The Antibacterial Efficacy of Far-UVC Light: A Combined-Method Study Exploring the Effects of Experimental and Bacterial Variables on Dose–Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Methods
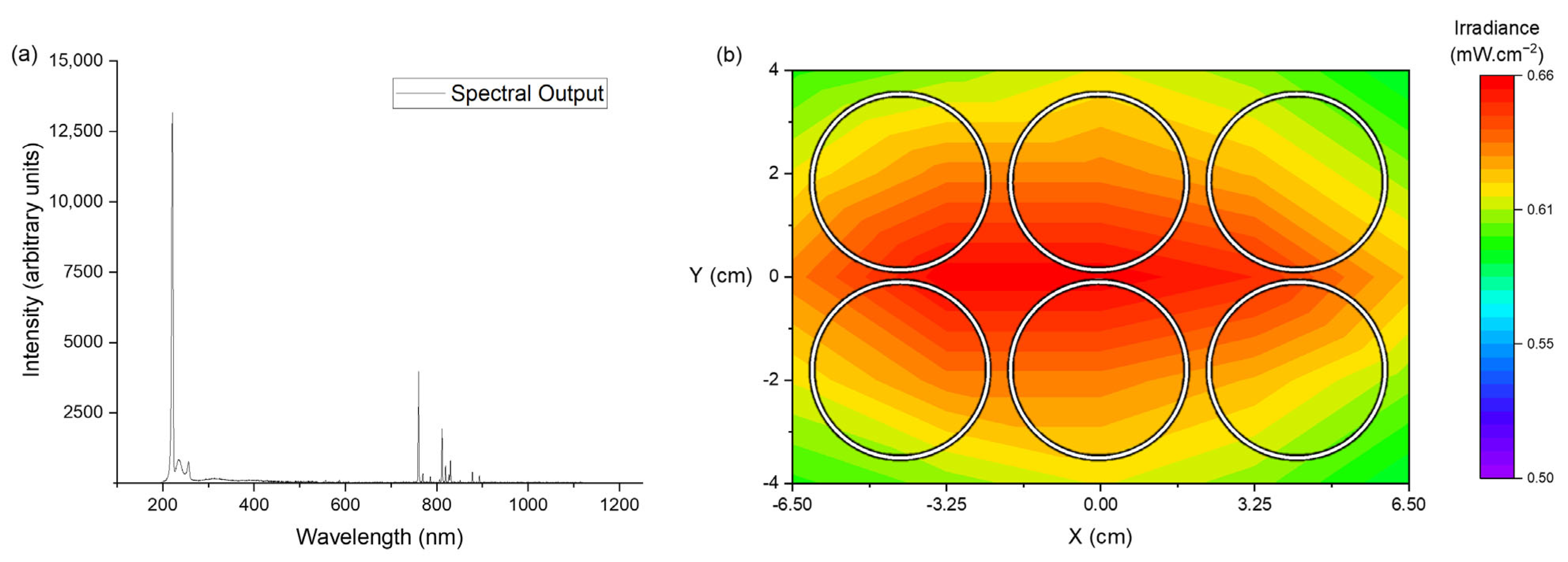
2.1.1. Far-UVC Light Source
2.1.2. Preparation of Bacterial Suspensions
2.1.3. Far-UVC Irradiation of Bacteria in Liquid Suspension
2.2. Literature Review
2.3. Statistical Analysis
3. Results
3.1. Experimental Analysis of the Germicidal Efficacy of Far-UVC Light
3.1.1. Effect of Cell Density on Far-UVC Bactericidal Efficacy
3.1.2. Far-UVC 1-Log10 Inactivation Dose (ID) for Differing Cell Densities
3.2. Literature Review
3.2.1. Comparison of Far-UVC 1-Log10 Inactivation Dose
3.2.2. Effect of Bacterial Cell Density on Far-UVC 1-Log10 Inactivation Dose
3.2.3. Effect of Irradiance on Far-UVC 1-Log10 Inactivation Dose
4. Discussion
4.1. Influence of Gram Type on Susceptibility to Far-UVC Inactivation
4.2. Influence of Genus/Species on Susceptibility to Far-UVC Inactivation
4.3. Influence of Cell Form on Susceptibility to Far-UVC Inactivation
4.4. Influence of Bacterial Cell Density on Susceptibility to Far-UVC Inactivation
4.5. Influence of Irradiance on Susceptibility to Far-UVC Inactivation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eadie, E.; Hiwar, W.; Fletcher, L.; Tidswell, E.; O’Mahoney, P.; Buonanno, M.; Welch, D.; Adamson, C.S.; Brenner, D.J.; Noakes, C.; et al. Far-UVC (222 nm) Efficiently Inactivates an Airborne Pathogen in a Room-sized Chamber. Sci. Rep. 2022, 12, 4373. [Google Scholar] [CrossRef]
- Hessling, M.; Haag, R.; Sicks, B. Review of Microbial Touchscreen Contamination for the Determination of Reasonable Ultraviolet Disinfection Doses. GMS Hyg. Infect. Control 2021, 16, Doc30. [Google Scholar] [CrossRef]
- Taylor, W.; Camilleri, E.; Craft, D.L.; Korza, G.; Granados, M.R.; Peterson, J.; Szczpaniak, R.; Weller, S.K.; Moeller, R.; Douki, T.; et al. DNA Damage Kills Bacterial Spores and Cells Exposed to 222-Nanometer UV Radiation. Appl. Environ. Microbiol. 2020, 86, e03039-19. [Google Scholar] [CrossRef]
- Narita, K.; Asano, K.; Naito, K.; Ohashi, H.; Sasaki, M.; Morimoto, Y.; Igarashi, T.; Nakane, A. Ultraviolet C Light with Wavelength of 222 nm Inactivates a Wide Spectrum of Microbial Pathogens. J. Hosp. Infect. 2020, 105, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, H.; Nomura, T.; Nazmul, T.; Omori, K.; Shigemoto, N.; Sakaguchi, T.; Ohge, H. Effectiveness of 222-nm Ultraviolet Light on Disinfecting SARS-CoV-2 Surface Contamination. Am. J. Infect. Control 2021, 49, 299–301. [Google Scholar] [CrossRef]
- Oh, C.; Sun, P.P.; Araud, E.; Nguyen, T.H. Mechanism and Efficacy of Virus Inactivation by a Microplasma UV Lamp Generating Monochromatic UV Irradiation at 222 nm. Water Res. 2020, 186, 116386. [Google Scholar] [CrossRef] [PubMed]
- Clauß, M. Higher Effectiveness of Photoinactivation of Bacterial Spores, UV Resistant Vegetative Bacteria and Mold Spores with 222 nm Compared to 254 nm Wavelength. Acta Hydrochim. Hydrobiol. 2006, 34, 525–532. [Google Scholar] [CrossRef]
