Exploration of the Binding Site of Arachidonic Acid in gp63 of Leishmania mexicana and in Orthologous Proteins in Clinically Important Parasites
Abstract
1. Introduction
2. Materials and Methods
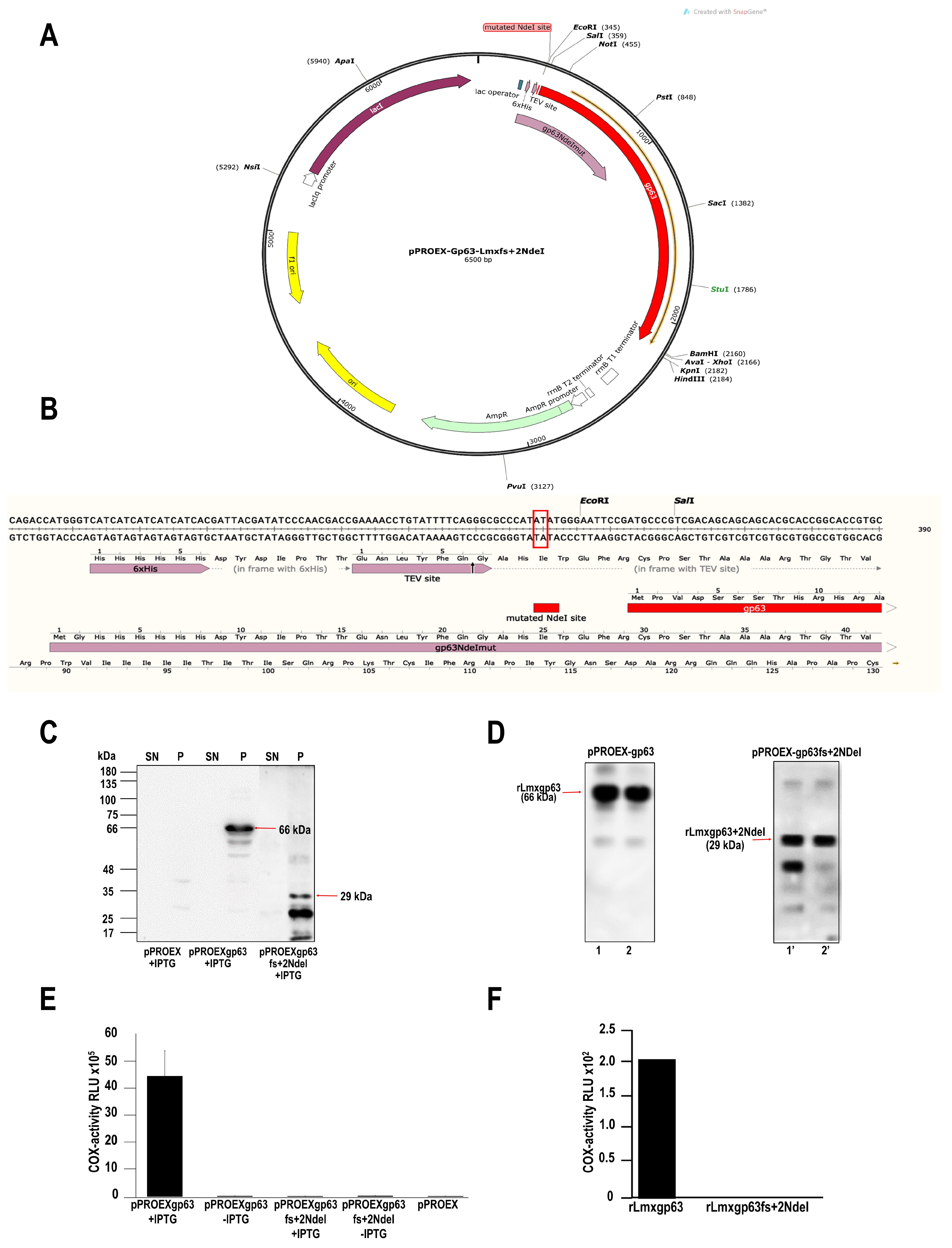
2.1. Production of a Mutant without COX Activity: Mutation by Frameshift of the gp63 Gene of L. mexicana
2.2. Induction and Purification of Recombinant Proteins from Leishmania Mexicana gp63
2.3. Parasites
2.4. Obtaining Soluble Fractions from Membrane Components of Enamoeba spp.
2.5. COX Activity
2.6. In Vitro Encystation and Fluorescence Microscopy
2.7. Bioinformatics Analysis
2.7.1. Bioinformatic Analyses of Local Alignment Search
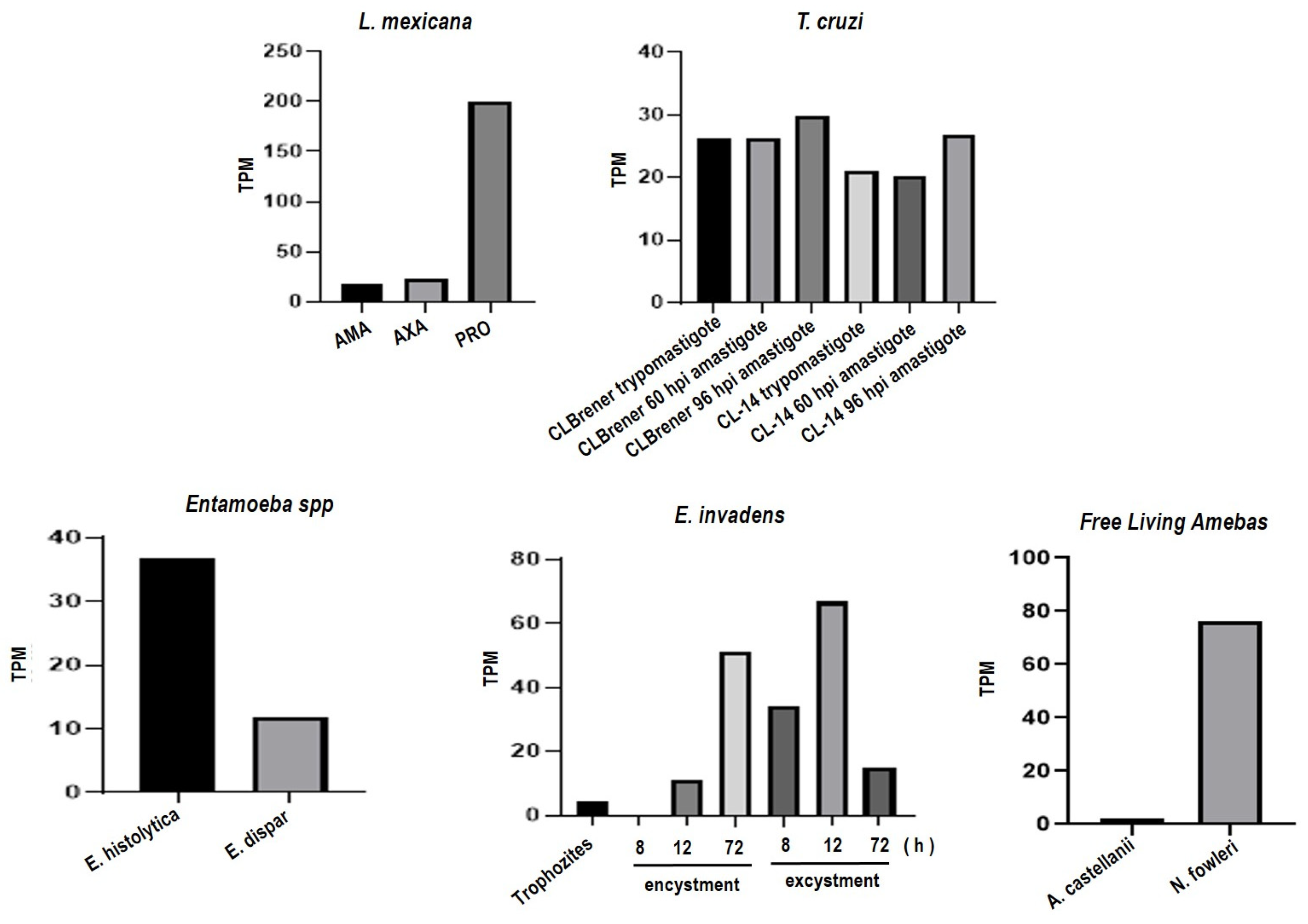
2.7.2. Data Analysis from RNA-Seq Repositories
2.7.3. Search in the Gene Ontology Database
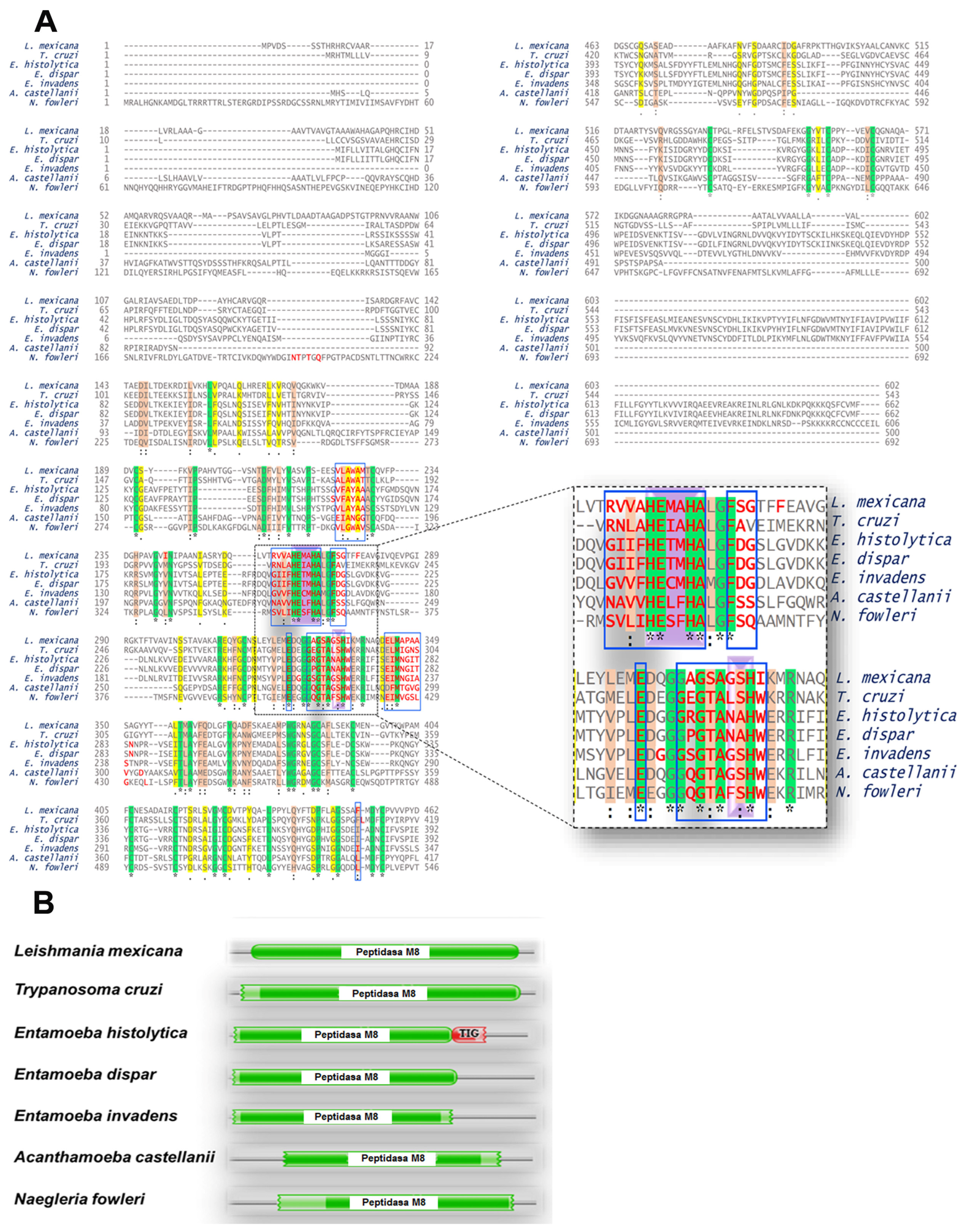
2.7.4. Multiple Alignments
2.7.5. Conserved Domains
2.7.6. Phylogenetic Tree
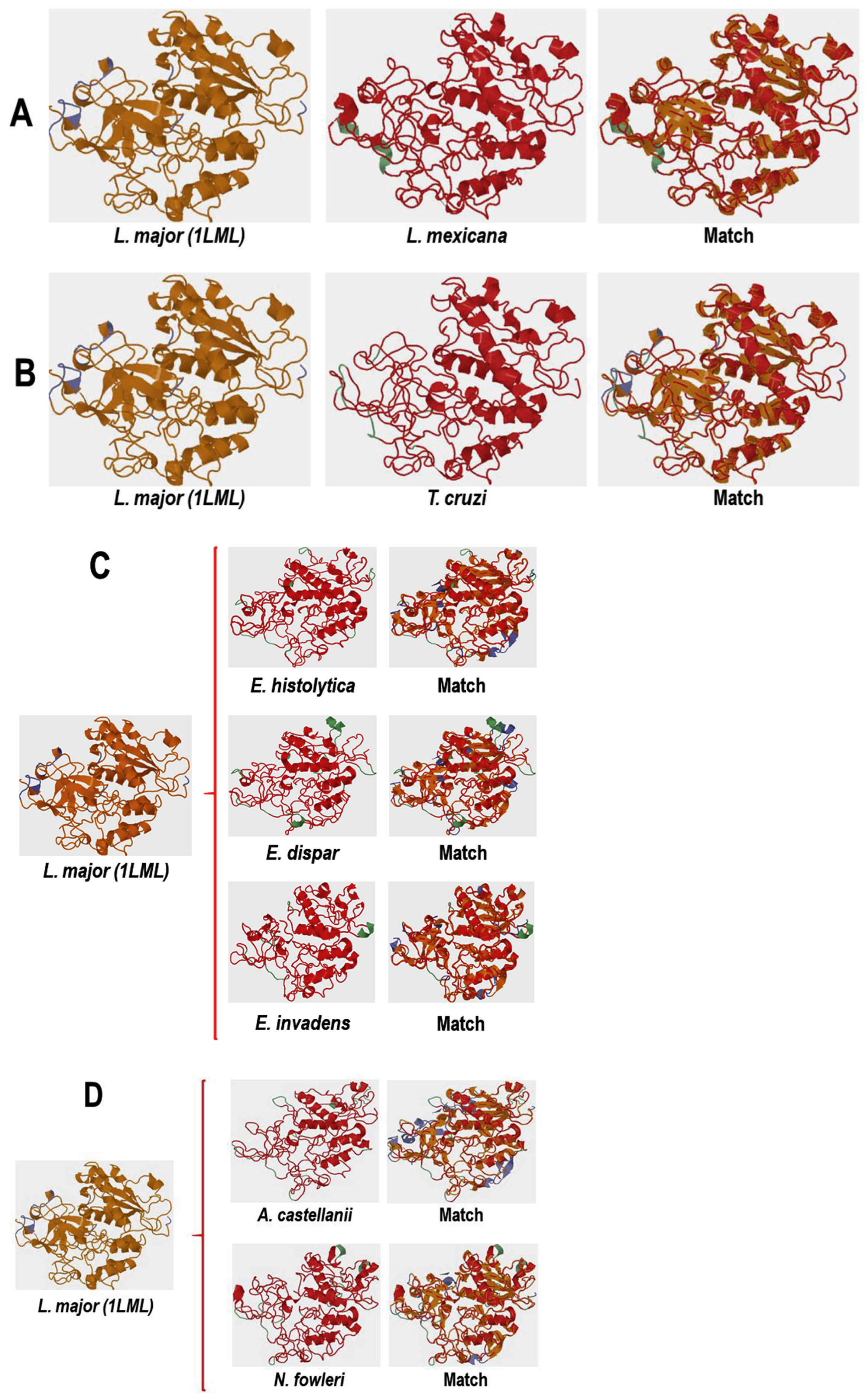
2.8. Comparative Analysis of the Three-Dimensional Structures of L. mexicana gp63 and Proteins with COX-like Activity from Different Protozoan Parasites
2.9. Comparative Analysis of the Amino Acid Sequences of the Proteins with COX-like Activity Bound to the AA Structure
2.10. Statistical Analysis
3. Results
3.1. The gp63 of L. mexicana Is Responsible for Cyclooxygenase (COX)-like Activity
Obtaining the gp63 Mutant
3.2. Trypanosoma cruzi, Entamoeba histolytica, Entamoeba dispar, Entamoeba invadens, Acanthamoeba castellanii and Naegleria fowleri Contain Proteins Orthologous to gp63 of L. mexicana
Bioinformatic Analysis for the Identification of Orthologous Proteins
3.3. Orthologous Proteins Have Characteristics of Metalloproteinases
Ontology of Genes Orthologous to gp63 of L. mexicana
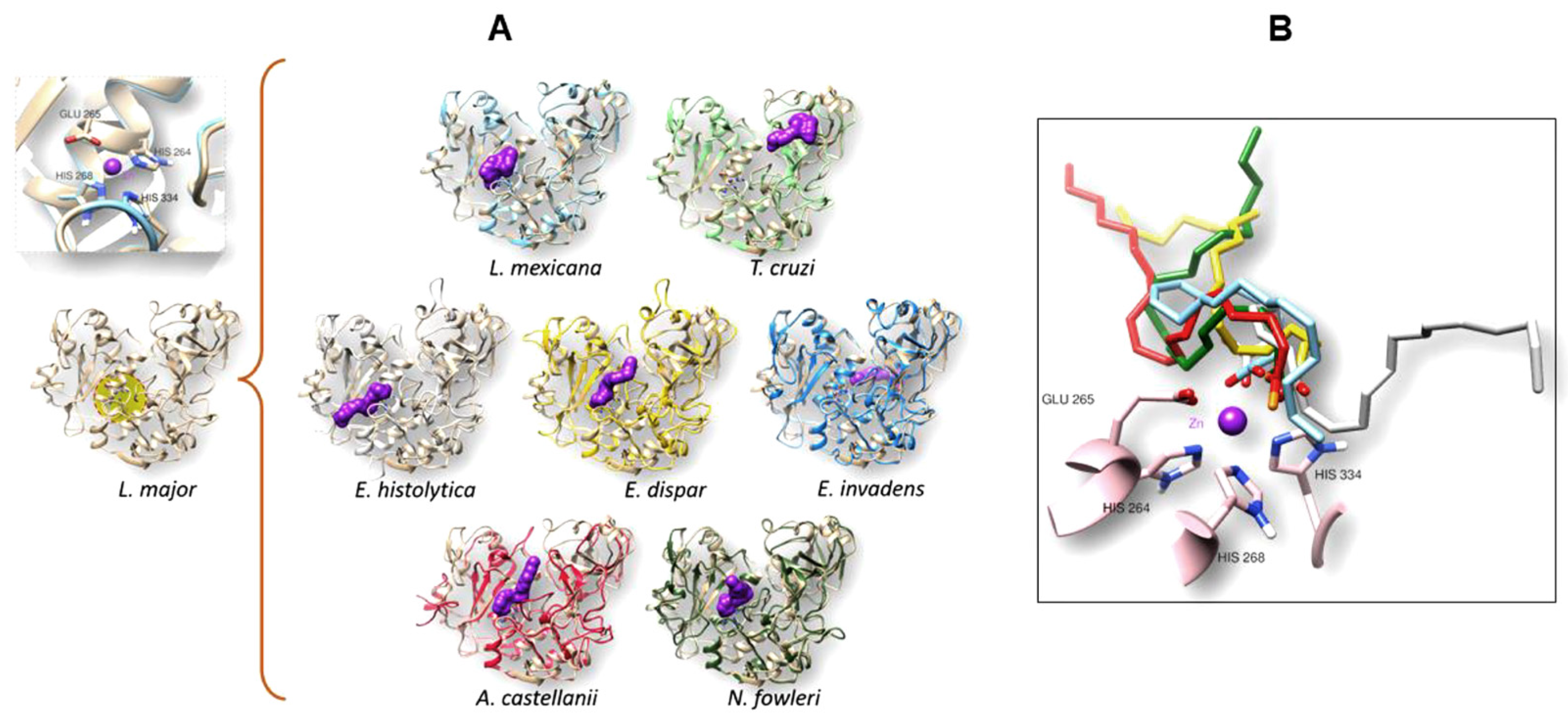
