistance to fluoroquinolones has been identified as the main cause of the global diffusion of the RT027 due to the selective pressure exerted by the widespread use of these antibiotics in healthcare settings. A whole-genome sequence (WGS) analysis has demonstrated that two genetically distinct lineages of RT 027 independently acquired resistance to fluoroquinolones and successively spread: one in the USA, South Korea, and Switzerland and the other in the UK, Europe, and Australia [154]. Due to their high rate of sporulation, increased expression of TcdA and TcdB, production of the binary toxin CDT, and resistance to antibiotics, RT 027 strains have been associated with an overall increase in CDIs’ incidence, severity, rates of recurrence, and mortality [121,155,156,157,158].
RT 078 emerged shortly after RT027 as a cause of infection in hospitals and in the community, with rates of severe diarrhea and an attributable mortality similar to those of RT 027 [159]. C. difficile RT 078 is able to infect/colonize livestock and its zoonotic potential is supported by a recent genomic analysis that demonstrated the spread of RT 078 clones across multiple continents, often without any association with the healthcare system, suggesting a zoonotic/anthroponotic long-range dissemination in the community [160].
New RTs can evolve from each other, constituting different C. difficile lineages [161]. In fact, recent types, such as RT 176 and RT 244, which have been found to be genetically related to RT 027 and have the same characteristics of virulence, constitute the RT 027 lineage [162,163]. Similarly, several types have been identified as related to RT 078, such as RT 126, RT 127, RT 033, and RT 288, constituting the RT 078 lineage [160].
The emergence of highly virulent types has led to the need to understand the possible impact of these new types on the gut microbiota community in order to develop more precise and sustainable approaches to CDIs’ prevention and treatment.
10. Impact of Highly Virulent C. difficile Types on Gut Microbiota
The recent increase in CDI cases in the community, without previous antibiotic treatment [164], suggests that C. difficile might also colonize the gut in the presence of non-dysbiotic microbiota and that this pathogen might be able to affect the gut microbiota’s composition. This hypothesis has been supported by the results obtained in a recent study, which indicate that C. difficile is able to alter the level of gene transcription and the metabolomic profile of the gut microbiota, causing a restructuring of the landscape of nutrients in the intestine and promoting persistent infection [165].
Robinson et al., using fecal minibioreactor arrays, have demonstrated that C. difficile colonization significantly perturbs the intestinal microbiota’s composition, particularly when certain C. difficile types are used [166]. In fact, it has been observed that RT 027 strains have an ecological advantage not only over other C. difficile types but also over the gut microbiota’s components due to their capability to exploit the limited nutrients available. A recent study has demonstrated that toxin-producing C. difficile colonization causes an alteration in the nutrient pool and that the inflammation induced by C. difficile RT 0127 toxin activity leads to alterations in the components of the gut microbial community, with a suppression of members from the family Bacteroidaceae and the increased growth of C. difficile [14].
Skraban et al. have analyzed the presence of bacterial, fungal, and archaeal microbiota in fecal samples from healthy individuals and patients with a request for C. difficile testing, considering the presence or absence of C. difficile RT 027 or non-RT 027 in this last group [167]. The results indicate an association between RT 027 strains and low levels of microbiota diversity, suggesting that this type of CDI is potentially capable of affecting the composition of the gut microbiota and, therefore, increasing its own colonization potential.
Interestingly, Horvat et al. have evaluated the effects of C. difficile on the gut microbiota using six different C. difficile strains belonging to RT 027 and to RT 014/020 for the co-cultivation of fecal microbiota in an in vitro model [168]. A general decrease in the microbial diversity of the fecal microbiota has been observed with both C. difficile types. Although the changes in the microbiota’s composition due to RT 027 and RT 014/020 are different, many of these alterations occur within the phylum Firmicutes, with a general decrease in commensals with potential protection functions (i.e., Lactobacillus, Clostridium_XIVa) and an increase in opportunistic pathogens (i.e., Enterococcus). Furthermore, fecal microbiota co-cultivated with other bacteria (Escherichia coli, Salmonella Enteritidis, or Staphylococcus epidermidis) show different patterns of alterations, supporting the hypothesis that C. difficile effects are specific. In another investigation, the same authors analyzed the interactions between different C. difficile RTs and two types of fecal microbiota (healthy and dysbiotic) using an in vitro batch model [169]. The results have indicated that, although strains belonging to all the RTs tested show a higher frequency of sporulation in the presence of dysbiotic fecal microbiota, the growth is strain-dependent.
The results obtained by several authors indicate a decrease in the richness and diversity of the fecal microbiota of both adults and children when co-cultured with different C. difficile types, with a decrement in the butyrate-producing members of Ruminococcaceae and Lachnospiraceae and an increase in Proteobacteria members [167,169,170,171,172]. In addition, important metabolic alterations were reported in the microbiota of children when co-cultured with different C. difficile types. In particular, higher concentrations of acetic acid, butyric acid, phenylacetic acid, p-hydroxyphenylacetic acid, and tyramine were detected when the fecal microbiota were co-cultured with C. difficile RT 027 and RT 176, which are associated with a great perturbation in the microbiota’s composition and C. difficile overgrowth [170].
All these data suggest that alterations in the gut microbiota of patients with CDI could be directly influenced and maintained by C. difficile, with effects that differentiate between the different types.
11. Microbial Interventions to Restore Gut Microbiota
Restoring the gut microbiota to a healthy state, or eubiosis, is an important goal for therapeutic and protective approaches to CDI. Microbial interventions can be divided into fecal microbiota transplantation (FMT), the use of probiotics, live biotherapeutic products (LBPs), prebiotics, and postbiotics [20,173] (Figure 3).
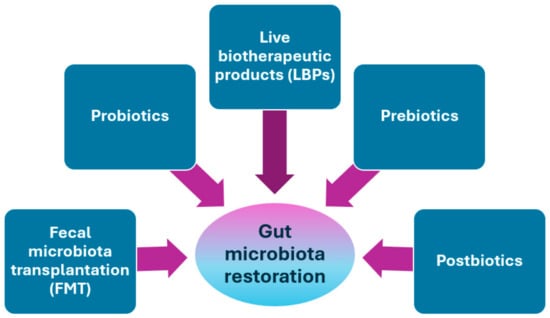
Figure 3.
Treatments for gut microbiota restoration in patients with CDI.
11.1. Fecal Microbiota Transplantation (FMT)
The efficacy of the gut microbiota’s restoration in the treatment of CDI has been demonstrated by the high percentage of clinical cure obtained with fecal microbiota transplantations (FMTs). An FMT consists of restoring the microbial diversity and the metabolic landscape of the gut microbiota of a patient with CDI through the delivery of feces from a healthy subject [174].
A first study, performed in 2013 by van Nood et al. and including patients with CDI treated with oral vancomycin followed by bowel lavage and a nasoduodenal infusion of feces solution from a donor; or with vancomycin combined with bowel lavage; or only with vancomycin, demonstrated that 81% of patients who received donor feces had a complete resolution of their symptoms after the first treatment [175].
A subsequent study demonstrated that employing an FMT via colonoscopy yields superior results in treating recurrent CDI compared to a nasoduodenal infusion, with a 90% of success rate versus a vancomycin regimen [176]. Although an FMT colonoscopy is commonly used, oral FMT capsules show an efficacy in treating recurrences of CDI that is comparable to colonoscopy, regardless of whether frozen or lyophilized stool is used for capsule preparation [177]. The use of frozen or lyophilized stool for FMT is supported by a recent study, which showed that the marginal decrease in relative efficacy from fresh FMT (93% efficacy) to frozen FMT (88% efficacy) and lyophilized FMT (83% efficacy) can be outweighed by the accessibility and ease of use of frozen or lyophilized preparations [178]. In addition, colonoscopy or oral capsules as administration routes for FMTs have an efficacy that overcomes enemas or nasogastric tubes [179].
An FMT is considered generally safe since its adverse events (loose stool, abdominal discomfort, or diarrhea), which occur in up to 50% of patients, are transient or self-limiting [180,181].
More recently, a new methodology known as washed microbiota transplantation (WMT), in which the stool specimen from a donor is automatically filtered and washed, has reduced the incidence rate of fever due to bacterial translocation across the ulcerated intestinal mucosa from 19.4% (manually prepared FMT) to 2.7% (WMT) [182]. In a multicenter study performed in China, patients affected by CDI were treated with WMT by a colonic transendoscopic enteral tube introduced via colonoscopy and secured in the colon with an endoclip that allowed for repeated procedures, obtaining a clinical cure rate of 90.7% [183].
Even though FMT demonstrates superiority in the treatment of recurrent CDI, there are still critical aspects that have limited its diffusion, such as a lack of dedicated structures, difficulties in the recruitment/selection/screening of donors, and the safety of patients. Internationally, panels of experts are involved in its methodological implementation, the standardization of stool banks, and quality assurance to guarantee accessibility to FMT worldwide [184,185].
Gut Microbiota Changes in FMT Patients
Unlike vancomycin and fidaxomicin monotherapies, which inhibit the immediate growth of C. difficile, FMT induces a durable treatment response, sustained through several mechanisms, including C. difficile’s competition with the FMT-delivered microbiota, the restoration of secondary bile acid metabolism, the stimulation of the mucosal immune system, and the repair of the gut barrier [186,187].
Several studies have observed that the composition of the fecal samples of pre-FMT patients shows a reduced relative abundance of bacterial families from the Bacteroidetes and Firmicutes phyla, such as Bacteroidaceae, Clostridiaceae, Eubacteriaceae, Lachnospiraceae, Prevotellaceae, and Ruminococcaceae, and an increase in the relative abundance of the Proteobacteria phylum, in particular of the Gammaproteobacteria, which are restored after FMT [188,189,190,191,192].