- Navarathna, T.; Jinadatha, C.; Corona, B.A.; Coppin, J.D.; Choi, H.; Bennett, M.R.; Ghamande, G.S.; Williams, M.D.; Keene, R.E.; Chatterjee, P. Efficacy of a Filtered Far-UVC Handheld Disinfection Device in Reducing the Microbial Bioburden of Hospital Surfaces. Am. J. Infect. Control. 2023, 51, 1406–1410. [Google Scholar] [CrossRef]
- Hessling, M.; Sicks, B.; Lau, B. Far-UVC Radiation for Disinfecting Hands or Gloves? Pathogens 2023, 12, 213. [Google Scholar] [CrossRef]
- Yin, R.; Anderson, C.E.; Zhao, J.; Boehm, A.B.; Mitch, W.A. Controlling Contaminants Using a Far-UVC-based Advanced Oxidation Process for Potable Reuse. Nat. Water 2023, 1, 555–562. [Google Scholar] [CrossRef]
- Zhao, J.; Payne, E.M.; Liu, B.; Shang, C.; Blatchley, E.R.; Mitch, W.A.; Yin, R. Making Waves: Opportunities and Challenges of Applying Far-UVC Radiation in Controlling Micropollutants in Water. Water Res. 2023, 241, 120169. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, B.; Zhao, J.; Tang, Z.; Li, W.; He, C.; Xia, D.; Linden, K.G.; Yin, R. Rapid Inactivation of Fungal Spores in Drinking Water by Far-UVC Photolysis of Free Chlorine. Environ. Sci. Technol. 2023, 57, 21876–21887. [Google Scholar] [CrossRef]
- Mesquita, P.J.B.d.; Sokas, R.K.; Rice, M.B.; Nardell, E.A. Far-UVC: Technology Update with an Untapped Potential to Mitigate Airborne Infections. Ann. Am. Thorac. Soc. 2023, 20, 1700–1702. [Google Scholar] [CrossRef]
- Wang, M.; Lai, A.C.K. Experimental Study on Enhancing Single-pass Air Disinfection Efficacy for Portable Disinfection Systems with Different Far-UVC Sources. Build Environ. 2024, 261, 111742. [Google Scholar] [CrossRef]
- Buonanno, M.; Ponnaiya, B.; Welch, D.; Stanislauskas, M.; Randers-Pehrson, G.; Smilenov, L.; Lowy, F.D.; Owens, D.M.; Brenner, D.J. Germicidal Efficacy and Mammalian Skin Safety of 222-nm UV Light. Radiat. Res. 2017, 187, 483–491. [Google Scholar] [CrossRef]
- Tavares, R.S.N.; Adamoski, D.; Girasole, A.; Lima, E.N.; da Silva Justo-Junior, A.; Domingues, R.; Silveira, A.C.C.; Marques, R.E.; de Carvalho, M.; Ambrosio, A.L.B.; et al. Different Biological Effects of Exposure to Far-UVC (222 nm) and Near-UVC (254 nm) Irradiation. J. Photochem. Photobiol. B 2023, 243, 112713. [Google Scholar] [CrossRef] [PubMed]
- Narita, K.; Asano, K.; Morimoto, Y.; Igarashi, T.; Nakane, A. Chronic Irradiation with 222-nm UVC Light Induces Neither DNA Damage nor Epidermal Lesions in Mouse Skin, Even at High Doses. PLoS ONE 2018, 13, e0201259. [Google Scholar] [CrossRef] [PubMed]
- Narita, K.; Asano, K.; Morimoto, Y.; Igarashi, T.; Hamblin, M.R.; Dai, T.; Nakane, A. Disinfection and Healing Effects of 222-nm UVC Light on Methicillin-resistant Staphylococcus aureus Infection in Mouse Wounds. J. Photochem. Photobiol. B 2018, 178, 10–18. [Google Scholar] [CrossRef]
- Kaidzu, S.; Sugihara, K.; Sasaki, M.; Nishiaki, A.; Igarashi, T.; Tanito, M. Evaluation of Acute Corneal Damage Induced by 222-nm and 254-nm Ultraviolet Light in Sprague–Dawley Rats. Free Radic. Res. 2019, 53, 611–617. [Google Scholar] [CrossRef]
