Oral Microbiota and the Risk of Gastrointestinal Cancers—A Narrative Literature Review
Abstract
1. Introduction
2. Methods
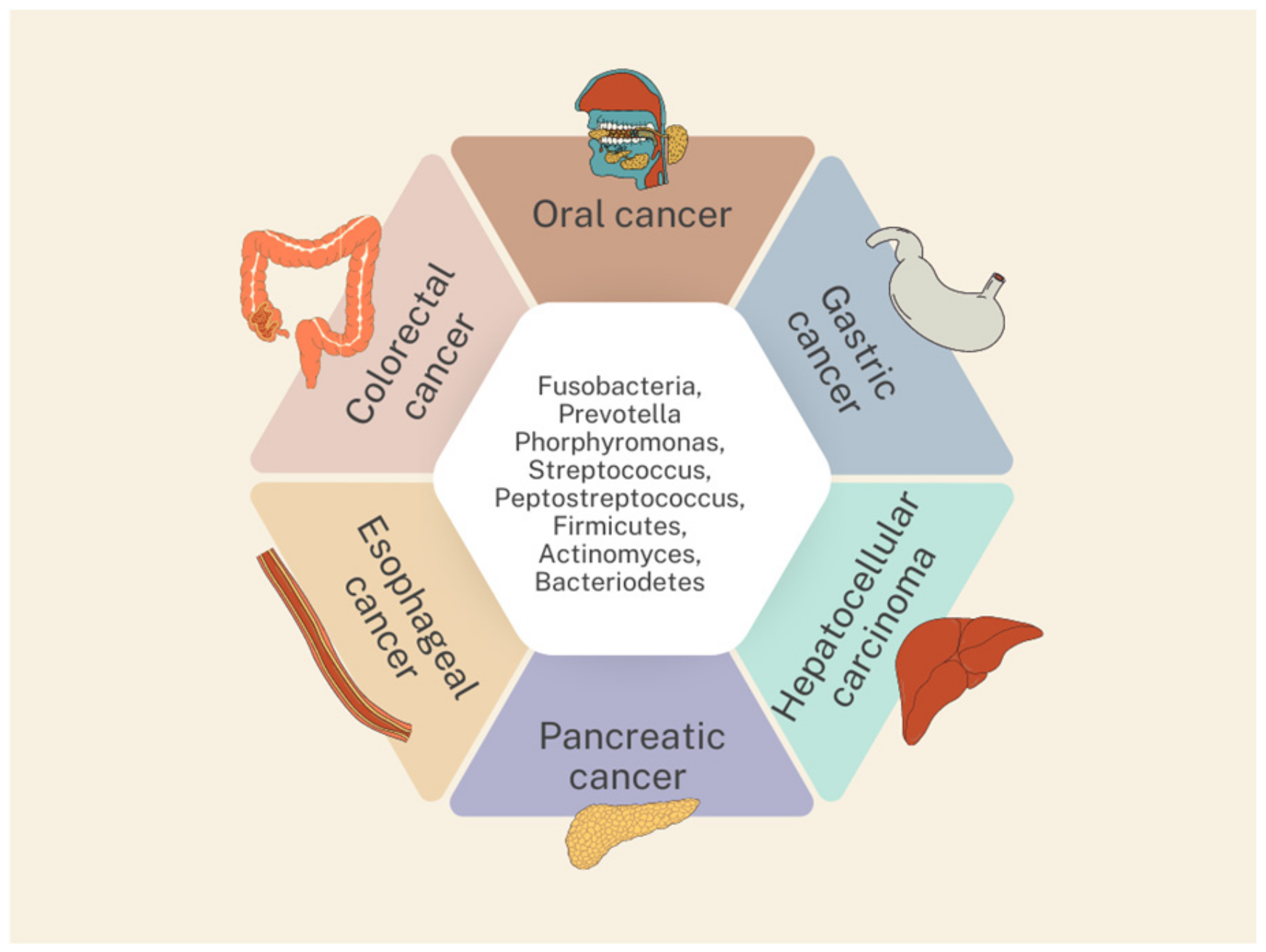
3. Oral Microbiota and Gastrointestinal Cancers
3.1. Oral Cancer
3.2. Esophageal Cancer
3.3. Gastric Cancer
3.4. Colorectal Cancer
3.5. Pancreatic Cancer
3.6. Hepatocellular Carcinoma
4. The Limitations of This Review and Suggestions for Future Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Le Bars, P.; Matamoros, S.; Montassier, E.; Le Vacon, F.; Potel, G.; Soueidan, A.; Jordana, F.; de La Cochetière, M.F. The oral cavity microbiota: Between health, oral disease, and cancers of the aerodigestive tract. Can. J. Microbiol. 2017, 63, 475–492. [Google Scholar] [CrossRef] [PubMed]
- HOMD. Human Oral Microbiome Database. Available online: https://www.ehomd.org/ (accessed on 23 July 2010).