A recent study investigated the gut microbiota changes that occurred in patients treated with vancomycin followed by FMT to compare the differences between responders and non-responders to this treatment [193]. Although no significant differences have been observed immediately after an FMT, after one week, responders have shown a gut microbiota similar to healthy donors, depleted of Enterobacteriaceae and enriched in Lachnospiraceae and Ruminococcaceae (Figure 4A).
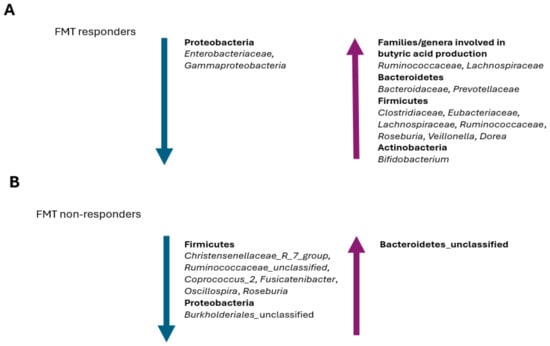
Figure 4.
Prevalent changes observed in the gut microbiota of patients with recurrent CDI that (A) positively respond to FMT and (B) negatively respond to FMT.
In a recent study by Tian et al., a reduction in the abundance of butyrate-producing bacteria of the Firmicutes phylum, including Christensenellaceae_R_7_group, Ruminococcaceae_unclassified, Coprococcus_2, Fusicatenibacter, Oscillospira, and Roseburia, was observed in the FMT non-responders [194] (Figure 4B). In fact, butyrate stabilizes the expression level of hypoxia-inducible factor-1 (HIF1) and can improve the intestinal barrier, prevent bacterial translocation, and reduce the local inflammatory response [195,196]. Thian et al. have also observed that both patients prior to an FMT and FMT non-responders show a reduction in their relative abundance of Burkholderiales_unclassified, Coprococcus_2, and Oscillospira, while an enrichment in Bacteroidetes_unclassified has been observed in FMT non-responders [194] (Figure 4A,B). Since Burkholderia has an important negative impact on CDI pathogenesis by inhibiting the Rho-glucosylation activity of C. difficile TcdB, its reduction or depletion led to a higher probability of treatment failure in FMT patients [197]. Similarly, C. difficile metabolic activities require the consumption of the succinic acid that is produced by Bacteroides [103]; therefore, an enrichment in Bacteroidetes_unclassified in FMT non-responders could potentially favor the recurrence of CDI (Figure 4B).
The recovery of both Roseburia and Veillonella, which are able to inhibit C. difficile, is usually correlated with a positive response to an FMT treatment and CDI recurrence resistance [198,199] (Figure 4A). In fact, a higher relative abundance of certain bacteria, including Veillonella, in the gut microbiota of the recipient prior to an FMT treatment seems to favor a positive response to the treatment itself [194]. Interestingly, the relative abundance of Veillonella gradually reduces with age and its depletion is probably related to higher rates of CDI and CDI recurrences in these patients [200]. Therefore, a Veillonella transplant should not be ignored in the FMT treatment of this patient population.
Bifidobacterium, Dorea, and Ruminococcaceae have mucinophilic properties similar to Bacteroides and C. difficile [194]; therefore, it is possible that, in FMT responders, these bacteria can promote the displacement of C. difficile through niche competition, preventing CDI recurrences. In general, Oscillospira is less abundant in the gut microbiota of patients with gastrointestinal inflammatory diseases, such as inflammatory bowel disease [191,201]. The depletion of Oscillopora in the gut microbiota of donors, as well as the absence of Burkholderiales_unclassified and Coprococcus_2, may be an important reason for the low effectiveness of FMT, since successful colonization of the FMT recipient’s intestine is largely due to the phylogeny of the gut microorganisms of donors [202].
Aggarwala et al., applying the Strainer algorithm, have tracked sequenced bacterial strains in metagenomic sequencing datasets from donors and FMT patients up to 5 years post-FMT [192]. The results obtained have demonstrated that >70% of strains from donors and <25% of strains from non-relapsing FMT recipients have been retained 5 years post-FMT. These data suggest that FMT represents a durable therapeutic treatment that determines a semipermanent alteration of the recipient’s gut microbiota after a single administration. The authors also observed that the engraftment efficacy of FMT donors’ microbiota differed by bacterial species [192]. Some species were able to stably engraft in the gut of recipients, such as those belonging to Bacteroidales, which were always engrafted. In fact, Bacteroidales have a higher transmission efficiency compared to other bacteria [203]. Contrastingly, Firmicutes have reduced and more difficult intestinal colonization, with complex adaptive activity that occurs at the expense of spore-forming activity, favoring intestinal colonization by non-spore-forming Firmicutes, such as Coprococcus_2 and Oscillospira, whose depletion is associated with poor prognosis in FMT patients [204,205].
11.2. Probiotics
Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [206]. Probiotics can be used to prevent the onset of dysbiosis or as therapeutic agents for an ongoing condition of dysbiosis (Figure 3).
Probiotics belong to bacterial species that are normal components of the human gut microbiota, designated as “Generally Regarded As Safe (GRAS)”, which are able to resist the gastric environment (bile and pancreatic secretions) and to be active and vital for a reasonable period in the intestine [207]. The ability to hydrolyze bile salts is often included as a criterion for the selection of probiotic strains.
The most commonly used probiotic microorganisms include Lactic acid bacteria, Bifidobacteria, Enterococci, Probionibacteria, Bacillus spp., the Escherichia coli strain Nissle 1917, and the yeast Saccharomyces boulardii [208,209]. Most of these probiotics act through competitive exclusion, modulating the gut microbiota and restoring a healthy intestinal microbial community, with a reduction in cholesterolemia, inflammation, diarrhea episodes, bowel discomfort, and a suppression of colon cancer and allergic states.
Several studies have evaluated the use of antibiotics in combination with probiotics for recurrent CDIs. In one randomized controlled trial, S. boulardii was used in combination with standard antibiotics for CDIs in patients with primary or recurrent CDI, showing that those receiving S. boulardii had fewer recurrences in comparison to those treated with a placebo [210]. In another study, patients with CDI recurrences were treated with high-dose vancomycin (2 g/d), low-dose vancomycin (500 mg/d), or metronidazole (1 g/d), in combination with S. boulardii (1 g/d), with a reduction in recurrences in the group treated with high-dose vancomycin followed by S. boulardii [211].
Wullt et al. have observed that patients with recurrent CDI treated with metronidazole for 10 days, followed by a dose of Lactobacillus plantarum, show fewer recurrences compared to those treated with a placebo, supporting L. plantarum as a probiotic that could be useful in preventing CDI, as also observed in other studies [212,213,214]. Also, a probiotic mixture including Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium lactis Bi-07, and B. lactis Bl-04 has been administered to patients with mild to moderate CDI, significantly improving infection duration and diarrhea severity compared to a placebo [215].
Recent studies have reported an association between the use of Lacticaseibacillus rhamnosus as a probiotic and increased butyrate production [216,217,218]. Furthermore, other studies have investigated C. butryicum, which promotes resistance to C. difficile infection, as a possible probiotic for CDI [56,57,58,59].
Although several studies have investigated the effects of probiotics on C. difficile colonization, growth, or toxin production, considering probiotics as an adjunctive therapy for CDI in combination with antibiotic treatment [219,220,221,222,223,224], their role in preventing CDI is still unclear. For these reasons, the current guidelines do not recommend the use of probiotics for primary CDI or CDI recurrence prevention [225,226].
Finally, the potential adverse events seen with probiotics remain crucial. In this respect, an association between the use of probiotics and increased mortality has been observed in a randomized, double-blind, placebo-controlled trial investigating the use of probiotics to prevent predicted severe pancreatitis [227]. Although there is evidence that probiotics can prevent CDI and that better therapeutic outcomes are obtained using probiotics, it is still not possible to know if these various probiotics are universally safe for every subject.
11.3. Live Biotherapeutic Products (LBPs)
Ready-to-prescribe formulations of live biotherapeutic products (LBPs), which are medicinal products containing live microorganisms (bacteria or yeasts) for human use, are considered a simplified option for daily CDI clinical practice (Figure 3).
In 2022, the US Food and Drug Administration (FDA) approved RBX2660, marketed as “REBYOTA™, for preventing CDI recurrences in adults who have undergone standard antibiotic treatment. RBX2660 consists of a consortium of live microbes from human feces, delivered as an enema. The efficacy of RBX2660 has been demonstrated in a randomized, double-blind, placebo-controlled phase III trial involving 267 adult patients with one or more CDI recurrences, with a success rate (defined as the absence of C. difficile-associated diarrhea within 8 weeks of the study treatment) of 70.6%, compared to 57.5% for the placebo group [228]. A metabolomic analysis has demonstrated a shift from a majority of primary bile acids to predominantly secondary bile acids when the RBX2660 is used [229].
The advantages of RBX2660 are its efficacy in reducing the recurrence of CDI, its cost-effectiveness, and safety profile, while the disadvantages include side effects (mild gastrointestinal complaints), its storage, and the need for specialized handling. Furthermore, the mode of administration may pose logistical challenges for some facilities and patients [230].