- Coohill, T.P. Virus-Cell Interactions as Probes for Vacuum-Ultraviolet Radiation Damage and Repair. Photochem. Photobiol. 1986, 44, 359–363. [Google Scholar] [CrossRef]
- Scarpa, F.; Branda, F.; Petrosillo, N.; Ciccozzi, M. On the SARS-CoV-2 Variants. Infect. Dis. Rep. 2024, 16, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Buonanno, M.; Kleiman, N.J.; Welch, D.; Hashmi, R.; Shuryak, I.; Brenner, D.J. 222 nm Far-UVC Light Markedly Reduces the Level of Infectious Airborne Virus in an Occupied Room. Sci. Rep. 2024, 14, 6722. [Google Scholar] [CrossRef] [PubMed]
- Buonanno, M.; Welch, D.; Shuryak, I.; Brenner, D.J. Far-UVC Light (222 nm) Efficiently and Safely Inactivates Airborne Human Coronaviruses. Sci. Rep. 2020, 10, 10285. [Google Scholar] [CrossRef] [PubMed]
- Song, B.M.; Lee, G.H.; Han, H.J.; Yang, J.H.; Lee, E.G.; Gu, H.; Park, H.K.; Ryu, K.; Kim, J.; Kang, S.M.; et al. Ultraviolet-C Light at 222 nm Has a High Disinfecting Spectrum in Environments Contaminated by Infectious Pathogens, Including SARS-CoV-2. PLoS ONE 2023, 18, e0294427. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial Infections: Epidemiology, Prevention, Control and Surveillance. Asian Pac. J. Trop. Biomed 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Opal, S.M.; Garber, G.E.; LaRosa, S.P.; Maki, D.G.; Freebairn, R.C.; Kinasewitz, G.T.; Dhainaut, J.-F.; Yan, S.B.; Williams, M.D.; Graham, D.E.; et al. Systemic Host Responses in Severe Sepsis Analyzed by Causative Microorganism and Treatment Effects of Drotrecogin Alfa (Activated). Clin. Infect. Dis. 2003, 37, 50–58. [Google Scholar] [CrossRef]
- Blanco, J.; Muriel-Bombín, A.; Sagredo, V.; Taboada, F.; Gandía, F.; Tamayo, L.; Collado, J.; García-Labattut, Á.; Carriedo, D.; Valledor, M.; et al. Incidence, Organ Dysfunction and Mortality in Severe Sepsis: A Spanish Multicentre Study. Crit. Care 2008, 12, R158. [Google Scholar] [CrossRef]
- Treacher, D.F.; Brown, K.A. The Basic Science of Sepsis. Surgery 2009, 27, 465–469. [Google Scholar] [CrossRef]
- Phua, J.; Ngerng, W.J.; See, K.C.; Tay, C.K.; Kiong, T.; Lim, H.F.; Chew, M.Y.; Yip, H.S.; Tan, A.; Khalizah, H.J.; et al. Characteristics and Outcomes of Culture-negative versus Culture-positive Severe Sepsis. Crit. Care 2013, 17, R202. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Kolář, M. Bacterial Infections, Antimicrobial Resistance and Antibiotic Therapy. Life 2022, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- MacIsaac, S.A.; Rauch, K.D.; Prest, T.; Simons, R.M.; Gagnon, G.A.; Stoddart, A.K. Improved Disinfection Performance for 280 nm LEDs Over 254 nm Low-pressure UV Lamps in Community Wastewater. Sci. Rep. 2023, 13, 7576. [Google Scholar] [CrossRef]
- Stewart, C.F.; Tomb, R.M.; Ralston, H.J.; Armstrong, J.; Anderson, J.G.; MacGregor, S.J.; Atreya, C.D.; Maclean, M. Violet-blue 405-nm Light-based Photoinactivation for Pathogen Reduction of Human Plasma Provides Broad Antibacterial Efficacy Without Visible Degradation of Plasma Proteins. Photochem. Photobiol. 2022, 98, 504–512. [Google Scholar] [CrossRef]