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, S.; Zhang, T.; Chen, X. Association between oral microflora and gastrointestinal tumors (Review). Oncol. Rep. 2021, 46, 160. [Google Scholar] [CrossRef] [PubMed]
- Mascitti, M.; Togni, L.; Troiano, G.; Caponio, V.C.A.; Gissi, D.B.; Montebugnoli, L.; Procaccini, M.; Lo Muzio, L.; Santarelli, A. Beyond Head and Neck Cancer: The Relationship Between Oral Microbiota and Tumour Development in Distant Organs. Front. Cell Infect. Microbiol. 2019, 9, 232. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Zhang, Y.; Niu, Q.; Fan, W.; Huang, F.; He, H. Oral microbiota and gastrointestinal cancer. Onco Targets Ther. 2019, 12, 4721–4728. [Google Scholar] [CrossRef]
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human Gut Microbiota and Gastrointestinal Cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49. [Google Scholar] [CrossRef]
- Binder Gallimidi, A.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef]
- Michaud, D.S. Role of bacterial infections in pancreatic cancer. Carcinogenesis 2013, 34, 2193–2197. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Eslami, H.; Kafil, H.S. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed. Pharmacother. 2017, 89, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Nosho, K.; Sukawa, Y.; Adachi, Y.; Ito, M.; Mitsuhashi, K.; Kurihara, H.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Igarashi, H.; et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 2016, 22, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W. Commentary: Oral bacteria as drivers for colorectal cancer. J. Periodontol. 2014, 85, 1155–1157. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.Y.; Yu, J.; Wong, S.H.; Peppelenbosch, M.P.; Fuhler, G.M. The gastrointestinal microbiota and its role in oncogenesis. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Srinivasprasad, V.; Dineshshankar, J.; Sathiyajeeva, J.; Karthikeyan, M.; Sunitha, J.; Ragunathan, R. Liaison between micro-organisms and oral cancer. J. Pharm. Bioallied Sci. 2015, 7, S354–S360. [Google Scholar] [CrossRef]
- Marttila, E.; Uittamo, J.; Rusanen, P.; Lindqvist, C.; Salaspuro, M.; Rautemaa, R. Acetaldehyde production and microbial colonization in oral squamous cell carcinoma and oral lichenoid disease. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 116, 61–68. [Google Scholar] [CrossRef]
- Muto, M.; Hitomi, Y.; Ohtsu, A.; Shimada, H.; Kashiwase, Y.; Sasaki, H.; Yoshida, S.; Esumi, H. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: Implications for carcinogenesis in upper aerodigestive tract. Int. J. Cancer 2000, 88, 342–350. [Google Scholar] [CrossRef]
- Park, S.Y.; Hwang, B.O.; Lim, M.; Ok, S.H.; Lee, S.K.; Chun, K.S.; Park, K.K.; Hu, Y.; Chung, W.Y.; Song, N.Y. Oral-Gut Microbiome Axis in Gastrointestinal Disease and Cancer. Cancers 2021, 13, 2124. [Google Scholar] [CrossRef]
- Sulaiman, Y.; Pacauskienė, I.M.; Šadzevičienė, R.; Anuzyte, R. Oral and Gut Microbiota Dysbiosis Due to Periodontitis: Systemic Implications and Links to Gastrointestinal Cancer: A Narrative Review. Medicina 2024, 60, 1416. [Google Scholar] [CrossRef]
- Raoul, P.; Maccauro, V.; Cintoni, M.; Scarpellini, E.; Ianiro, G.; Gasbarrini, A.; Mele, M.C.; Rinninella, E. Microbiota-Gastric Cancer Interactions and the Potential Influence of Nutritional Therapies. Int. J. Mol. Sci. 2024, 25, 1679. [Google Scholar] [CrossRef]