In 2023, the FDA approved SER-109 (VOWST™), which consists of purified encapsulated Firmicute spores that are administered orally over three consecutive days. Its efficacy has been demonstrated in a randomized, double-blind, placebo-controlled phase III trial involving 182 patients with three or more CDI recurrences, with a SER-109 success rate of 88% compared to 60% with the placebo [231]. In the study, mild to moderate gastrointestinal adverse events were observed after the SER-109 treatment. Interestingly, SER-109 species have been detected in treated patients after a week, and they have been associated with bile-acid profiles known to inhibit the germination of C. difficile spores.
Ongoing clinical trials are investigating other LBPs based on non-pathogenic strains, such as VE303 and NTCD-M3, which can be orally administered [232,233]. VE303, from Vedanta Biosciences, includes eight strains of non-pathogenic commensal Clostridia that are grown in clonal stool banks and encapsulated for oral delivery. The results from a phase II, randomized, double-blind, placebo-controlled, dose-ranging study involving adult patients with one or more recurrences of CDI and patients with primary CDI at risk of recurrences have demonstrated that high-dose VE303 prevents CDI recurrences in both groups of patients compared to the placebo (13.8% vs. 45.5%, p = 0.006) [234]. NTCD-M3 is a novel live LBP that consists of spores of the nontoxigenic C. difficile strain M3. A phase II clinical trial involving 173 patients with primary CDI or recurrent CDI has demonstrated that NTCD-M3 is able to prevent CDI recurrences when administered shortly after the antibiotic treatment of a primary CDI, with 11% of CDI recurrences in the NTCD-M3 group of patients compared to 30% in the placebo group [235].
11.4. Prebiotics
The current prebiotic concept refers to non-digestible compounds that, through their metabolization by microorganisms colonizing the gut, modulate the composition and the activity of the gut microbiota, conferring beneficial effects to the host [234] (Figure 3).
Prebiotics, which can be in natural or synthesized forms (inulin, oligosaccharides, lactulose, pyrodextrins, dietary fibers, and resistant starches, etc.) [234], are used as a complementary therapy for the treatment of various diseases, such as inflammatory bowel disease, Parkinson’s disease, or obesity [235].
The combination of a prebiotic with a probiotic to favor and enhance the colonization and growth of certain microorganisms is known as a “synbiotic” [235]. Rätsep et al. have demonstrated that the combination of xylitol and L. plantarum Inducia is able to suppress the germination of C. difficile spores and their outgrowth into vegetative forms in hamsters, reducing the colonization of the gut by this pathogen [236]. This therapeutic approach includes the use of the synbiotic during antimicrobial therapy for the prevention of CDI and to reduce recurrences. Valdés-Varela et al. have evaluated the in vitro inhibitory potential of four bifidobacterial strains, co-cultured with different prebiotics as carbon sources, on C. difficile growth and on the toxicity of C. difficile culture supernatants toward human intestinal epithelial cells HT29 [237]. Their co-culture with B. longum IPLA20022 or B. breve IPLA20006 in the presence of two commercial short-chain fructooligosaccharides (scFOSs) has been demonstrated to significantly reduce the growth of C. difficile and the toxicity of C. difficile supernatants. These results suggest that the optimal prebiotic substrate is probably strain-specific, so further investigations will be necessary to determine the best prebiotic substrate for different probiotic strains and the effects of synbiotics on the human intestinal microbiota.
11.5. Postbiotics
Salminen et al. defined a postbiotic as a “preparation of inanimate microorganisms and/or their components that confers a health benefit to the host” [238]. Postbiotics can act indirectly, modulating the microbiome’s composition, or directly, regulating the immune system [239] (Figure 3).
Currently, postbiotics derived from the industrial fermentation of the Lactobacillus or Bifidobacterium bacterial genera or from yeasts, such as Saccharomyces cerevisiae, are commonly metabolites or polysaccharides with antioxidant, anti-inflammatory, or immunomodulatory properties [240,241]. Novel postbiotics include whole inactivated bacteria cells, such as pasteurized Akkermansia muciniphila, which can improve glycemia and obesity rates [242].
Postbiotics represent a new option for preventing/treating CDIs. In fact, probiotics from various lactic acid bacteria, when co-cultured with toxigenic C. difficile, can reduce the cytotoxic effects of toxins on various cell lines, as well as the level of IL-8 and tumor necrosis factor α (TNF-α) released by intestinal epithelial cells when infected by C. difficile [242,243]. The lyophilized cell-free supernatant from bacteriocin-like-producing Enterococcus faecalis has been demonstrated to act as a postbiotic, with antibacterial activity against C. difficile and the capacity to inhibit the germination of spores [242].
Probiotics research is a rapidly evolving field due to their safety, especially for vulnerable patients, their longer shelf life, and their possibility of being used in topical formulations [243,244].
12. Conclusions
A C. difficile infection (CDI) is typically the result of perturbations in the gut microbiota’s composition, mainly due to the use of antibiotics. The gut microbiota has a key role in resisting C. difficile; therefore, alterations in its microbial structure create conditions conducive to C. difficile’s colonization and infection. Data on the gut microbiota’s composition indicate a decrease in species richness/diversity in both asymptomatic C. difficile carriers and patients with CDI. However, the presence or absence of certain bacteria taxa often appears more impactful in C. difficile colonization or in the development of a CDI than their diversity or species richness alone. Furthermore, recent data support the hypothesis that the gut microbiota can modulate a CDI’s severity, but also that C. difficile, particularly highly virulent types, such as RT 027, can directly influence and maintain alterations in the gut microbiota of patients with CDIs.
Accumulating data suggest that gut microbiota manipulations are an important therapeutic strategy for the prevention and treatment of CDIs. A variety of microbiota-based approaches have been developed, with some already adopted in clinical practice (i.e., FMT) and some still under investigation. However, all these approaches have still face challenges related to their standardization and safety concerns that deserve further investigation.
The increasing volume of information and the development of precise and in-depth datasets on the gut microbiota composition profiles of patients and healthy individuals will allow us to predict responses to CDI treatment and develop personalized microbiota therapies, which could be decisive for the appropriate prevention and management of CDIs in the near future.
Funding
This research received no external funding.
Acknowledgments
I would like to show my gratitude to Fabrizio Barbanti for his precious support and constant assistance throughout the writing of this manuscript.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Nasiri, M.J.; Goudarzi, M.; Hajikhani, B.; Ghazi, M.; Goudarzi, H.; Pouriran, R. Clostridioides (Clostridium) difficile infection in hospitalized patients with antibiotic-associated diarrhea: A systematic review and meta-analysis. Anaerobe 2018, 50, 32–37. [Google Scholar] [CrossRef]
- Crobach, M.J.T.; Vernon, J.J.; Loo, V.G.; Kong, L.Y.; Péchiné, S.; Wilcox, M.H.; Kuijper, E.J. Understanding Clostridium difficile colonization. Clin. Microbiol. Rev. 2018, 31, e00021-17. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.P.; Lee, J.C.; Lin, H.J.; Liu, H.C.; Wu, Y.H.; Tsai, P.J.; Ko, W.C. Clinical impact of Clostridium difficile colonization. J. Microbiol. Immunol. Infect. 2015, 48, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Leffler, D.A.; Lamont, J.T. Clostridium difficile infection. N. Engl. J. Med. 2015, 372, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Rupnik, M. Heterogeneity of large clostridial toxins: Importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 2008, 32, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Lacy, D.B. The role of toxins in Clostridium difficile infection. FEMS Microbiol. Rev. 2017, 41, 723–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Monaghan, T.; Yadegar, A.; Louie, T.; Kao, D. Insights into the evolving epidemiology of Clostridioides difficile infection and treatment: A global perspective. Antibiotics 2023, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Pedroza Matute, S.; Iyavoo, S. Exploring the gut microbiota: Lifestyle choices, disease associations, and personal genomics. Front. Nutr. 2023, 10, 1225120. [Google Scholar] [CrossRef] [PubMed]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel. Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Ufondu, A.; Lee, K.; Jayaraman, A. Emerging computational tools and models for studying gut microbiota composition and function. Curr. Opin. Biotechnol. 2020, 66, 301–331. [Google Scholar] [CrossRef] [PubMed]