- Meurle, T.; Knaus, J.; Barbano, A.; Hoenes, K.; Spellerberg, B.; Hessling, M. Photoinactivation of Staphylococci with 405 nm Light in a Trachea Model with Saliva Substitute at 37 °C. Healthcare 2021, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Vollmerhausen, T.L.; Conneely, A.; Bennett, C.; Wagner, V.E.; Victor, J.C.; O’Byrne, C.P. Visible and UVA Light as a Potential Means of Preventing Escherichia coli Biofilm Formation in Urine and on Materials Used in Urethral Catheters. J. Photochem. Photobiol. B 2017, 170, 295–303. [Google Scholar] [CrossRef]
- Claus, H. Ozone Generation by Ultraviolet Lamps. Photochem. Photobiol. 2021, 97, 471–476. [Google Scholar] [CrossRef]
- Tomb, R.M.; White, T.A.; Coia, J.E.; Anderson, J.G.; MacGregor, S.J.; Maclean, M. Review of the Comparative Susceptibility of Microbial Species to Photoinactivation Using 380-480 nm Violet-Blue Light. Photochem. Photobiol. 2018, 94, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Mukaka, M.M. Statistics Corner: A Guide to Appropriate Use of Correlation Coefficient in Medical Research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar] [PubMed]
- Gierke, A.M.; Hessling, M. Investigation on Potential ESKAPE Surrogates for 222 and 254 nm Irradiation Experiments. Front. Microbiol. 2022, 13, 942708. [Google Scholar] [CrossRef]
- Matafonova, G.G.; Batoev, V.B.; Astakhova, S.A.; Gómez, M.; Christofi, N. Efficiency of KrCl Excilamp (222 nm) for Inactivation of Bacteria in Suspension. Lett. Appl. Microbiol. 2008, 47, 508–513. [Google Scholar] [CrossRef]
- Sun, W.; Jing, Z.; Zhao, Z.; Yin, R.; Santoro, D.; Mao, T.; Lu, Z. Dose–Response Behavior of Pathogens and Surrogate Microorganisms Across the Ultraviolet-C Spectrum: Inactivation Efficiencies, Action Spectra, and Mechanisms. Environ. Sci. Technol. 2023, 57, 10891–10900. [Google Scholar] [CrossRef]
- Jing, Z.; Lu, Z.; Santoro, D.; Zhao, Z.; Huang, Y.; Ke, Y.; Wang, X.; Sun, W. Which UV Wavelength is the Most Effective for Chlorine-Resistant Bacteria in Terms of the Impact of Activity, Cell Membrane and DNA? Chem. Eng. J. 2022, 447, 137584. [Google Scholar] [CrossRef]
- Kang, J.W.; Kim, S.S.; Kang, D.H. Inactivation Dynamics of 222 nm Krypton-Chlorine Excilamp Irradiation on Gram-positive and Gram-negative Foodborne Pathogenic Bacteria. Food Res. Int. 2018, 109, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, K.; Jangra, R.; Ish, A.; Jain, N.; Prakash, R. A Dielectric Barrier Discharge Based Low Pressure Narrow Band Far UV-C 222 nm Excimer Lamp and its Efficiency Analysis. Phys. Scr. 2024, 99, 025018. [Google Scholar] [CrossRef]
- Matsumoto, T.; Hoshiai, T.; Tatsuno, I.; Hasegawa, T. Action Spectra of Bacteria and Purification of Pollutant Water at Faucets Using a Water Waveguide Method. Water 2022, 14, 1394. [Google Scholar] [CrossRef]
- Chen, H.; Moraru, C.I. Exposure to 222 nm Far UV-C Effectively Inactivates Planktonic Foodborne Pathogens and Inhibits Biofilm Formation. Innov. Food Sci. Emerg. Technol. 2023, 87, 103411. [Google Scholar] [CrossRef]