- Camañes-Gonzalvo, S.; Montiel-Company, J.M.; Lobo-de-Mena, M.; Safont-Aguilera, M.J.; Fernández-Diaz, A.; López-Roldán, A.; Paredes-Gallardo, V.; Bellot-Arcís, C. Relationship between oral microbiota and colorectal cancer: A systematic review. J. Periodontal Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Elghannam, M.T.; Hassanien, M.H.; Ameen, Y.A.; Turky, E.A.; ELattar, G.M.; ELRay, A.A.; ELTalkawy, M.D. Helicobacter pylori and oral-gut microbiome: Clinical implications. Infection 2024, 52, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, X.; Zou, A.; Mai, Z.; Huang, Z.; Sun, L.; Zhao, J. circIGHG-Induced Epithelial-to-Mesenchymal Transition Promotes Oral Squamous Cell Carcinoma Progression via miR-142-5p/IGF2BP3 Signaling. Cancer Res. 2021, 81, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.H.; Iyer, N.G.; Tan, M.H.; Edgren, G. Changing epidemiology of oral squamous cell carcinoma of the tongue: A global study. Head. Neck. 2017, 39, 297–304. [Google Scholar] [CrossRef]
- Komlós, G.; Csurgay, K.; Horváth, F.; Pelyhe, L.; Németh, Z. Periodontitis as a risk for oral cancer: A case-control study. BMC Oral Health 2021, 21, 640. [Google Scholar] [CrossRef]
- Tuominen, H.; Rautava, J. Oral Microbiota and Cancer Development. Pathobiology 2021, 88, 116–126. [Google Scholar] [CrossRef]
- Guo, Z.C.; Jing, S.L.; Jia, X.Y.; Elayah, S.A.; Xie, L.Y.; Cui, H.; Tu, J.B.; Na, S.J. Porphyromonas gingivalis promotes the progression of oral squamous cell carcinoma by stimulating the release of neutrophil extracellular traps in the tumor immune microenvironment. Inflamm. Res. 2024, 73, 693–705. [Google Scholar] [CrossRef]
- Guo, Z.C.; Jing, S.L.; Jumatai, S.; Gong, Z.C. Porphyromonas gingivalis promotes the progression of oral squamous cell carcinoma by activating the neutrophil chemotaxis in the tumour microenvironment. Cancer Immunol. Immunother. 2023, 72, 1523–1539. [Google Scholar] [CrossRef]
- Wen, L.; Mu, W.; Lu, H.; Wang, X.; Fang, J.; Jia, Y.; Li, Q.; Wang, D.; Wen, S.; Guo, J.; et al. Porphyromonas gingivalis Promotes Oral Squamous Cell Carcinoma Progression in an Immune Microenvironment. J. Dent. Res. 2020, 99, 666–675. [Google Scholar] [CrossRef]
- Lan, Z.; Zou, K.L.; Cui, H.; Zhao, Y.Y.; Yu, G.T. Porphyromonas gingivalis suppresses oral squamous cell carcinoma progression by inhibiting MUC1 expression and remodeling the tumor microenvironment. Mol. Oncol. 2024, 18, 1174–1188. [Google Scholar] [CrossRef]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Yeh, Y.M.; Yu, H.Y.; Chin, C.Y.; Hsu, C.W.; Liu, H.; Huang, P.J.; Hu, S.N.; Liao, C.T.; Chang, K.P.; et al. Oral Microbiota Community Dynamics Associated With Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Su, S.C.; Chang, L.C.; Huang, H.D.; Peng, C.Y.; Chuang, C.Y.; Chen, Y.T.; Lu, M.Y.; Chiu, Y.W.; Chen, P.Y.; Yang, S.F. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis 2021, 42, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Winckler, B.; Lu, M.; Cheng, H.; Yuan, Z.; Yang, Y.; Jin, L.; Ye, W. Oral Microbiota and Risk for Esophageal Squamous Cell Carcinoma in a High-Risk Area of China. PLoS ONE 2015, 10, e0143603. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, T.; Yan, Y.; Zhang, Y.; Li, Z.; Wang, Y.; Yang, J.; Xia, Y.; Xiao, H.; Han, H.; et al. Alterations of Oral Microbiota in Chinese Patients With Esophageal Cancer. Front. Cell Infect. Microbiol. 2020, 10, 541144. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Z.; Sun, K.; Li, M.X.; Qi, Y.J. Upper gastrointestinal tract microbiota with oral origin in relation to oesophageal squamous cell carcinoma. Ann. Med. 2023, 55, 2295401. [Google Scholar] [CrossRef]
- Hu, J.; Han, S.; Chen, Y.; Ji, Z. Variations of Tongue Coating Microbiota in Patients with Gastric Cancer. Biomed. Res. Int. 2015, 2015, 173729. [Google Scholar] [CrossRef]