- Alagiakrishnan, K.; Morgadinho, J.; Halverson, T. Approach to the diagnosis and management of dysbiosis. Front. Nutr. 2024, 11, 1330903. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.R.; Pike, C.M.; Parsons, R.J.; Rivera, A.J.; Foley, M.H.; McLaren, M.R.; Montgomery, S.A.; Theriot, C.M. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat. Commun. 2021, 12, 462. [Google Scholar] [CrossRef] [PubMed]
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: Pathogenesis and host defence. Nat. Rev. Microbiol. 2016, 14, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Loo, V.G.; Bourgault, A.M.; Poirier, L.; Lamothe, F.; Michaud, S.; Turgeon, N.; Toye, B.; Beaudoin, A.; Frost, E.H.; Gilca, R.; et al. Host and pathogen factors for Clostridium difficile infection and colonization. N. Engl. J. Med. 2011, 365, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Schäffler, H.; Breitrück, A. Clostridium difficile—From colonization to infection. Front. Microbiol. 2018, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Maida, M.; Burisch, J.; Simonelli, C.; Hold, G.; Ventimiglia, M.; Gasbarrini, A.; Cammarota, G. Efficacy of different faecal microbiota transplantation protocols for Clostridium difficile infection: A systematic review and meta-analysis. United Eur. Gastroenterol. J. 2018, 6, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, T.; Sun, J.; Liu, N. Fecal microbiota transplantation and health outcomes: An umbrella review of meta-analyses of randomized controlled trials. Front. Cell. Infect. Microbiol. 2022, 12, 899845. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Zahirović, A.; Bizjak, M.; Rupnik, M.; Štrukelj, B.; Berlec, A. New treatment approaches for Clostridioides difficile infections: Alternatives to antibiotics and fecal microbiota transplantation. Gut Microbes 2024, 16, 2337312. [Google Scholar] [CrossRef]
- Gonzales-Luna, A.J.; Carlson, T.J.; Garey, K.W. Gut microbiota changes associated with Clostridioides difficile infection and its various treatment strategies. Gut Microbes 2023, 15, 2223345. [Google Scholar] [CrossRef]
- Dixit, K.; Chaudhari, D.; Dhotre, D.; Shouche, Y.; Saroj, S. Restoration of dysbiotic human gut microbiome for homeostasis. Life Sci. 2021, 278, 119622. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Engevik, M.A.; Spinler, J.K.; Versalovic, J. Healthy human gastrointestinal microbiome: Composition and function after a decade of Exploration. Dig. Dis. Sci. 2020, 65, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The gut microbiota in the first decade of life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef]
- Schloissnig, S.; Arumugam, M.; Sunagawa, S.; Mitreva, M.; Tap, J.; Zhu, A.; Waller, A.; Mende, D.R.; Kultima, J.R.; Martin, J.; et al. Genomic variation landscape of the human gut microbiome. Nature 2013, 493, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, F. A possible perspective about the compositional models, evolution, and clinical meaning of human enterotypes. Microorganisms 2021, 9, 2341. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Ning, K. Stereotypes about enterotype: The old and new ideas. Genom. Proteom. Bioinform. 2019, 17, 4–12. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M.; Tilg, H.; van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Kachrimanidou, M.; Tsintarakis, E. Insights into the role of human gut microbiota in Clostridioides difficile infection. Microorganisms 2020, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Giel, J.L.; Sorg, J.A.; Sonenshein, A.L.; Zhu, J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE 2020, 5, e8740. [Google Scholar] [CrossRef]
- Antunes, L.C.M.; Finlay, B.B. A comparative analysis of the effect of antibiotic treatment and enteric infection on intestinal homeostasis. Gut Microbes 2011, 2, 105–108. [Google Scholar] [CrossRef]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.M.; Yalcinkaya, N.; Wu, Q.; Swennes, A.; Tessier, M.E.; Roberts, P.; Miyajima, F.; Savidge, T.; Sorg, J.A. Bile acid-independent protection against Clostridioides difficile infection. PLoS Pathog. 2021, 17, e1010015. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: Role of secondary bile acids. Cell Chem. Biol. 2019, 26, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Thanissery, R.; Winston, J.A.; Theriot, C.M. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 2017, 45, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Studer, N.; Desharnais, L.; Beutler, M.; Brugiroux, S.; Terrazos, M.A.; Menin, L.; Schürch, C.M.; McCoy, K.D.; Kuehne, S.A.; Minton, N.P.; et al. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell Infect. Microbiol. 2016, 6, 191. [Google Scholar] [CrossRef] [PubMed]
- Chen See, J.R.; Leister, J.; Wright, J.R.; Kruse, P.I.; Khedekar, M.V.; Besch, C.E.; Kumamoto, C.A.; Madden, G.R.; Stewart, D.B.; Lamendella, R. Clostridioides difficile infection is associated with differences in transcriptionally active microbial communities. Front. Microbiol. 2024, 15, 1398018. [Google Scholar] [CrossRef] [PubMed]
- Revolinski, S.L.; Munoz-Price, L.S. Clostridium difficile exposures, colonization, and the microbiome: Implications for prevention. Infect. Control. Hosp. Epidemiol. 2018, 39, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, R.D. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun. 1984, 45, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.M.; Sorg, J.A. Gut associated metabolites and their roles in Clostridioides difficile pathogenesis. Gut Microbes 2022, 14, 2094672. [Google Scholar] [CrossRef]
- O’Reilly, C.; O’Connor, P.M.; O’Sullivan, Ó.; Rea, M.C.; Hill, C.; Ross, R.P. Impact of nisin on Clostridioides difficile and microbiota composition in a faecal fermentation model of the human colon. J. Appl. Microbiol. 2022, 132, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. Clostridioides difficile infection: Microbe-microbe interactions and live biotherapeutics. Front. Microbiol. 2023, 14, 1182612. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.D.; Theriot, C.M. Contribution of inhibitory metabolites and competition for nutrients to colonization resistance against Clostridioides difficile by commensal Clostridium. Microorganisms 2021, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Moya, A.; Gosalbes, M.J.; Latorre, A. Colonization resistance of the gut microbiota against Clostridium difficile. Antibiotics 2015, 4, 337–357. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, e23. [Google Scholar] [CrossRef] [PubMed]
- Umesaki, Y.; Setoyama, H.; Matsumoto, S.; Imaoka, A.; Itoh, K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 1999, 67, 3504–3511. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Ariyoshi, T.; Kuroki, Y.; Eguchi, S.; Higashi, S.; Mori, T.; Nonogaki, T.; Iwasaki, K.; Yamashita, M.; Asai, N.; et al. Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci. Rep. 2021, 11, 15007. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Nagao-Kitamoto, H.; Kitamoto, S.; Kim, C.H.; Kamada, N. The butyrate-producing bacterium Clostridium butyricum suppresses Clostridioides difficile infection via neutrophil- and antimicrobial cytokine–dependent but GPR43/109a-independent mechanisms. J. Immunol. 2021, 206, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Osaki, T.; Hanawa, T.; Kurata, S.; Sugiyama, E.; Takahashi, M.; Tanaka, M.; Taguchi, H.; Kamiya, S. Establishment of an endogenous Clostridium difficile rat infection model and evaluation of the effects of Clostridium butyricum MIYAIRI 588 probiotic strain. Front. Microbiol. 2018, 9, 1264. [Google Scholar] [CrossRef]
- Lee, J.C.; Chiu, C.W.; Tsai, P.J.; Lee, C.C.; Huang, I.H.; Ko, W.C.; Hung, Y.P. Clostridium butyricum therapy for mild-moderate Clostridioides difficile infection and the impact of diabetes mellitus. Biosci. Microbiota Food Health 2022, 41, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Artacho, A.; Ott, S.J.; Moya, A.; Gosalbes, M.J.; Latorre, A. Structural and functional changes in the gut microbiota associated to Clostridium difficile infection. Front. Microbiol. 2014, 5, 335. [Google Scholar]
- Dai, Z.L.; Wu, G.; Zhu, W.Y. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768–1786. [Google Scholar] [CrossRef] [PubMed]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, e4548. [Google Scholar] [CrossRef] [PubMed]
- Rojo, D.; Gosalbes, M.J.; Ferrari, R.; Pérez-Cobas, A.E.; Hernández, E.; Oltra, R.; Buesa, J.; Latorre, A.; Barbas, C.; Ferrer, M.; et al. Clostridium difficile heterogeneously impacts intestinal community architecture but drives stable metabolome responses. ISME J. 2015, 9, 2206–2220. [Google Scholar] [CrossRef] [PubMed]
- Furuya-Kanamori, L.; Marquess, J.; Yakob, L.; Riley, T.V.; Paterson, D.L.; Foster, N.F.; Huber, C.A.; Clements, A.C. Asymptomatic Clostridium difficile colonization: Epidemiology and clinical implications. BMC Infect. Dis. 2015, 15, 516. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Soto Ocana, J.; Zackular, J.P. From nursery to nursing home: Emerging concepts in Clostridioides difficile pathogenesis. Infect. Immun. 2020, 88, e00934-19. [Google Scholar] [CrossRef] [PubMed]
- Shirley, D.A.; Tornel, W.; Warren, C.A.; Moonah, S. Clostridioides difficile infection in children: Recent updates on epidemiology, diagnosis, therapy. Pediatrics 2023, 52, e2023062307. [Google Scholar] [CrossRef] [PubMed]
- Semon, A.K.; Keenan, O.; Zackular, J.P. Clostridioides difficile and the microbiota early in life. J. Pediatr. Infect. Dis. Soc. 2021, 10 (Suppl. 3), S3–S7. [Google Scholar] [CrossRef]