- Rufyikiri, A.S.; Martinez, R.; Addo, P.W.; Wu, B.-S.; Yousefi, M.; Malo, D.; Orsat, V.; Vidal, S.M.; Fritz, J.H.; MacPherson, S.; et al. Germicidal Efficacy of Continuous and Pulsed Ultraviolet-C Radiation on Pathogen Models and SARS-CoV-2. Photochem. Photobiol. Sci. 2024, 23, 339–354. [Google Scholar] [CrossRef]
- Ivanova, I.; Svilenska, T.; Kurz, B.; Grobecker, S.; Maisch, T.; Berneburg, M.; Kamenisch, Y. Improved Spectral Purity of 222-nm Irradiation Eliminates Detectable Cyclobutylpyrimidine Dimers Formation in Skin Reconstructs even at High and Repetitive Disinfecting Doses. Photochem. Photobiol. 2022, 98, 1149–1156. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, B.; He, C.; Xia, D.; Yin, R. Nitrate Protects Microorganisms and Promotes Formation of Toxic Nitrogenous Byproducts during Water Disinfection by Far-UVC Radiation. Environ. Sci. Technol. 2023, 57, 9064–9074. [Google Scholar] [CrossRef]
- Clauß, M.; Mannesmann, R.; Kolch, A. Photoreactivation of Escherichia coli and Yersinia enterolytica after Irradiation with a 222 nm Excimer Lamp Compared to a 254 nm Low-pressure Mercury Lamp. Acta Hydrochim. Hydrobiol. 2005, 33, 579–584. [Google Scholar] [CrossRef]
- Neo, Y.; Hashimoto, G.; Koike, R.; Ohhara, T.; Matsumoto, T. Solid-State Far-Ultraviolet C Light Sources for the Disinfection of Pathogenic Microorganisms Using Graphene Nanostructure Field Emitters. Global Chall 2023, 7, 2200236. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, Y.; Gan, J.; Yu, X.; Wang, L. Superiority of UV222 Radiation by in Situ Aquatic Electrode KrCl Excimer in Disinfecting Waterborne Pathogens: Mechanism and Efficacy. J. Hazard Mater. 2023, 452, 131292. [Google Scholar] [CrossRef]
- Kang, J.W.; Kang, D.H. The Synergistic Bactericidal Mechanism of Simultaneous Treatment with a 222-nanometer Krypton-Chlorine Excilamp and a 254- nanometer Low-pressure Mercury Lamp. Appl. Environ. Microbiol. 2019, 85, e01952-18. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Bright, K.; Ikner, L.; Ley, C.; Seyedi, S.; Gerba, C.P.; Sobsey, M.D.; Piper, P.; Linden, K.G. UV Inactivation of Common Pathogens and Surrogates Under 222 nm Irradiation from KrCl* Excimer Lamps. Photochem. Photobiol. 2023, 99, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Bohrerova, Z.; Linden, K.G. Assessment of DNA Damage and Repair in Mycobacterium terrae after Exposure to UV Irradiation. J. Appl. Microbiol. 2006, 101, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, K.; Kaidzu, S.; Sasaki, M.; Ichioka, S.; Takayanagi, Y.; Shimizu, H.; Sano, I.; Hara, K.; Tanito, M. One-year Ocular Safety Observation of Workers and Estimations of Microorganism Inactivation Efficacy in the Room Irradiated with 222-nm Far Ultraviolet-C Lamps. Photochem. Photobiol. 2023, 99, 967–974. [Google Scholar] [CrossRef]