- Yamamura, K.; Baba, Y.; Miyake, K.; Nakamura, K.; Shigaki, H.; Mima, K.; Kurashige, J.; Ishimoto, T.; Iwatsuki, M.; Sakamoto, Y.; et al. Fusobacterium nucleatum in gastroenterological cancer: Evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol. Lett. 2017, 14, 6373–6378. [Google Scholar] [CrossRef]
- Yu, G.; Torres, J.; Hu, N.; Medrano-Guzman, R.; Herrera-Goepfert, R.; Humphrys, M.S.; Wang, L.; Wang, C.; Ding, T.; Ravel, J.; et al. Molecular Characterization of the Human Stomach Microbiota in Gastric Cancer Patients. Front. Cell Infect. Microbiol. 2017, 7, 302. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, M.; Salazar, C.R.; Hays, R.; Bedi, S.; Chen, Y.; Li, Y. Chronic Periodontal Disease, Periodontal Pathogen Colonization, and Increased Risk of Precancerous Gastric Lesions. J. Periodontol. 2017, 88, 1124–1134. [Google Scholar] [CrossRef]
- Ogaya, Y.; Kadota, T.; Hamada, M.; Nomura, R.; Nakano, K. Characterization of the unique oral microbiome of children harboring Helicobacter pylori in the oral cavity. J. Oral Microbiol. 2024, 16, 2339158. [Google Scholar] [CrossRef] [PubMed]
- Wongsuwanlert, M.; Teanpaisan, R.; Pahumunto, N.; Kaewdech, A.; Ruangsri, P.; Sunpaweravong, S. Prevalence and virulence factors of Helicobacter pylori isolated from oral cavity of non-disease, gastritis, and gastric cancer patients. J. Dent. Sci. 2024, 19, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, A.; Dighe, O.R.; Lamture, Y. Role of Helicobacter pylori in Gastric Carcinoma: A Review. Cureus 2023, 15, e37205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Kong, C.; Yang, Y.; Cai, S.; Li, X.; Cai, G.; Ma, Y. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics 2020, 10, 11595–11606. [Google Scholar] [CrossRef] [PubMed]
- Tortora, S.C.; Agurto, M.G.; Martello, L.A. The oral-gut-circulatory axis: From homeostasis to colon cancer. Front. Cell Infect. Microbiol. 2023, 13, 1289452. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef]
- Bergsten, E.; Mestivier, D.; Donnadieu, F.; Pedron, T.; Barau, C.; Meda, L.T.; Mettouchi, A.; Lemichez, E.; Gorgette, O.; Chamaillard, M.; et al. Parvimonas micra, an oral pathobiont associated with colorectal cancer, epigenetically reprograms human colonocytes. Gut Microbes 2023, 15, 2265138. [Google Scholar] [CrossRef]
- Russo, E.; Gloria, L.D.; Nannini, G.; Meoni, G.; Niccolai, E.; Ringressi, M.N.; Baldi, S.; Fani, R.; Tenori, L.; Taddei, A.; et al. From adenoma to CRC stages: The oral-gut microbiome axis as a source of potential microbial and metabolic biomarkers of malignancy. Neoplasia 2023, 40, 100901. [Google Scholar] [CrossRef]
- Mo, S.; Ru, H.; Huang, M.; Cheng, L.; Mo, X.; Yan, L. Oral-Intestinal Microbiota in Colorectal Cancer: Inflammation and Immunosuppression. J. Inflamm. Res. 2022, 15, 747–759. [Google Scholar] [CrossRef]
- Nearing, J.T.; DeClercq, V.; Langille, M.G.I. Investigating the oral microbiome in retrospective and prospective cases of prostate, colon, and breast cancer. NPJ Biofilms Microbiomes 2023, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Ferrarese, R.; Zuppardo, R.A.; Puzzono, M.; Mannucci, A.; Amato, V.; Ditonno, I.; Patricelli, M.G.; Raucci, A.R.; Clementi, M.; Elmore, U.; et al. Oral and Fecal Microbiota in Lynch Syndrome. J. Clin. Med. 2020, 9, 2735. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Rezasoltani, S.; Aghdaei, H.A.; Jasemi, S.; Gazouli, M.; Dovrolis, N.; Sadeghi, A.; Schlüter, H.; Zali, M.R.; Sechi, L.A.; Feizabadi, M.M. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers 2022, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Sexton, R.E.; Uddin, M.H.; Bannoura, S.; Khan, H.Y.; Mzannar, Y.; Li, Y.; Aboukameel, A.; Al-Hallak, M.N.; Al-Share, B.; Mohamed, A.; et al. Connecting the Human Microbiome and Pancreatic Cancer. Cancer Metastasis Rev. 2022, 41, 317–331. [Google Scholar] [CrossRef]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef]