- Tonooka, T.; Sakata, S.; Kitahara, M.; Hanai, M.; Ishizeki, S.; Takada, M.; Sakamoto, M.; Benno, Y. Detection and quantification of four species of the genus Clostridium in infant feces. Microbiol. Immunol. 2005, 49, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, M.; Baharav, N.; Melzer, E.; Regev-Yochay, G.; Yahav, D. Screening for asymptomatic Clostridioides difficile carriage among hospitalized patients: A narrative review. Infect. Dis. Ther. 2023, 12, 2223–2240. [Google Scholar] [CrossRef]
- De Roo, A.C.; Regenbogen, S.E. Clostridium difficile infection: An epidemiology update. Clin. Colon. Rectal. Surg. 2020, 33, 49–57. [Google Scholar] [CrossRef]
- Kyne, L.; Warny, M.; Qamar, A.; Kelly, C.P. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 2000, 342, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, M.H.; Gerding, D.N.; Poxton, I.R.; Kelly, C.; Nathan, R.; Birch, T.; Cornely, O.A.; Rahav, G.; Bouza, E.; Lee, C.; et al. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N. Engl. J. Med. 2017, 376, 305–317. [Google Scholar] [CrossRef]
- Dieterle, M.G.; Young, V.B. Reducing recurrence of C. difficile infection. Cell 2017, 169, 375. [Google Scholar] [CrossRef] [PubMed]
- Blixt, T.; Gradel, K.O.; Homann, C.; Seidelin, J.B.; Schonning, K.; Lester, A.; Houlind, J.; Stangerup, M.; Gottlieb, M.; Knudsen, J.D. Asymptomatic carriers contribute to nosocomial Clostridium difficile infection: A cohort study of 4508 patients. Gastroenterology 2017, 152, 1031–1041.e2. [Google Scholar] [CrossRef] [PubMed]
- Lanzas, C.; Dubberke, E.R.; Lu, Z.; Reske, K.A.; Gröhn, Y.T. Epidemiological model for Clostridium difficile transmission in healthcare settings. Infect. Control. Hosp. Epidemiol. 2011, 32, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Charis, A.; Ning Yu, M.; Lockhart, M.C.; McGuigan, C.C.; Wiuff, C.; Davey, P.G.; Donnan, P.T. Community-associated Clostridium difficile infection among older people in Tayside, Scotland, is associated with antibiotic exposure and care home residence: Cohort study with nested case–control. J. Antimicrob. Chemother. 2013, 68, 2927–2933. [Google Scholar]
- Wilcox, M.H.; Mooney, L.; Bendall, R.; Settle, C.D.; Fawley, W.N. A case-control study of community-associated Clostridium difficile infection. J. Antimicrob. Chemother. 2008, 62, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Zacharioudakis, I.M.; Zervou, F.N.; Pliakos, E.E.; Ziakas, P.D.; Mylonakis, E. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: A systematic review and meta-analysis. Am. J. Gastroenterol. 2015, 110, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Nissle, K.; Kopf, D.; Rösler, A. Asymptomatic and yet C. difficile-toxin positive? Prevalence and risk factors of carriers of toxigenic Clostridium difficile among geriatric in-patients. BMC Geriatr. 2016, 16, 185. [Google Scholar] [CrossRef]
- Rousseau, C.; Levenez, F.; Fouqueray, C.; Doré, J.; Collignon, A.; Lepage, P. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J. Clin. Microbiol. 2011, 49, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Sakata, S.; Tonooka, T.; Ishizeki, S.; Takada, M.; Sakamoto, M.; Fukuyama, M.; Benno, Y. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol. Lett. 2005, 243, 417–423. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere 2019, 3, e00041-18. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef]
- Tun, H.M.; Peng, Y.; Chen, B.; Konya, T.B.; Morales-Lizcano, N.P.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; et al. Ethnicity associations with Food sensitization are mediated by gut microbiota development in the first year of life. Gastroenterology 2021, 161, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Liu, X.; Jia, X.; Cheng, Y.; Luo, Y.; Yuan, L.; Wang, Y.; Zhao, C.; Guo, S.; Li, L.; et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci. Rep. 2014, 4, 7485. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Lepage, P.; Jolivet, S.; Delannoy, J.; Mesa, V.; Ancel, P.Y.; Rozé, J.C.; Butel, M.J.; Barbut, F.; Aires, J. Gut microbiota diversity of preterm neonates is associated with Clostridioides difficile colonization. Front. Cell Infect. Microbiol. 2022, 12, 907323. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.Y.; Zhang, H.; Brannan, L.E.; Carman, R.J.; Boone, J.H. Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Microbiome 2016, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Th, L.; Dong, D.; Jiang, C.; Li, Z.; Wang, X.; Peng, Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 2015, 34, 1–7. [Google Scholar]
- Vázquez-Cuesta, S.; Villar, L.; García, N.L.; Fernández, A.I.; Olmedo, M.; Alcalá, L.; Marín, M.; Muñoz, P.; Bouza, E.; Reigadas, E. Characterization of the gut microbiome of patients with Clostridioides difficile infection, patients with non-C. difficile diarrhea, and C. difficile-colonized patients. Front. Cell Infect. Microbiol. 2023, 13, 1130701. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.T.; Ducarmon, Q.R.; Terveer, E.M.; Harmanus, C.; Sanders, I.M.J.G.; Verduin, K.M.; Kuijper, E.J.; Zwittink, R.D. The bacterial gut microbiota of adult patients infected, colonized or noncolonized by Clostridioides difficile. Microorganisms 2020, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.; Miller, M.A.; Edens, T.J.; Mehrotra, S.; Dewar, K.; Manges, A.R. Bloom and bust: Intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Henderickx, J.G.E.; Crobach, M.J.T.; Terveer, E.M.; Smits, W.K.; Kuijper, E.J.; Zwittink, R.D. Fungal and bacterial gut microbiota differ between Clostridioides difficile colonization and infection. Microbiome Res. Rep. 2023, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, M.; Meng, J.; Wang, L.; Chen, M. A review of the interaction between diet composition and gut microbiota and its impact on associated disease. J. Future Foods 2024, 4, 221–232. [Google Scholar] [CrossRef]
- Britton, R.A.; Young, V.B. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 2014, 146, 1547–1553. [Google Scholar] [CrossRef] [PubMed]
- Vila, A.V.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.M.A.E.; Masclee, A.A.M.; Fu, J.; et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Hryckowian, A.J.; Treuren, W.V.; Smits, S.A.; Davis, N.M.; Gardner, J.O.; Bouley, D.M.; Sonnenburg, J.L. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat. Microbiol. 2018, 3, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Lange, K.; Buerger, M.; Stallmach, A.; Bruns, T. Effects of antibiotics on gut microbiota. Dig. Dis. 2016, 34, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, D.; Nigam, P.S. Antibiotic-therapy-induced gut dysbiosis affecting gut microbiota-brain axis and cognition: Restoration by intake of probiotics and synbiotics. Int. J. Mol Sci. 2023, 24, 3074. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Neulinger, S.C.; Heinsen, F.A.; Knecht, C.; Schilhabel, A.; Schmitz, R.A.; Zimmermann, A.; dos Santos, V.M.; Ferrer, M.; Rosenstiel, P.C.; et al. Effects of beta-lactam antibiotics and fluoroquinolones on human gut microbiota in relation to Clostridium difficile associated diarrhea. PLoS ONE 2014, 9, e89417. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; de Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota—A systematic review. J. Infect. 2019, 79, 471–489. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, K.; Khanna, S. Gut microbiome and Clostridioides difficile infection: A closer look at the microscopic interface. Ther. Adv. Gastroenterol. 2021, 14, 1756284821994736. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.; O’Neill, F.J.; Wilcox, M.H. Effects of cefotaxime and desacetylcefotaxime upon Clostridium difficile proliferation and toxin production in a triple-stage chemostat model of the human gut. J. Antimicrob. Chemother. 2003, 52, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Altman, K.W.; Chhaya, V.; Hammer, N.D.; Pavlova, S.; Vesper, B.J.; Tao, L.; Radosevich, J.A. Effect of proton pump inhibitor pantoprazole on growth and morphology of oral Lactobacillus strains. Laryngoscope 2008, 118, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Clooney, A.G.; Bernstein, C.N.; Leslie, W.D.; Vagianos, K.; Sargent, M.; Laserna-Mendieta, E.J.; Claesson, M.J.; Targownik, L.E. A comparison of the gut microbiome between long-term users and non-users of proton pump inhibitors. Aliment. Pharmacol. Ther. 2016, 43, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Indiani, C.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.F.C.; Darrieux, M.; Parisotto, T.M. Childhood obesity and Firmicutes/Bacteroidetes ratio in the gut microbiota: A systematic review. Child Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Buddle, J.E.; Fagan, R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence 2023, 14, 2150452. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Johanesen, P.A.; Carter, G.P.; Rose, E.; Lyras, D. Clostridium difficile virulence factors: Insights into an anaerobic spore-forming pathogen. Gut Microbes 2014, 5, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Hundsberger, T.; Leukel, P.; Sauerborn, M.; Eichel-Streiber, C.V. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 1996, 181, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.H.; Tang, Y.J.; Silva, J., Jr. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 2000, 18, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Kordus, S.L.; Thomas, A.K.; Lacy, D.B. Clostridioides difficile toxins: Mechanisms of action and antitoxin therapeutics. Nat. Rev. Microbiol. 2022, 20, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Skinner, A.M.; Phillips, S.T.; Merrigan, M.M.; O’Leary, K.J.; Sambol, S.P.; Siddiqui, F.; Peterson, L.R.; Gerding, D.N.; Johnson, S. The relative role of toxins A and B in the virulence of Clostridioides difficile. J. Clin. Med. 2020, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Gerding, D.N.; Johnson, S.; Rupnik, M.; Aktories, K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes 2014, 5, 15–27. [Google Scholar] [CrossRef]