- Aquino de Muro, M.; Shuryak, I.; Uhlemann, A.C.; Tillman, A.; Seeram, D.; Zakaria, J.; Welch, D.; Erde, S.M.; Brenner, D.J. The Abundance of the Potential Pathogen Staphylococcus hominis in the Air Microbiome in a Dental Clinic and its Susceptibility to Far-UVC Light. MicrobiologyOpen 2023, 12, e1348. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.W.; Hong, H.N.; Kang, D.H. Application of a Krypton-Chlorine Excilamp to Control Alicyclobacillus acidoterrestris Spores in Apple Juice and Identification of its Sporicidal Mechanism. Appl. Environ. Microbiol. 2020, 86, e00159-20. [Google Scholar] [CrossRef]
- Beck, S.E.; Wright, H.B.; Hargy, T.M.; Larason, T.C.; Linden, K.G. Action Spectra for Validation of Pathogen Disinfection in Medium-pressure Ultraviolet (UV) Systems. Water Res. 2015, 70, 27–37. [Google Scholar] [CrossRef]
- Wang, D.; Oppenländer, T.; El-Din, M.G.; Bolton, J.R. Comparison of the Disinfection Effects of Vacuum-UV (VUV) and UV Light on Bacillus subtilis Spores in Aqueous Suspensions at 172, 222 and 254 nm. Photochem. Photobiol. 2010, 86, 176–181. [Google Scholar] [CrossRef]
- Cetinkaya, Y.; Falk, P.; Mayhall, C.G. Vancomycin-resistant Enterococci. Clin. Microbiol. Rev. 2000, 13, 686–707. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, 10–1128. [Google Scholar] [CrossRef]
- Baron, E.J. Classification. In Medical Microbiology, 4th ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Setlow, P. Resistance of Spores of Bacillus Species to Ultraviolet Light. Environ. Mol. Mutagen. 2001, 38, 97–104. [Google Scholar] [CrossRef]
- Abshire, R.L.; Dunton, H. Resistance of Selected Strains of Pseudomonas aeruginosa to Low-density Ultraviolet Radiation. Appl. Environ. Microbiol. 1981, 41, 1419–1423. [Google Scholar] [CrossRef]
- Chatzisymeon, E. Inactivation of Bacteria in Seafood Processing Water by Means of UV Treatment. J. Food Eng. 2016, 173, 1–7. [Google Scholar] [CrossRef]
- Muranyi, P.; Wunderlich, J.; Heise, M. Sterilization Efficiency of a Cascaded Dielectric Barrier Discharge. J. Appl. Microbiol. 2007, 103, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Coohill, T.P.; Sagripanti, J.-L. Overview of the Inactivation by 254 nm Ultraviolet Radiation of Bacteria with Particular Relevance to Biodefense. Photochem. Photobiol. 2008, 84, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Peleg, M. Microbial Dose-Response Curves and Disinfection Efficacy Models Revisited. Food Eng. Rev. 2021, 13, 305–321. [Google Scholar] [CrossRef]
- Blatchley, E., III.; Meeusen, A.; Aronson, A. Inactivation of Bacillus Spores by Ultraviolet or Gamma Radiation. J. Environ. Eng. 2005, 131, 1245–1252. [Google Scholar] [CrossRef]
- Wells, W.F. Airborne Contagion and Air Hygiene; Harvard University Press: Cambridge, MA, USA, 1955. [Google Scholar]
- Pousty, D.; Hofmann, R.; Gerchman, Y.; Mamane, H. Wavelength-dependent Time–dose Reciprocity and Stress Mechanism for UV-LED Disinfection of Escherichia coli. J. Photochem. Photobiol. B 2021, 217, 112129. [Google Scholar] [CrossRef]