3.4. The gp63 of L. mexicana, and the Orthologous Proteins Present in T. cruzi, Entamoeba histolytica, Entamoeba dispar, E. invadens, A. castellanii, and N. fowleri, Have a Probable Binding Site for Arachidonic Acid
3.4.1. Conformational Analysis of gp63
3.4.2. Molecular Docking Analysis
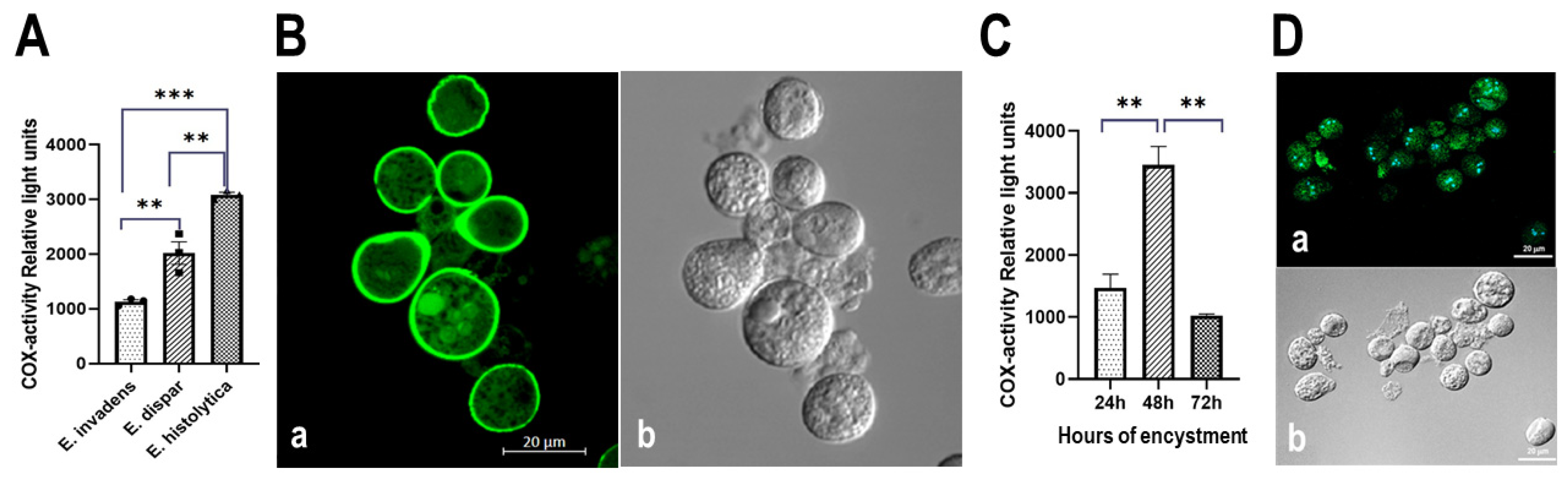
3.5. The Entamoeba Genus Contains a COX-like Activity Which Is Present during the Encystment Process of E. invadens
3.5.1. Determination of COX Activity from Soluble Fractions of E. histolytica, E. dispar and E. invadens, Using Exogenous AA and the Commercial COX Activity Kit
3.5.2. Detection of COX Activity during the Encystment of E. invadens Trophozoites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bosetti, F. Arachidonic Acid Metabolism in Brain Physiology and Pathology: Lessons from Genetically Altered Mouse Models. J. Neurochem. 2007, 102, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Balsinde, J.; Winstead, M.V.; Dennis, E.A. Phospholipase A2 Regulation of Arachidonic Acid Mobilization. FEBS Lett. 2002, 531, 2–6. [Google Scholar] [CrossRef]
- Kudo, I.; Murakami, M. Prostaglandin E Synthase, a Terminal Enzyme for Prostaglandin E2 Biosynthesis. BMB Rep. 2005, 38, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Rouzer, C.A.; Marnett, L.J. Cyclooxygenases: Structural and Functional Insights. J. Lipid Res. 2009, 50, S29–S34. [Google Scholar] [CrossRef] [PubMed]
- Dey, I.; Keller, K.; Belley, A.; Chadee, K. Identification and Characterization of a Cyclooxygenase-like Enzyme from Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 2003, 100, 13561–13566. [Google Scholar] [CrossRef]
- Lejeune, M.; Moreau, F.; Chadee, K. Prostaglandin E2 Produced by Entamoeba histolytica Signals via EP4 Receptor and Alters Claudin-4 to Increase Ion Permeability of Tight Junctions. Am. J. Pathol. 2011, 179, 807–818. [Google Scholar] [CrossRef]
