Is Intestinal Microbiota Fully Restored After Chickens Have Recovered from Coccidiosis?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chicken Model of Coccidiosis
2.2. Bacterial DNA Isolation and 16S rRNA Gene Sequencing
2.3. Bioinformatics
2.4. Statitical Analysis
3. Results
3.1. Growth and Pathological Response to E. maxima Infection
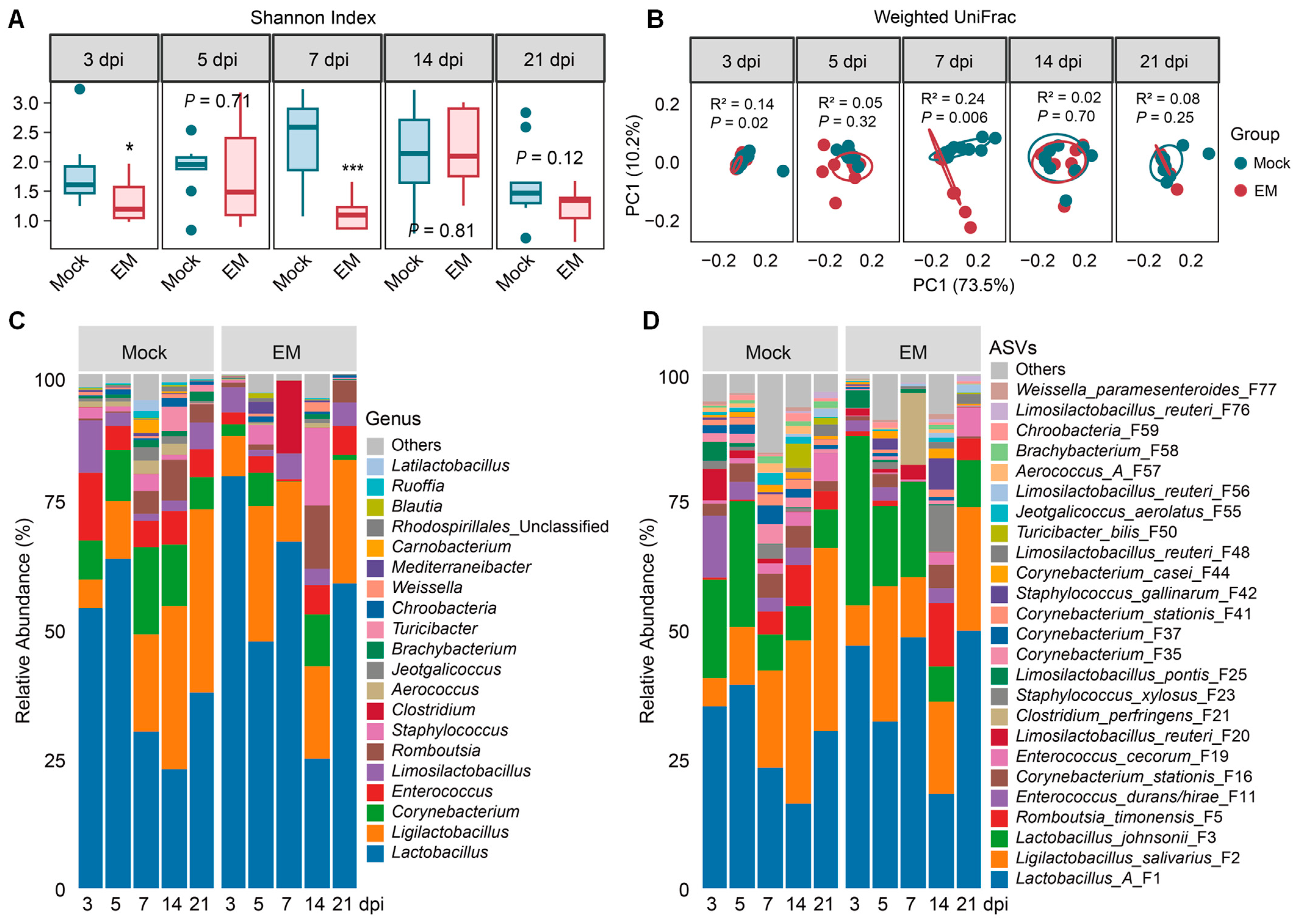
3.2. Ileal Microbiome Dynamics in Response to E. maxima Infection
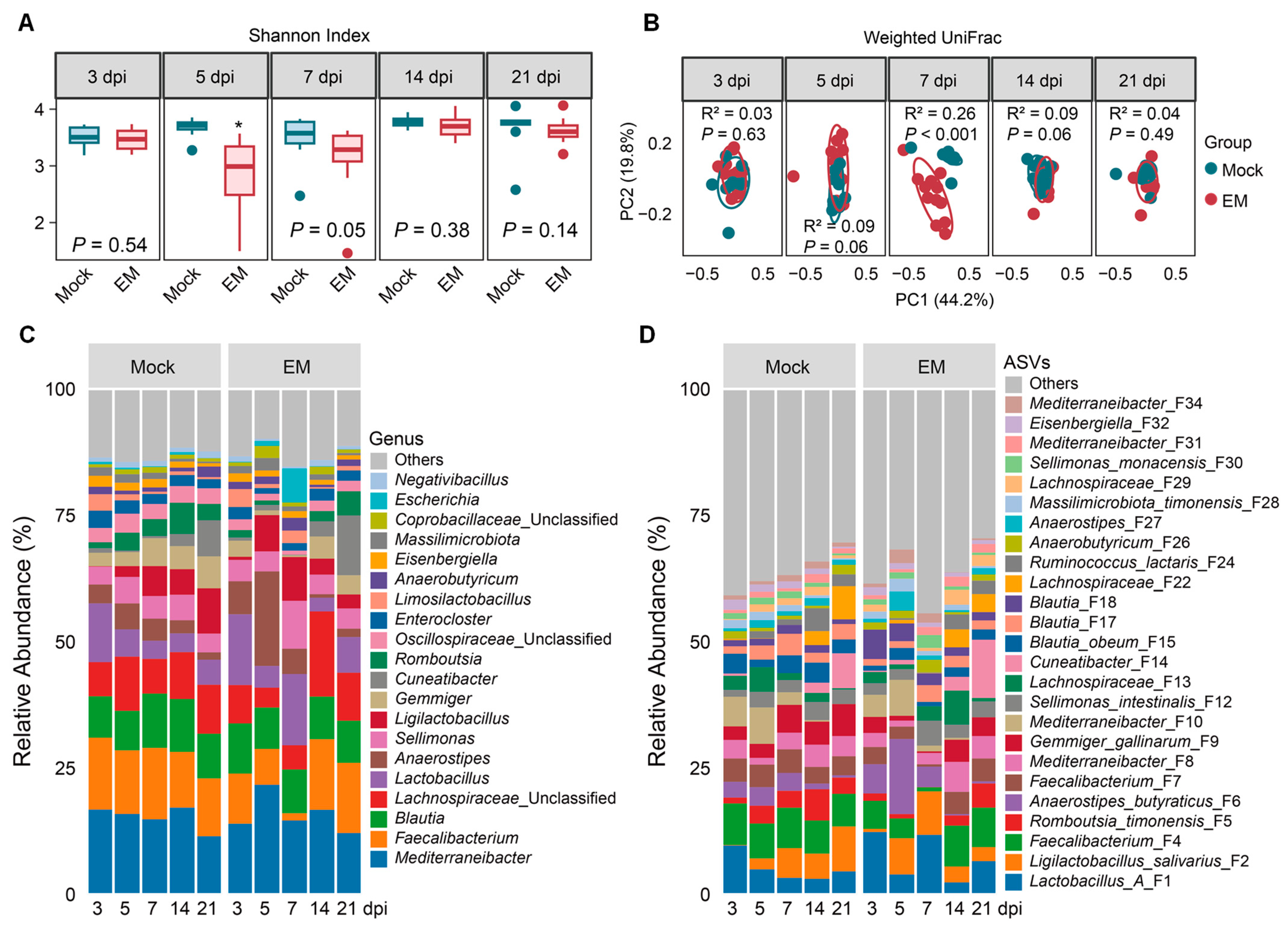
3.3. Cecal Microbiome Dynamics in Response to E. maxima Infection
4. Discussion
4.1. No Compensatory Weight Gain of Chickens After Recovery from Coccidiosis
4.2. Enrichment of Lactic Acid Bacteria in Response to E. maxima Infection
4.3. Diminishment of SCFA-Producing Bacteria in Response to E. maxima Infection
4.4. Transient Blooming of Pathobionts in Response to E. maxima Infection
4.5. Restoration of the Intestinal Microbiota After Recovery from Coccidiosis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sorbara, M.T.; Pamer, E.G. Microbiome-based therapeutics. Nat. Rev. Microbiol. 2022, 20, 365–380. [Google Scholar] [CrossRef]
- Lee, J.Y.; Tsolis, R.M.; Baumler, A.J. The microbiome and gut homeostasis. Science 2022, 377, eabp9960. [Google Scholar] [CrossRef]
- Fassarella, M.; Blaak, E.E.; Penders, J.; Nauta, A.; Smidt, H.; Zoetendal, E.G. Gut microbiome stability and resilience: Elucidating the response to perturbations in order to modulate gut health. Gut 2021, 70, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Barratt, M.J.; Nuzhat, S.; Ahsan, K.; Frese, S.A.; Arzamasov, A.A.; Sarker, S.A.; Islam, M.M.; Palit, P.; Islam, M.R.; Hibberd, M.C.; et al. Bifidobacterium infantis treatment promotes weight gain in Bangladeshi infants with severe acute malnutrition. Sci. Transl. Med. 2022, 14, eabk1107. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, K.Y.; Meng, T.Q.; Ye, Z.; Guo, S.M.; Li, Z.M.; Xiong, C.L.; Yin, Y.; Li, H.G.; Zhou, L.Q. Gut microbiota may not be fully restored in recovered COVID-19 patients after 3-month recovery. Front. Nutr. 2021, 8, 638825. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, P.; Hu, D. Advancements in understanding chicken coccidiosis: From Eimeria biology to innovative control strategies. One Health Adv. 2024, 2, 6. [Google Scholar] [CrossRef]