- Jacob, J.A. Study Links Periodontal Disease Bacteria to Pancreatic Cancer Risk. JAMA 2016, 315, 2653–2654. [Google Scholar] [CrossRef]
- Wei, M.Y.; Shi, S.; Liang, C.; Meng, Q.C.; Hua, J.; Zhang, Y.Y.; Liu, J.; Zhang, B.; Xu, J.; Yu, X.J. The microbiota and microbiome in pancreatic cancer: More influential than expected. Mol. Cancer. 2019, 18, 97. [Google Scholar] [CrossRef]
- Tan, Q.; Ma, X.; Yang, B.; Liu, Y.; Xie, Y.; Wang, X.; Yuan, W.; Ma, J. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes 2022, 14, 2073785. [Google Scholar] [CrossRef]
- Wei, A.L.; Li, M.; Li, G.Q.; Wang, X.; Hu, W.M.; Li, Z.L.; Yuan, J.; Liu, H.Y.; Zhou, L.L.; Li, K.; et al. Oral microbiome and pancreatic cancer. World J. Gastroenterol. 2020, 26, 7679–7692. [Google Scholar] [CrossRef] [PubMed]
- Saba, E.; Farhat, M.; Daoud, A.; Khashan, A.; Forkush, E.; Menahem, N.H.; Makkawi, H.; Pandi, K.; Angabo, S.; Kawasaki, H.; et al. Oral bacteria accelerate pancreatic cancer development in mice. Gut 2024, 73, 770–786. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Li, S.; He, M.; Ao, Z.; Wang, J.; Wu, Q.; Wang, Q. Oral Pathogenic Bacteria and the Oral-Gut-Liver Axis: A New Understanding of Chronic Liver Diseases. Diagnostics 2023, 13, 3324. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Ren, Z.; Li, A.; Zhang, H.; Jiang, J.; Xu, S.; Luo, Q.; Zhou, K.; Sun, X.; Zheng, S.; et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci. Rep. 2016, 6, 33142. [Google Scholar] [CrossRef]
- Yang, J.; He, Q.; Lu, F.; Chen, K.; Ni, Z.; Wang, H.; Zhou, C.; Zhang, Y.; Chen, B.; Bo, Z.; et al. A distinct microbiota signature precedes the clinical diagnosis of hepatocellular carcinoma. Gut Microbes 2023, 15, 2201159. [Google Scholar] [CrossRef]
| Type of Cancer | Incidence (World Rank) | Mortality (World Rank) | 5-Year Prevalence (Europe) |
|---|---|---|---|
| Oral cancer | 16th | 15th | 18.5% |
| Esophageal cancer | 11th | 7th | 10% |
| Gastric cancer | 5th | 5th | 11.6% |
| Colorectal cancer | 3rd | 2nd | 29.4% |
| Pancreatic cancer | 12th | 6th | 26.7% |
| Hepatocellular carcinoma | 6th | 3rd | 8.7% |
| Mechanism | Significant Pathogens | Related Signaling Pathways | |
|---|---|---|---|
| Chronic inflammation | Porphyromonas gingivalis | Inducing chronic inflammation and oncometabolite production by activating anti-apoptotic proteins, growth factors, and cytokines that foster cancer growth and dissemination. | [1,8,9,10,11,12] |
| Fusobacterium nucleatum | Lipopolysaccharides (LPSs) can activate response through Toll-like receptors (TLRs), including TLR2 and TLR4, which can inhibit apoptosis and promote tumor growth. | ||
| Inhibition of immune response | Fusobacterium nucleatum | Expansion of myeloid immune cells that inhibit T-cell proliferation and induce T-cell apoptosis. | [1,8,13] |
| Porphyromonas gingivalis | Protecting tumor cells from NK cells and immune cells through interaction between Fap2 protein and inhibitory immunoreceptor TIGIT (T-cell immunoreceptor with Ig and ITIM domains) on NK and T cells. | ||
| Interference with signaling pathways | Fusobacterium nucleatum | Binding to epithelial (E) cadherin via FadA, leading to growth stimulation of human cancer cells. | [8,14,15] |