- Estrada-Figueroa, L.A.; Díaz-Gandarilla, J.A.; Hernández-Ramírez, V.I.; Arrieta-González, M.M.; Osorio-Trujillo, C.; Rosales-Encina, J.L.; Toledo-Leyva, A.; Talamás-Rohana, P. Leishmania mexicana Gp63 Is the Enzyme Responsible for Cyclooxygenase (COX) Activity in This Parasitic Protozoa. Biochimie 2018, 151, 73–84. [Google Scholar] [CrossRef]
- Ramamoorthy, R.; Donelson, J.E.; Paetz, K.E.; Maybodi, M.; Roberts, S.C.; Wilson, M.E. Three Distinct RNAs for the Surface Protease Gp63 Are Differentially Expressed during Development of Leishmania donovani chagasi Promastigotes to an Infectious Form. J. Biol. Chem. 1992, 267, 1888–1895. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J. [13] Evolutionary Families of Metallopeptidases. Methods Enzymol. 1995, 248, 183–228. [Google Scholar]
- El-Sayed, N.M.A.; Donelson, J.E. African Trypanosomes Have Differentially Expressed Genes Encoding Homologues of the Leishmania GP63 Surface Protease. J. Biol. Chem. 1997, 272, 26742–26748. [Google Scholar] [CrossRef]
- Grandgenett, P.M.; Coughlin, B.C.; Kirchhoff, L.V.; Donelson, J.E. Differential Expression of GP63 Genes in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2000, 110, 409–415. [Google Scholar] [CrossRef] [PubMed]
- LaCount, D.J.; Gruszynski, A.E.; Grandgenett, P.M.; Bangs, J.D.; Donelson, J.E. Expression and Function of the Trypanosoma brucei Major Surface Protease (GP63) Genes. J. Biol. Chem. 2003, 278, 24658–24664. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.; Davis, P.H.; Tillack, M.; Matthiesen, J.; Lotter, H.; Stanley, S.L.; Tannich, E.; Bruchhaus, I. Differences in the Transcriptome Signatures of Two Genetically Related Entamoeba histolytica Cell Lines Derived from the Same Isolate with Different Pathogenic Properties. BMC Genom. 