- Papatheodorou, P.; Minton, N.P.; Aktories, K.; Barth, H. An updated view on the cellular uptake and mode-of-action of Clostridioides difficile toxins. Adv. Exp. Med. Biol. 2024, 1435, 219–247. [Google Scholar] [PubMed]
- Aktories, K.; Wegner, A. Mechanisms of the cytopathic action of actin-ADP-ribosylating toxins. Mol Microbiol. 1992, 6, 2905–2908. [Google Scholar] [CrossRef] [PubMed]
- Schwan, C.; Stecher, B.; Tzivelekidis, T.; van Ham, M.; Rohde, M.; Hardt, W.D.; Wehland, J.; Aktories, K. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009, 5, e1000626. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.J.; Ballard, J.D. Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol. Mol. Biol. Rev. 2013, 77, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Lanis, J.M.; Barua, S.; Ballard, J.D. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 2010, 6, e1001061. [Google Scholar] [CrossRef]
- Theriot, C.M.; Young, V.B. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu. Rev. Microbiol. 2015, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Young, V.B. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes 2014, 5, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.M.; Rogers, M.A.; Ring, C.; Mogle, J.; Petrosino, J.P.; Young, V.B.; Aronoff, D.M.; Schloss, P.D. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 2014, 5, e01021-14. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.D.; Ann, H.W.; Lee, W.J.; Kim, J.H.; Seong, H.; Kim, J.H.; Ahn, J.Y.; Jeong, S.J.; Ku, N.S.; Yeom, J.S.; et al. Characteristics of faecal microbiota in Korean patients with Clostridioides difficile-associated diarrhea. Infect. Chemother. 2019, 51, 365–375. [Google Scholar] [CrossRef]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Maier, L.; Hardt, W.D. ’Blooming’ in the gut: How dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 2013, 11, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Yi, J.; Kim, J.H.; Lee, S.; Moon, H.W. Composition of gut microbiota in patients with toxigenic Clostridioides (Clostridium) difficile: Comparison between subgroups according to clinical criteria and toxin gene load. PLoS ONE 2019, 14, e0212626. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Taminiau, B.; Rodriguez, C.; Daube, G. Gut microbiota composition associated with Clostridioides difficile colonization and infection. Pathogens 2022, 11, 781. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Arguello, E.S.; Jenq, R.R.; Littmann, E.; Kim, G.J.; Miller, L.C.; Ling, L.; Figueroa, C.; Robilotti, E.; Perales, M.A.; et al. Protective factors in the intestinal microbiome against Clostridium difficile infection in recipients of allogeneic hematopoietic stem cell transplantation. J. Infect. Dis. 2017, 215, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Fishbein, S.R.; Robinson, J.I.; Hink, T.; Reske, K.A.; Newcomer, E.P.; Burnham, C.D.; Henderson, J.P.; Dubberke, E.R.; Dantas, G. Multi-omics investigation of Clostridioides difficile-colonized patients reveals pathogen and commensal correlates of C. difficile pathogenesis. eLife 2022, 11, e72801. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Labbe, A.; Loo, V.G.; Atherton, J.K.; Behr, M.A.; Masson, L.; Tellis, P.A.; Brousseau, R. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridium difficile-associated disease. J. Infect. Dis. 2010, 202, 1877–1884. [Google Scholar] [CrossRef]
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The gut bacterial community potentiates Clostridioides difficile infection severity. mBio 2022, 13, e0118322. [Google Scholar] [CrossRef] [PubMed]
- Magdy Wasfy, R.; Mbaye, B.; Borentain, P.; Tidjani Alou, M.; Murillo Ruiz, M.L.; Caputo, A.; Andrieu, C.; Armstrong, N.; Million, M.; Gerolami, R. Ethanol-producing Enterocloster bolteae is enriched in chronic hepatitis B-associated gut dysbiosis: A case-control culturomics Study. Microorganisms 2023, 11, 2437. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Berkell, M.; Mysara, M.; Xavier, B.B.; van Werkhoven, C.H.; Monsieurs, P.; Lammens, C.; Ducher, A.; Vehreschild, M.J.G.T.; Goossens, H.; de Gunzburg, J.; et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 2021, 12, 2241. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Montassier, E.; Schmidt, B.; Patel, R.; Knights, D.; Pardi, D.S.; Kashyap, P. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment. Pharmacol. Ther. 2016, 44, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Gazzola, A.; Panelli, S.; Corbella, M.; Merla, C.; Comandatore, F.; De Silvestri, A.; Piralla, A.; Zuccaro, V.; Bandi, C.; Marone, P.; et al. Microbiota in Clostridioides difficile-associated diarrhea: Comparison in recurrent and non-recurrent infections. Biomedicines 2020, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Chen, C.; Bobr, A.; Yao, D.; Lu, Y.; Nelson, V.M.; Sadowsky, M.J.; Khoruts, A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G310–G319. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, J.R.; Kearney, S.; Li, N.; Bogart, E.; Bullock, K.; Gerber, G.K.; Bry, L.; Clish, C.B.; Alm, E.; Korzenik, J.R. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 2016, 43, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.K.; Brensinger, C.M.; Wu, Q.; Lewis, J.D. Increasing incidence of multiply recurrent Clostridium difficile infection in the United States: A cohort study. Ann. Intern. Med. 2017, 167, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Davies, K.; Barbut, F. Ribotypes and new virulent strains across Europe. Adv. Exp. Med. Biol. 2024, 1435, 151–168. [Google Scholar] [PubMed]
- McDonald, L.C.; Killgore, G.E.; Thompson, A.; Owens, R.C.; Kazakova, S.V.; Sambol, S.P.; Johnson, S.; Gerding, D.N. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 2005, 353, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Beldavs, Z.G.; Winston, L.G.; Dumyati, G.; Holzbauer, S.; Dunn, J.; Farley, M.M.; Lyons, C.; Johnston, H.; Phipps, E.; et al. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin. Infect. Dis. 2014, 58, 1394–1400. [Google Scholar]
- Bauer, M.P.; Notermans, D.W.; van Benthem, B.H.; Brazier, J.S.; Wilcox, M.H.; Rupnik, M.; Monnet, D.L.; van Dissel, J.T.; Kuijper, E.J.; Group, E.S. Clostridium difficile infection in Europe: A hospital-based survey. Lancet 2011, 377, 63–73. [Google Scholar] [CrossRef]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar]
- Curry, S.R.; Marsh, J.W.; Muto, C.A.; O’Leary, M.M.; Pasculle, A.W.; Harrison, L.H. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 2007, 45, 215–221. [Google Scholar] [CrossRef]
- Warny, M.; Pepin, J.; Fang, A.; Killgore, G.; Thompson, A.; Brazier, J.; Frost, E.; McDonald, L.C. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 2005, 366, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Meléndez, A.; Morfin-Otero, R.; Villarreal-Treviño, L.; Baines, S.D.; Camacho-Ortíz, A.; Garza-González, E. Molecular epidemiology of predominant and emerging Clostridioides difficile ribotypes. J. Microbiol. Methods 2020, 175, 105974. [Google Scholar] [CrossRef]
- Spigaglia, P.; Mastrantonio, P.; Barbanti, F. Antibiotic resistances of Clostridioides difficile. Adv. Exp. Med. Biol. 2024, 1435, 169–198. [Google Scholar]
- Goorhuis, A.; Bakker, D.; Corver, J.; Debast, S.B.; Harmanus, C.; Notermans, D.W.; Bergwerff, A.A.; Dekker, F.W.; Kuijper, E.J. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 2008, 47, 1162–1170. [Google Scholar] [CrossRef]
- Knight, D.R.; Kullin, B.; Androga, G.O.; Barbut, F.; Eckert, C.; Johnson, S.; Spigaglia, P.; Tateda, K.; Tsai, P.J.; Riley, T.V. Evolutionary and genomic insights into Clostridioides difficile sequence type 11: A diverse zoonotic and antimicrobial-resistant lineage of global One Health importance. mBio 2019, 10, e00446-19. [Google Scholar] [CrossRef] [PubMed]
- Stabler, R.A.; Dawson, L.F.; Valiente, E.; Cairns, M.D.; Martin, M.J.; Donahue, E.H.; Riley, T.V.; Songer, J.G.; Kuijper, E.J.; Dingle, K.E.; et al. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS ONE 2012, 7, e31559. [Google Scholar] [CrossRef]
- Nyc, O.; Pituch, H.; Matejkova, J.; Obuch-Woszczatynski, P.; Kuijper, E.J. Clostridium difficile PCR ribotype 176 in the Czech Republic and Poland. Lancet 2011, 377, 1407. [Google Scholar] [CrossRef] [PubMed]
- Valiente, E.; Dawson, L.F.; Cairns, M.D.; Stabler, R.A.; Wren, B.W. Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J. Med. Microbiol. 2012, 61, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Khanna, S. Community-acquired Clostridium difficile infection: An increasing public health threat. Infect. Drug Resist. 2014, 7, 63–72. [Google Scholar] [PubMed]
- Jenior, M.L.; Leslie, J.L.; Young, V.B.; Schloss, P.D. Clostridium difficile alters the structure and metabolism of distinct cecal microbiomes during initial infection to promote sustained colonization. mSphere 2018, 3, e00261-18. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.; Auchtung, J.; Collins, J.; Britton, R. Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates. Infect. Immun. 2014, 82, 2815–2825. [Google Scholar] [CrossRef] [PubMed]