- Matsumoto, T.; Tatsuno, I.; Yoshida, Y.; Tomita, M.; Hasegawa, T. Time-dose Reciprocity Mechanism for the Inactivation of Escherichia coli Explained by a Stochastic Process with Two Inactivation Effects. Sci. Rep. 2022, 12, 22588. [Google Scholar] [CrossRef] [PubMed]






| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Wavelengths ≥ 200, ≤230 nm | Wavelengths ≤ 199, ≥231 nm |
| Transparent liquid suspension | Multiple wavelengths used in combination |
| Opaque or nutritious liquid suspension | |
| Bacteria on surfaces Bacteria in aerosols Pulsed light sources Cocktails of multiple bacteria | |
| Uncommon or genetically modified mutant strains |
| Pearson Correlation Coefficient | Relationship Designation |
|---|---|
| 0.00 to 0.30 | Negligible correlation |
| 0.30 to 0.50 | Weak positive or negative |
| 0.50 to 0.70 | Moderate positive or negative |
| 0.70 to 0.90 | Strong positive or negative |
| 0.90 to 1.00 | Very strong positive or negative |
| Far-UVC 1-Log10 Inactivation Dose (mJ·cm−2) | ||||||
|---|---|---|---|---|---|---|
| 101 CFU·mL−1 | 102 CFU·mL−1 | 103 CFU·mL−1 | 105 CFU·mL−1 | 107 CFU·mL−1 | Mean ± SD | |
| E. coli | 6.38 | 4.80 | 4.95 | 3.91 | 3.09 | 4.63 ± 1.23 |
| E. faecium | 17.16 | 10.88 | 7.95 | 7.87 | 11.30 | 11.03 ± 3.78 |
| P. aeruginosa | 3.11 | 2.05 | 3.09 | 5.90 | 5.66 | 3.96 ± 1.72 |
| S. aureus | 6.91 | 4.03 | 4.13 | 5.55 | 10.91 | 6.30 ± 2.83 |
| Bacterial Species | Gram Type | Cell Form | Median Far-UVC 1-log10 ID (mJ·cm−2) | No. of Data Points (from One of More Studies) | References |
|---|---|---|---|---|---|
| A. kookii | Negative | Veg. cells | 2.93 | 1 | [39] |
| B. cereus | Positive | Veg. cells | 49.31 | 8 | [4,7,40] |
| B. subtilis | Positive | Veg. cells | 4.68 | 10 | [3,40,41,42] |
| C. jejuni | Negative | Veg. cells | 1.02 | 1 | [4] |
| C. sporogenes | Positive | Veg. cells | 5.56 | 1 | [4] |
| E. mundtii | Positive | Veg. cells | 6.90 | 1 | [39] |
| E. coli | Negative | Veg. cells | 4.32 | 22 | [4,39,40,41,43,44,45,46,47,48,49,50,51,52,53,54] |
| L. pneumophila | Negative | Veg. cells | 1.67 | 1 | [54] |
| L. monocytogenes | Positive | Veg. cells | 1.71 | 4 | [43,46,53,54] |
| M. fortuitum | Positive | Veg. cells | 3.18 | 2 | [41,42] |
| M. smegmatis | Positive | Veg. cells | 10.45 | 1 | [54] |
| M. terrae | Positive | Veg. cells | 5.25 | 1 | [55] |
| Pantoea spp. | Negative | Veg. cells | 3.73 | 1 | [42] |
| P. aeruginosa | Negative | Veg. cells | 3.43 | 7 | [4,7,41,42,45,46] |
| P. fluorescens | Negative | Veg. cells | 2.01 | 1 | [39] |
| S. enterica | Negative | Veg. cells | 2.27 | 4 | [4,43,53,54] |
| S. aureus | Positive | Veg. cells | 4.31 | 20 | [3,4,7,40,41,43,44,46,48,52,54,56] |
| S. carnosus | Positive | Veg. cells | 3.28 | 1 | [39] |
| S. epidermidis | Positive | Veg. cells | 5.14 | 3 | [45,57] |
| S. hominis | Positive | Veg. cells | 4.85 | 1 | [57] |
| Stenotrophomonas spp. | Negative | Veg. cells | 3.73 | 1 | [42] |
| S. pyogenes | Positive | Veg. cells | 33.14 | 6 | [40] |
| Y. enterolytica | Negative | Veg. cells | 2.20 | 1 | [50] |
| A. acidoterrestris | Positive | Endospores | 6.97 | 1 | [58] |
| B. cereus | Positive | Endospores | 20.86 | 3 | [3,4,7] |