2010, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.E.; Sateriale, A.; Bessoff, K.E.; Huston, C.D. Control of Entamoeba histolytica Adherence Involves Metallosurface Protease 1, an M8 Family Surface Metalloprotease with Homology to Leishmanolysin. Infect. Immun. 2012, 80, 2165–2176. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ramírez, V.I.; Estrada-Figueroa, L.A.; Medina, Y.; Lizarazo-Taborda, M.R.; Toledo-Leyva, A.; Osorio-Trujillo, C.; Morales-Mora, D.; Talamás-Rohana, P. A Monoclonal Antibody against a Leishmania mexicana COX-like Enzymatic Activity Also Recognizes Similar Proteins in Different Protozoa of Clinical Importance. Parasitol. Res. 2023, 122, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A New Medium for the Axenic Cultivation of Entamoeba histolytica and Other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Sippl, M.J.; Wiederstein, M. A Note on Difficult Structure Alignment Problems. Bioinformatics 2008, 24, 426–427. [Google Scholar] [CrossRef] [PubMed]
- Mercado-Camargo, J.; Cervantes-Ceballos, L.; Vivas-Reyes, R.; Pedretti, A.; Serrano-García, M.L.; Gómez-Estrada, H. Homology Modeling of Leishmanolysin (Gp63) from Leishmania panamensis and Molecular Docking of Flavonoids. ACS Omega 2020, 5, 14741–14749. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Yao, C.; Donelson, J.E.; Wilson, M.E. The Major Surface Protease (MSP or GP63) of Leishmania sp. Biosynthesis, Regulation of Expression, and Function. Mol. Biochem. Parasitol. 2003, 132, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wagh, P.K.; Peace, B.E.; Waltz, S.E. Met-Related Receptor Tyrosine Kinase Ron in Tumor Growth and Metastasis. Adv. Cancer Res. 2008, 100, 1–33. [Google Scholar] [PubMed]
- Hasan, M.M.; Teixeira, J.E.; Lam, Y.-W.; Huston, C.D. Coactosin Phosphorylation Controls Entamoeba histolytica Cell Membrane Protrusions and Cell Motility. mBio 2020, 11, e00660-20. [Google Scholar] [CrossRef] [PubMed]