- Mesa-Pineda, C.; Navarro-Ruiz, J.L.; Lopez-Osorio, S.; Chaparro-Gutierrez, J.J.; Gomez-Osorio, L.M. Chicken Coccidiosis: From the Parasite Lifecycle to Control of the disease. Front. Vet. Sci. 2021, 8, 787653. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.P.; Knox, J.; Dehaeck, B.; Huntington, B.; Rathinam, T.; Ravipati, V.; Ayoade, S.; Gilbert, W.; Adebambo, A.O.; Jatau, I.D.; et al. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020, 51, 115. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Osorio, S.; Chaparro-Gutierrez, J.J.; Gomez-Osorio, L.M. Overview of poultry Eimeria life cycle and host-parasite interactions. Front. Vet. Sci. 2020, 7, 384. [Google Scholar] [CrossRef] [PubMed]
- Hauck, R. Interactions between parasites and the bacterial microbiota of chickens. Avian Dis. 2017, 61, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Madlala, T.; Okpeku, M.; Adeleke, M.A. Understanding the interactions between Eimeria infection and gut microbiota, towards the control of chicken coccidiosis: A review. Parasite 2021, 28, 48. [Google Scholar] [CrossRef]
- Miska, K.B.; Campos, P.M.; Cloft, S.E.; Jenkins, M.C.; Proszkowiec-Weglarz, M. Temporal changes in jejunal and ileal microbiota of broiler chickens with clinical coccidiosis (Eimeria maxima). Animals 2024, 14, 2976. [Google Scholar] [CrossRef]
- Liu, J.; Guo, J.; Whitmore, M.A.; Tobin, I.; Kim, D.M.; Zhao, Z.; Zhang, G. Dynamic response of the intestinal microbiome to Eimeria maxima-induced coccidiosis in chickens. Microbiol. Spectr. 2024, 12, e0082324. [Google Scholar] [CrossRef]
- Yang, Q.; Whitmore, M.A.; Robinson, K.; Lyu, W.; Zhang, G. Butyrate, forskolin, and lactose synergistically enhance disease resistance by inducing the expression of the genes involved in innate host defense and barrier function. Antibiotics 2021, 10, 1175. [Google Scholar] [CrossRef]