- Skraban, J.; Dzeroski, S.; Zenko, B.; Mongus, D.; Gangl, S.; Rupnik, M. Gut microbiota patterns associated with colonization of different Clostridium difficile ribotypes. PLoS ONE 2013, 8, e58005. [Google Scholar] [CrossRef] [PubMed]
- Horvat, S.; Mahnic, A.; Breskvar, M.; Dzeroski, S.; Rupnik, M. Evaluating the effect of Clostridium difficile conditioned medium on fecal microbiota community structure. Sci. Rep. 2017, 27, 16448. [Google Scholar] [CrossRef]
- Horvat, S.; Rupnik, M. Interactions between Clostridioides difficile and fecal microbiota in vitro batch model: Growth, sporulation, and microbiota changes. Front. Microbiol. 2018, 9, 1633. [Google Scholar] [CrossRef] [PubMed]
- Horvat, S.; Mahnic, A.; Makuc, D.; Pečnik, K.; Plavec, J.; Rupnik, M. Children gut microbiota exhibits a different composition and metabolic profile after in vitro exposure to Clostridioides difficile and increases its sporulation. Front. Microbiol. 2022, 13, 1042526. [Google Scholar] [CrossRef]
- Vincent, C.; Manges, A.R. Antimicrobial use, human gut microbiota and Clostridium difficile colonization and infection. Antibiotics 2015, 4, 230–253. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, I.M.; Chifiriuc, M.C.; Pircalabioru, G.G.; Filip, R.; Bolocan, A.; Lazăr, V.; Diţu, L.M.; Bleotu, C. Gut dysbiosis and Clostridioides difficile infection in neonates and adults. Front. Microbiol. 2022, 12, 651081. [Google Scholar] [CrossRef] [PubMed]
- Stallhofer, J.; Steube, A.; Katzer, K.; Stallmach, A. Microbiota-based therapeutics as new standard-of-care treatment for recurrent Clostridioides difficile infection. Visc. Med. 2024, 40, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Wortelboer, K.; Nieuwdorp, M.; Herrema, H. Fecal microbiota transplantation beyond Clostridioides difficile infections. EBioMedicine 2019, 44, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Masucci, L.; Ianiro, G.; Bibbo, S.; Dinoi, G.; Costamagna, G.; Sanguinetti, M.; Gasbarrini, A. Randomised clinical trial: Faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 2015, 41, 835–843. [Google Scholar] [CrossRef]
- Du, C.; Luo, Y.; Walsh, S.; Grinspan, A. Oral fecal microbiota transplant capsules are safe and effective for recurrent Clostridioides difficile infection: A systematic review and meta-analysis. J. Clin. Gastroenterol. 2021, 55, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Gangwani, M.K.; Aziz, M.; Aziz, A.; Priyanka, F.; Weissman, S.; Phan, K.; Dahiya, D.S.; Ahmed, Z.; Sohail, A.H.; Lee-Smith, W.; et al. Fresh versus frozen versus lyophilized fecal microbiota transplant for recurrent Clostridium difficile infection: A systematic review and network meta-analysis. J. Clin. Gastroenterol. 2023, 57, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Ramai, D.; Zakhia, K.; Fields, P.J.; Ofosu, A.; Patel, G.; Shahnazarian, V.; Lai, J.K.; Dhaliwal, A.; Reddy, M.; Chang, S. Fecal microbiota transplantation (FMT) with colonoscopy is superior to enema and nasogastric tube while comparable to capsule for the treatment of recurrent Clostridioides difficile infection: A systematic review and meta-analysis. Dig. Dis. Sci. 2021, 66, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Hvas, C.L.; Dahl Jørgensen, S.M.; Jørgensen, S.P.; Storgaard, M.; Lemming, L.; Hansen, M.M.; Erikstrup, C.; Dahlerup, J.F. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent Clostridium difficile infection. Gastroenterology 2019, 156, 1324–1332.e3. [Google Scholar] [CrossRef] [PubMed]
- Rokkas, T.; Gisbert, J.P.; Gasbarrini, A.; Hold, G.L.; Tilg, H.; Malfertheiner, P.; Megraud, F.; O’Morain, C.A. A network meta-analysis of randomized controlled trials exploring the role of fecal microbiota transplantation in recurrent Clostridium difficile infection. United Eur. Gastroenterol. J. 2019, 7, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, W.; Li, P.; Wen, Q.; Cui, B.; Zhang, F. Washed preparation of faecal microbiota changes the transplantation related safety, quantitative method and delivery. Microb. Biotechnol. 2022, 15, 2439–2449. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ai, R.J.; Xu, J.; Wen, Q.; Pan, H.Q.; Zhang, Z.H.; Ning, W.; Fang, Y.; Ding, D.F.; Wang, Q.; et al. Washed microbiota transplantation for Clostridioides difficile infection: A national multicenter real-world study. J. Dig. Dis. 2023, 24, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Kelly, C.R.; Mullish, B.H.; Allegretti, J.R.; Kassam, Z.; Putignani, L.; Fischer, M.; Keller, J.J.; Costello, S.P.; et al. International consensus conference on stool banking for faecal microbiota transplantation in clinical practice. Gut 2019, 68, 2111–2121. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.J.; Ooijevaar, R.E.; Hvas, C.L.; Terveer, E.M.; Lieberknecht, S.C.; Högenauer, C.; Arkkila, P.; Sokol, H.; Gridnyev, O.; Mégraud, F.; et al. A standardised model for stool banking for faecal microbiota transplantation: A consensus report from a multidisciplinary UEG working group. United Eur. Gastroenterol. J. 2021, 9, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.A.; Babakhani, F.; Sears, P.; Nguyen, L.; Sorg, J.A. Both fidaxomicin and vancomycin inhibit outgrowth of Clostridium difficile spores. Antimicrob. Agents Chemother. 2013, 57, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Khoruts, A.; Sadowsky, M.J. Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Seekatz, A.M.; Aas, J.; Gessert, C.E.; Rubin, T.A.; Saman, D.M.; Bakken, J.S.; Young, V.B. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 2014, 5, e00893-14. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Chang, T.E.; Wang, Y.P.; Lee, K.C.; Lin, Y.T.; Chiou, J.J.; Huang, C.W.; Yang, U.C.; Li, F.Y.; Huang, H.C.; et al. Alteration of gut microbial composition associated with the therapeutic efficacy of fecal microbiota transplantation in Clostridium difficile infection. J. Formos. Med. Assoc. 2022, 121, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Shankar, V.; Hamilton, M.J.; Khoruts, A.; Kilburn, A.; Unno, T.; Paliy, O.; Sadowsky, M.J. Species and genus level resolution analysis of gut microbiota in Clostridium difficile patients following fecal microbiota transplantation. Microbiome 2014, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Vazquez-Baeza, Y.; Gonzalez, A.; Weiss, S.; Schmidt, B.; Muniz-Pedrogo, D.A.; Rainey, J.F.; Kammer, P.; Nelson, H.; Sadowsky, M.; et al. Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 2017, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Aggarwala, V.; Mogno, I.; Li, Z.; Yang, C.; Britton, G.J.; Chen-Liaw, A.; Mitcham, J.; Bongers, G.; Gevers, D.; Clemente, J.C.; et al. Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes. Nat. Microbiol. 2021, 6, 1309–1318. [Google Scholar] [CrossRef]
- Wei, S.; Bahl, M.I.; Baunwall, S.M.D.; Dahlerup, J.F.; Hvas, C.L.; Licht, T.R. Gut microbiota differs between treatment outcomes early after fecal microbiota transplantation against recurrent Clostridioides difficile infection. Gut Microbes 2022, 14, 2084306. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Cui, J.; Ye, C.; Zhao, J.; Yang, B.; Xu, Y.; Ji, S.; Wang, L.; Lv, X.; Ma, C.; et al. Depletion of butyrate-producing microbes of the Firmicutes predicts nonresponse to FMT therapy in patients with recurrent Clostridium difficile infection. Gut Microbes 2023, 15, 2236362. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Kaiser, T.; Vaughn, B.P.; Graiziger, C.T.; Hamilton, M.J.; Rehman, T.U.; Song, K.; Khoruts, A.; Sadowsky, M.J. Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation. Microbiome 2018, 6, 166. [Google Scholar] [CrossRef] [PubMed]
- Fachi, J.L.; Felipe, J.S.; Pral, L.P.; da Silva, B.K.; Corrêa, R.O.; de Andrade, M.C.P.; da Fonseca, D.M.; Basso, P.J.; Câmara, N.O.S.; de Sales, E.S.É.; et al. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep. 2019, 27, 750–761.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, J.; Gao, W.; Li, L.; Li, P.; Zhang, L.; Gong, Y.N.; Peng, X.; Xi, J.J.; Chen, S.; et al. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature 2014, 513, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, J.J.; Allegretti, J.R.; Gibson, T.E.; McClure, E.; Delaney, M.; Bry, L.; Gerber, G.K. Gut metabolites predict Clostridioides difficile recurrence. Microbiome 2022, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Pakpour, S.; Bhanvadia, A.; Zhu, R.; Amarnani, A.; Gibbons, S.M.; Gurry, T.; Alm, E.J.; Martello, L.A. Identifying predictive features of Clostridium difficile infection recurrence before, during, and after primary antibiotic treatment. Microbiome 2017, 5, 148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhong, H.; Li, Y.; Shi, Z.; Ren, H.; Zhang, Z.; Zhou, X.; Tang, S.; Han, X.; Lin, Y.; et al. Sex- and age-related trajectories of the adult human gut microbiota shared across populations of different ethnicities. Nat. Aging 2021, 1, 87–100. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.C.; Vatanen, T.; Cutfield, W.S.; O’Sullivan, J.M. The super-donor phenomenon in fecal microbiota transplantation. Front. Cell. Infect. Microbiol. 2019, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Valles-Colomer, M.; Bacigalupe, R.; Vieira-Silva, S.; Suzuki, S.; Darzi, Y.; Tito, R.Y.; Yamada, T.; Segata, N.; Raes, J.; Falony, G. Variation and transmission of the human gut microbiota across multiple familial generations. Nat. Microbiol. 2022, 7, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Almeida, A.; Kumar, N.; Vervier, K.; Adoum, A.T.; Viciani, E.; Dawson, N.J.R.; Forster, S.C.; Cormie, C.; Goulding, D.; et al. Host adaptation in gut Firmicutes is associated with sporulation loss and altered transmission cycle. Genome Biol. 2021, 22, 204. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.Y.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73 (Suppl. 2), 386S–392S. [Google Scholar] [CrossRef] [PubMed]