- Liechti, N.; Schürch, N.; Bruggmann, R.; Wittwer, M. Nanopore Sequencing Improves the Draft Genome of the Human Pathogenic Amoeba Naegleria fowleri. Sci. Rep. 2019, 9, 16040. [Google Scholar] [CrossRef]
- Siddiqui, R.; Lakhundi, S.; Iqbal, J.; Khan, N.A. Effect of Non-Steroidal Anti-Inflammatory Drugs on Biological Properties of Acanthamoeba castellanii Belonging to the T4 Genotype. Exp. Parasitol. 2016, 168, 45–50. [Google Scholar] [CrossRef]
- d’Avila-Levy, C.M.; Altoé, E.C.F.; Uehara, L.A.; Branquinha, M.H.; Santos, A.L.S. GP63 Function in the Interaction of Trypanosomatids with the Invertebrate Host: Facts and Prospects. Subcell Biochem. 2014, 74, 253–270. [Google Scholar]
- Clarke, M.; Lohan, A.J.; Liu, B.; Lagkouvardos, I.; Roy, S.; Zafar, N.; Bertelli, C.; Schilde, C.; Kianianmomeni, A.; Bürglin, T.R.; et al. Genome of Acanthamoeba castellanii Highlights Extensive Lateral Gene Transfer and Early Evolution of Tyrosine Kinase Signaling. Genome Biol. 2013, 14, R11. [Google Scholar] [CrossRef] [PubMed]
- Andrade, Y.M.F.d.S.; Castro, M.V.d.; Tavares, V.d.S.; Souza, R.d.S.O.; Faccioli, L.H.; Lima, J.B.; Sorgi, C.A.; Borges, V.M.; Araújo-Santos, T. Polyunsaturated Fatty Acids Alter the Formation of Lipid Droplets and Eicosanoid Production in Leishmania Promastigotes. Mem. Inst. Oswaldo Cruz 2023, 118, e220160. [Google Scholar] [CrossRef] [PubMed]


| Description | E Value | Query Cover % | Identity % | Accession |
|---|---|---|---|---|
| Leishmania mexicana (GP63, leishmanolysin) | 0.0 | 100 | 100 | XP_003872886.1 |
| Trypanosoma cruzi (surface protease GP63 putative) | 1 × 10−111 | 86 | 37.86 | XP_817808.1 |
| Entamoeba histolytica (cell surface protease GP63 putative) | 4 × 10−37 | 71 | 27.43 | XP_652632.1 |