| B. pumilus | Positive | Endospores | 12.11 | 2 | [59] |
| B. subtilis | Positive | Endospores | 9.33 | 3 | [3,47,60] |
| B. thuringiensis Al Hakam | Positive | Endospores | 10.57 | 1 | [3] |
| C. difficile | Positive | Endospores | 9.62 | 3 | [3,4] |
| C. pasteurianum | Positive | Endospores | 2.40 | 1 | [7] |
| C. sporogenes | Positive | Endospores | 15.41 | 1 | [4] |
| S. griseus | Positive | Endospores | 6.38 | 1 | [7] |
| T. vulgaris | Positive | Endospores | 13.75 | 1 | [7] |
| Bacterial Species | No. of Data Points | No. of Contributing Papers | r-Value | Relationship | p-Value |
|---|---|---|---|---|---|
| B. cereus | 8 | 3 | 0.807 | Strong Positive Correlation | 0.015 * |
| B. subtilis | 8 | 3 | 0.529 | Moderate Positive Correlation | 0.177 |
| L. monocytogenes | 3 | 3 | 0.838 | Strong Positive Correlation | 0.367 |
| S. aureus | 16 | 10 | 0.491 | Weak Positive Correlation | 0.054 |
| S. pyogenes | 6 | 1 | 0.976 | Very Strong Positive Correlation | 0.001 * |
| E. coli | 21 | 15 | 0.658 | Moderate Positive Correlation | 0.001 * |
| P. aeruginosa | 7 | 6 | 0.989 | Very Strong Positive Correlation | <0.001 * |
| S. enterica | 3 | 3 | −0.430 | Weak Negative Correlation | 0.717 |
| Species | No. of Data Points | No. of Contributing Papers | r-Value | Relationship | p-Value |
|---|---|---|---|---|---|
| B. cereus | 8 | 3 | −0.131 | Negligible Correlation | 0.757 |
| B. cereus (endospores) | 3 | 3 | 0.451 | Weak Positive Correlation | 0.702 |
| B. subtilis | 10 | 4 | 0.492 | Weak Positive Correlation | 0.148 |
| L. monocytogenes | 4 | 4 | 0.245 | Negligible Correlation | 0.755 |
| S. aureus | 19 | 11 | 0.097 | Negligible Correlation | 0.692 |
| E. coli | 21 | 15 | −0.020 | Negligible Correlation | 0.930 |
| P. aeruginosa | 7 | 6 | −0.181 | Negligible Correlation | 0.697 |
| S. enterica | 4 | 4 | 0.729 | Strong Positive Correlation | 0.271 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griffin, D.T.; Gourlay, T.; Maclean, M. The Antibacterial Efficacy of Far-UVC Light: A Combined-Method Study Exploring the Effects of Experimental and Bacterial Variables on Dose–Response. Pathogens 2024, 13, 698. https://doi.org/10.3390/pathogens13080698
Griffin DT, Gourlay T, Maclean M. The Antibacterial Efficacy of Far-UVC Light: A Combined-Method Study Exploring the Effects of Experimental and Bacterial Variables on Dose–Response. Pathogens. 2024; 13(8):698. https://doi.org/10.3390/pathogens13080698
Chicago/Turabian StyleGriffin, David T., Terence Gourlay, and Michelle Maclean. 2024. "The Antibacterial Efficacy of Far-UVC Light: A Combined-Method Study Exploring the Effects of Experimental and Bacterial Variables on Dose–Response" Pathogens 13, no. 8: 698. https://doi.org/10.3390/pathogens13080698
APA StyleGriffin, D. T., Gourlay, T., & Maclean, M. (2024). The Antibacterial Efficacy of Far-UVC Light: A Combined-Method Study Exploring the Effects of Experimental and Bacterial Variables on Dose–Response. Pathogens, 13(8), 698. https://doi.org/10.3390/pathogens13080698