- Liévin-Le Moal, V.; Servin, A.L. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: From probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin. Microbiol. Rev. 2014, 27, 167–199. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Surawicz, C.M.; Greenberg, R.N.; Fekety, R.; Elmer, G.W.; Moyer, K.A.; Melcher, S.A.; Bowen, K.E.; Cox, J.L.; Noorani, Z.; et al. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA 1994, 271, 913–918. [Google Scholar] [CrossRef]
- Surawicz, C.M.; McFarland, L.V.; Greenberg, R.N.; Rubin, M.; Fekety, R.; Mulligan, M.E.; Garcia, R.J.; Brandmarker, S.; Bowen, K.; Borjal, D.; et al. The search for a better treatment for recurrent Clostridium difficile disease: Use of high-dose vancomycin combined with Saccharomyces boulardii. Clin. Infect. Dis. 2000, 31, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Wullt, M.; Hagslatt, M.L.; Odenholt, I. Lactobacillus plantarum 299v for the treatment of recurrent Clostridium difficile-associated diarrhoea: A double-blind, placebo-controlled trial. Scand. J. Infect. Dis. 2003, 35, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Klarin, B.; Wullt, M.; Palmquist, I.; Molin, G.; Larsson, A.; Jeppsson, B. Lactobacillus plantarum 299v reduces colonisation of Clostridium difficile in critically ill patients treated with antibiotics. Acta Anaesthesiol. Scand. 2008, 52, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Kujawa-Szewieczek, A.; Adamczak, M.; Kwiecień, K.; Dudzicz, S.; Gazda, M.; Więcek, A. The effect of Lactobacillus plantarum 299v on the incidence of Clostridium difficile infection in high-risk patients treated with antibiotics. Nutrients 2015, 7, 10179–10188. [Google Scholar] [CrossRef] [PubMed]
- Barker, A.K.; Duster, M.; Valentine, S.; Hess, T.; Archbald-Pannone, L.; Guerrant, R.; Safdar, N. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J. Antimicrob. Chemother. 2017, 72, 3177–3180. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Sun, Y.; Mu, P.; Zheng, T.; Mu, H.; Deng, F.; Deng, Y.; Wen, J. Lactobacillus rhamnosus GG supplementation modulates the gut microbiota to promote butyrate production, protecting against deoxynivalenol exposure in nude mice. Biochem. Pharmacol. 2020, 175, 113868. [Google Scholar] [CrossRef] [PubMed]
- Carucci, L.; Nocerino, R.; Paparo, L.; De Filippis, F.; Coppola, S.; Giglio, V.; Cozzolino, T.; Valentino, V.; Sequino, G.; Bedogni, G.; et al. Therapeutic effects elicited by the probiotic Lacticaseibacillus rhamnosus GG in children with atopic dermatitis. The results of the ProPAD trial. Pediatr. Allergy Immunol. 2022, 33, e13836. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.W.; Tsai, P.J.; Lee, C.C.; Ko, W.C.; Hung, Y.P. Application of microbiome management in therapy for Clostridioides difficile Infections: From fecal microbiota transplantation to probiotics to microbiota-preserving antimicrobial agents. Pathogens 2021, 10, 649. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, J.Z.; Yap, C.; Lytvyn, L.; Lo, C.K.; Beardsley, J.; Mertz, D.; Johnston, B.C. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst. Rev. 2017, 12, CD006095. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zeng, L.; Yan, Z.; Jia, J.; Gao, J.; Wei, Y. The mechanisms and safety of probiotics against toxigenic Clostridium difficile. Expert. Rev. Anti. Infect. Ther. 2020, 18, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, J.Y.; Peng, X.; Xiao, K.Y.; Xu, Q.; Wang, C. Which probiotic has the best effect on preventing Clostridium difficile-associated diarrhea? A systematic review and network meta-analysis. J. Dig. Dis. 2020, 21, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Varela, L.; Gueimonde, M.; Ruas-Madiedo, P. Probiotics for prevention and treatment of Clostridium difficile infection. Adv. Exp. Med. Biol. 2018, 1050, 161–176. [Google Scholar] [PubMed]
- Plummer, S.; Weaver, M.A.; Harris, J.C.; Dee, P.; Hunter, J. Clostridium difficile pilot study: Effects of probiotic supplementation on the incidence of C. difficile diarrhoea. Int. Microbiol. 2004, 7, 59–62. [Google Scholar] [PubMed]
- Van Prehn, J.; Reigadas, E.; Vogelzang, E.H.; Bouza, E.; Hristea, A.; Guery, B.; Krutova, M.; Norén, T.; Allerberger, F.; Coia, J.E.; et al. European society of clinical microbiology and infectious diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin. Microbiol. Infect. 2021, 27 (Suppl. 2), S1–S21. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.R.; Fischer, M.; Allegretti, J.R.; LaPlante, K.; Stewart, D.B.; Limketkai, B.N.; Stollman, N.H. ACG clinical guidelines: Prevention, diagnosis, and treatment of Clostridioides difficile Infections. Am. J. Gastroenterol. 2021, 116, 1124–1147. [Google Scholar] [CrossRef] [PubMed]
- Besselink, M.G.; van Santvoort, H.C.; Buskens, E.; Boermeester, M.A.; van Goor, H.; Timmerman, H.M.; Nieuwenhuijs, V.B.; Bollen, T.L.; van Ramshorst, B.; Witteman, B.J.; et al. Probiotic prophylaxis in predicted severe acute pancreatitis: A randomised, double-blind, placebo-controlled trial. Lancet 2008, 371, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Assi, M.; Lee, C.; Yoho, D.; Louie, T.; Knapple, W.; Aguilar, H.; Garcia-Diaz, J.; Wang, G.P.; Berry, S.M.; et al. Efficacy and safety of RBX2660 in PUNCH CD3, a phase III, randomized, double-blind, placebo-controlled trial with a bayesian primary analysis for the prevention of recurrent Clostridioides difficile infection. Drugs 2022, 82, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Papazyan, R.; Ferdyan, N.; Srinivasan, K.; Gonzalez, C.; Shannon, W.D.; Blount, K.; Fuchs, B.C. Human fecal bile acid analysis after investigational microbiota-based live biotherapeutic delivery for recurrent Clostridioides difficile infection. Microorganisms 2023, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Doosetty, S.; Umeh, C.; Eastwood, W.; Samreen, I.; Penchala, A.; Kaur, H.; Chilinga, C.; Kaur, G.; Mohta, T.; Nakka, S.; et al. Efficacy of Fecal Microbiota (REBYOTA) in recurrent Clostridium difficile infections: A systematic review and meta-analysis. Cureus 2024, 16, e58862. [Google Scholar] [CrossRef] [PubMed]
- Feuerstadt, P.; Louie, T.J.; Lashner, B.; Wang, E.E.L.; Diao, L.; Bryant, J.A.; Sims, M.; Kraft, C.S.; Cohen, S.H.; Berenson, C.S.; et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 2022, 386, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Louie, T.; Golan, Y.; Khanna, S.; Bobilev, D.; Erpelding, N.; Fratazzi, C.; Carini, M.; Menon, R.; Ruisi, M.; Norman, J.M.; et al. VE303, a defined bacterial consortium, for prevention of recurrent Clostridioides difficile infection: A randomized clinical trial. JAMA 2023, 329, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Gerding, D.N.; Meyer, T.; Lee, C.; Cohen, S.H.; Murthy, U.K.; Poirier, A.; Van Schooneveld, T.C.; Pardi, D.S.; Ramos, A.; Barron, M.A.; et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: A randomized clinical trial. JAMA 2015, 313, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Delzenne, N.M.; Cani, P.D.; Walter, J. Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.; Montero, I.; Fernandez-Gosende, M.; Martinez, N.; Cantabrana, C.H.; Rios-Covian, D. Past, present, and future of microbiome-based therapies. Microbiome Res. Rep. 2024, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Rätsep, M.; Kõljalg, S.; Sepp, E.; Smidt, I.; Truusalu, K.; Songisepp, E.; Stsepetova, J.; Naaber, P.; Mikelsaar, R.H.; Mikelsaar, M. A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe 2017, 47, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Varela, L.; Hernández-Barranco, A.M.; Ruas-Madiedo, P.; Gueimonde, M. Effect of Bifidobacterium upon Clostridium difficile Growth and toxicity when co-cultured in different prebiotic Substrates. Front. Microbiol. 2016, 7, 738. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, N.; Feng, Y.; Zhou, M.; Li, H.; Zhang, X.; Ma, X. From probiotics to postbiotics: Concepts and applications. Anim. Res. One Health 2023, 1, 92–114. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, S.; Li, S.; Jiang, W.; Wang, J.; Xiao, J.; Chen, T.; Ma, J.; Khan, M.Z.; Wang, W.; et al. Unlocking the power of postbiotics: A revolutionary approach to nutrition for humans and animals. Cell Metab. 2024, 36, 725–744. [Google Scholar] [CrossRef] [PubMed]
- Mantziari, A.; Salminen, S.; Szajewska, H.; Malagón-Rojas, J.N. Postbiotics against pathogens commonly involved in pediatric infectious diseases. Microorganisms 2020, 8, 1510. [Google Scholar] [CrossRef] [PubMed]
- Luenglusontigit, P.; Sathapondecha, P.; Saengsuwan, P.; Surachat, K.; Boonserm, P.; Singkhamanan, K. Effects of postbiotic from bacteriocin-like inhibitory substance producing Enterococcus faecalis on toxigenic Clostridioides difficile. J. Health Sci. Med. Res. 2023, 41, e2023918. [Google Scholar] [CrossRef]
- Piqué, N.; Berlanga, M.; Miñana-Galbis, D. Health benefits of heat-killed (Tyndallized) probiotics: An overview. Int. J. Mol. Sci. 2019, 20, 2534. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.; Oliveira, A.L.; Oliveira, C.; Pintado, M.; Amaro, A.; Madureira, A.R. Current postbiotics in the cosmetic market—An update and development opportunities. Appl. Microbiol. Biotechnol. 2022, 106, 5879–5891. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).