| Entamoeba dispar SAW760 (uncharacterized protein EDI_037980) | 3 × 10−35 | 81 | 25.81 | XP_001740726.1 |
| Entamoeba invadens IP1(hypothetical protein EIN_174510) | 1 × 10−31 | 66 | 27.59 | XP_004184102.1 |
| Acanthamoeba castellanii str. Neff (leishmanolysin, putative) | 5 × 10−45 | 52 | 32.75 | XP_004337275.1 |
| Naegleria fowleri (unnamed protein product) | 3 × 10−30 | 52 | 28.25 | XP_0044566011.1 |
| Species | Gene Bank ID | Uniprot ID | TriTrypDB/ AmoebaDB | Name of Protein | Gene Ontology Predicted Functions |
|---|---|---|---|---|---|
| L. mexicana | XP_003872886.1 | E9AN57 | LmxM.10.0470 | GP63, leishmnolysin | GO: 0004222 metalloendopeptidase activity GO: 0005886 plasma membrane GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0016020 membrane GO: 0046872 metal ion binding |
| T. cruzi | XP_817808.1 | Q4DTV2 | TcCLB.508693.100 | Surface protease GP63 putative | GO: 0004222 metalloendopeptidase activity GO: 0005886 plasma membrane GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0016020 membrane GO: 0046872 metal ion binding |
| E. histolytica | XP_655632.1 | C4M655 | EH.042870 | Cell surface protease gp63puttive | GO: 0004222 metalloendopeptidase activity GO: 0005886 plasma membrane GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0008270 zinc ion binding membrane GO: 0016020 membrane |
| E. dispar | XP_001740726.1 | B0ERK0 | EDI_037980 | Hypothetical protein conserved | GO: 0004222 metalloendopeptidase activity GO: 0005886 plasma membrane GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0016020 membrane GO: 0016021 integral component of membrane |
| E. invadens | XP_004184102.1 | A0A0A1TW87 | EIN_174510 | Hypothetical protein | GO: 0004222 metalloendopeptidase activity GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0016020 membrane GO: 0016021 integral component of membrane |