| Local metabolism of carcinogens | Neisseria | Activating alcohol and smoking-related carcinogens such as acetaldehyde and nitrosamines. | [8,16,17,18] |
| Candida glabrata |
| Type of Cancer | The Authors of the Study | Year | Bacteria ↑ | Bacteria ↓ | |
|---|---|---|---|---|---|
| Oral cancer | Zhao et al. | 2017 | Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, Parvimonas | No information | [32] |
| Yang et al. | 2018 | Fusobacterium periodonticum, Parvimonas micra, Streptococcus constellatus, Haemophilus influenza, Filifactor alocis | Streptococcus, Haemophilus, Porphyromonas, Actinomyces | [33] | |
| Su et al. | 2021 | Fusobacterium | Streptococcus | [34] | |
| Esophageal cancer | Chen et al. | 2015 | Prevotella, Streptococcus, Porphyromonas | Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus, Cardiobacterium | [35] |
| Zhao et al. | 2020 | Firmicutes, Negatividcutes, Selenmonadales, Prevotellaceae | Protecobacteria, Betaproteobacteria, Neisseriales, Neisseriaceae, Neisseria | [36] | |
| Gao et al. | 2023 | Porphyromonas gingivalis, Fusobacterium nucleatum | No information | [37] | |
| Gastric cancer | Hu et al. | 2015 | Prevotella, Streptococcus, Veillonella, Actinomyces, Leptotrichia | Neisseria, Haemophilus, Porphyromonas, Fusobacterium | [38] |
| Yu et al. | 2017 | Bacteriodetes, Firmicutes, Fusobacteria, Spirochaetes | Proteobacteria | [40] | |
| Sun et al. | 2017 | Treponema denticola, Tannerella forsythia, Actinobacillus | No information | [41] | |
| Colorectal cancer | Flemer et al. | 2018 | Actinomyces, Rothia, Veillonella, Parvimonas micra, Peptostreptococcus stomatis, Dialister pneumosintes, Neisseria, Prevotella, Fusobacterium nucleatum | No information | [54] |
| Zhang et al. | 2020 | Fusobacterium, Prevotella, Veillonellla, Treponema, Phorphyromonas | No information | [45] | |
| Rezasoltani et al. | 2022 | Eubacterium, Bifidobacterium, Fusobacterium | No information | [55] | |
| Nearing et al. | 2023 | Fusobacterium peridonticum | Streptococcus | [52] | |
| Pancreatic cancer | Fan et al. | 2018 | Porphyromonas gingivalis | No information | [58] |
| Wei et al. | 2020 | Streptococcus, Leptotrichina | No information | [62] | |
| Tan et al. | 2022 | Porphyromonas gingivalis | No information | [61] | |
| Hepatocellular carcinoma | Lu et al. | 2016 | Oribacterium, Fusobacterium | No information | [65] |
| Yang et al. | 2023 | Firmicutes, Actinobacteria | No information | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knop-Chodyła, K.; Kochanowska-Mazurek, A.; Piasecka, Z.; Głaz, A.; Wesołek-Bielaska, E.W.; Syty, K.; Forma, A.; Baj, J. Oral Microbiota and the Risk of Gastrointestinal Cancers—A Narrative Literature Review. Pathogens 2024, 13, 819. https://doi.org/10.3390/pathogens13090819
Knop-Chodyła K, Kochanowska-Mazurek A, Piasecka Z, Głaz A, Wesołek-Bielaska EW, Syty K, Forma A, Baj J. Oral Microbiota and the Risk of Gastrointestinal Cancers—A Narrative Literature Review. Pathogens. 2024; 13(9):819. https://doi.org/10.3390/pathogens13090819
Chicago/Turabian StyleKnop-Chodyła, Kinga, Anna Kochanowska-Mazurek, Zuzanna Piasecka, Aneta Głaz, Ewelina Weronika Wesołek-Bielaska, Kinga Syty, Alicja Forma, and Jacek Baj. 2024. "Oral Microbiota and the Risk of Gastrointestinal Cancers—A Narrative Literature Review" Pathogens 13, no. 9: 819. https://doi.org/10.3390/pathogens13090819
APA StyleKnop-Chodyła, K., Kochanowska-Mazurek, A., Piasecka, Z., Głaz, A., Wesołek-Bielaska, E. W., Syty, K., Forma, A., & Baj, J. (2024). Oral Microbiota and the Risk of Gastrointestinal Cancers—A Narrative Literature Review. Pathogens, 13(9), 819. https://doi.org/10.3390/pathogens13090819