| A. castellanii | XP_004337275.1 | L8GQS8 | ACA1_29880 | Leishmanolysin putative | GO: 0004222 metalloendopeptidase activity GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0016020 membrane |
| N. fowleri | XP_044566011.1 | A0A6A5C651 | NF0068990 | Metalloendopeptidase zinc ion binding protein | GO: 0004222 metalloendopeptidase activity GO: 0006508 proteolysis GO: 0007155 cell adhesion GO: 0016020 membrane |
| Description | Length | QC% | TC% | Score | RMS | SI% |
|---|---|---|---|---|---|---|
| Leishmania mexicana | 465 | 100 | 98 | 465 | 0.07 | 81 |
| Trypanosoma cruzi | 444 | 95 | 97 | 442 | 0.44 | 39 |
| Entamoeba histolytica | 424 | 91 | 89 | 420 | 0.73 | 25 |
| Entamoeba dispar | 405 | 87 | 89 | 400 | 0.79 | 26 |
| Entamoeba invadens | 418 | 89 | 93 | 414 | 0.69 | 20 |
| Acanthamoeba castellanii | 377 | 81 | 91 | 373 | 0.73 | 31 |
| Naegleria fowleri | 432 | 93 | 90 | 428 | 0.68 | 28 |
| Organism | Interaction Residues on the Binding Site | H-Bond | Energy (kcal/mol) |
|---|---|---|---|
| L. mexicana | VAL223 SER222 ALA348 SER330 PRO347 ALA346 HIS334 SER333 SER273 HIS268 GLU265 TRP226 HIS264 | HIS264 | −11.16 |
| T. cruzi | GLU452 GLY453 LEU367 LEU368 TYR379 ARG416 PRO417 VAL419 TYR418 | TYR379 ARG416 | −8.84 |
| E. histolytica | ARG261 TYR161 TYR126 HIS210 GLU207 HIS206 THR282 ILE281 GLY280 HIS267 ALA266 THR263 GLY262 ASP215 | HIS206 | −9.57 |
| E. dispar | SER155 SER157 GLU392 TYR395 THR282 SER283 ILE281 GLY262 THR263 GLY280 ALA266 HIS267 HIS210 HIS206 GLU207 ALA162 ALA160 TYR161 PHE159 VAL158 | HIS267 GLU207 | −10.32 |
| E. invadens | LEU156 GLY157 PHE160 SER60 ILE232 VAL243 TYR64 ASP265 LYS440 PRO439 SER408 CYS443 ASN406 THR446 ARG153 PRO241 | --- | −8.92 |
| A. castellanii | GLU184 LEU417 ASN420 VAL298 VAL300 GLY279 GLY297 HIS284 HIS231 HIS235 GLU232 GLY189 ALA228 ASN188 ALA187 ILE186 GLU185 | --- | −11.02 |
| N. fowleri | GLY310 LEU312 GLU345 SER428 LYS431 LEU429 GLY430 GLY409 LEU434 GKY427 HIS414 SER413 HIS356 GLN204 HIS352 GLU353 THR200 GLY313 ASN199 VAL311 | --- | −10.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Ramírez, V.I.; Matus-Meza, A.-S.; Oviedo, N.; Magos-Castro, M.A.; Osorio-Trujillo, C.; Salazar-Villatoro, L.; Constantino-Jonapa, L.A.; Talamás-Rohana, P. Exploration of the Binding Site of Arachidonic Acid in gp63 of Leishmania mexicana and in Orthologous Proteins in Clinically Important Parasites. Pathogens 2024, 13, 718. https://doi.org/10.3390/pathogens13090718
Hernández-Ramírez VI, Matus-Meza A-S, Oviedo N, Magos-Castro MA, Osorio-Trujillo C, Salazar-Villatoro L, Constantino-Jonapa LA, Talamás-Rohana P. Exploration of the Binding Site of Arachidonic Acid in gp63 of Leishmania mexicana and in Orthologous Proteins in Clinically Important Parasites. Pathogens. 2024; 13(9):718. https://doi.org/10.3390/pathogens13090718
Chicago/Turabian StyleHernández-Ramírez, Verónica Ivonne, Audifás-Salvador Matus-Meza, Norma Oviedo, Marco Antonio Magos-Castro, Carlos Osorio-Trujillo, Lizbeth Salazar-Villatoro, Luis Alejandro Constantino-Jonapa, and Patricia Talamás-Rohana. 2024. "Exploration of the Binding Site of Arachidonic Acid in gp63 of Leishmania mexicana and in Orthologous Proteins in Clinically Important Parasites" Pathogens 13, no. 9: 718. https://doi.org/10.3390/pathogens13090718
APA StyleHernández-Ramírez, V. I., Matus-Meza, A.-S., Oviedo, N., Magos-Castro, M. A., Osorio-Trujillo, C., Salazar-Villatoro, L., Constantino-Jonapa, L. A., & Talamás-Rohana, P. (2024). Exploration of the Binding Site of Arachidonic Acid in gp63 of Leishmania mexicana and in Orthologous Proteins in Clinically Important Parasites. Pathogens, 13(9), 718. https://doi.org/10.3390/pathogens13090718