- Johnson, J.; Reid, W.M. Anticoccidial drugs: Lesion scoring techniques in battery and floor-pen experiments with chickens. Exp. Parasitol. 1970, 28, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Liu, J.; Whitmore, M.A.; Tobin, I.; Zhao, Z.; Zhang, G. Two intestinal microbiota-derived metabolites, deoxycholic acid and butyrate, synergize to enhance host defense peptide synthesis and alleviate necrotic enteritis. J. Anim. Sci. Biotechnol. 2024, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wang, Q.; Cole, J.R. Updated RDP taxonomy and RDP Classifier for more accurate taxonomic classification. Microbiol. Resour. Announc. 2024, 13, e01063-23. [Google Scholar] [CrossRef]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.-S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Freitas, L.; Sakomura, N.K.; Reis, M.P.; Mariani, A.B.; Lambert, W.; Andretta, I.; Letourneau-Montminy, M.P. Coccidiosis infection and growth performance of broilers in experimental trials: Insights from a meta-analysis including modulating factors. Poult. Sci. 2023, 102, 103021. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Inohara, N.; Nunez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef]
- Andre, A.C.; Debande, L.; Marteyn, B.S. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell. Microbiol. 2021, 23, e13338. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Wang, X.; Robinson, K.; Whitmore, M.A.; Stewart, S.N.; Zhao, J.; Zhang, G. Identification of an intestinal microbiota signature associated with the severity of necrotic enteritis. Front. Microbiol. 2021, 12, 703693. [Google Scholar] [CrossRef]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pasolli, E.; Ercolini, D. The food-gut axis: Lactic acid bacteria and their link to food, the gut microbiome and human health. FEMS Microbiol. Rev. 2020, 44, 454–489. [Google Scholar] [CrossRef]
- Deng, Z.; Hou, K.; Zhao, J.; Wang, H. The probiotic properties of lactic acid bacteria and their applications in animal husbandry. Curr. Microbiol. 2021, 79, 22. [Google Scholar] [CrossRef]
- Khurajog, B.; Disastra, Y.; Lawwyne, L.D.; Sirichokchatchawan, W.; Niyomtham, W.; Yindee, J.; Hampson, D.J.; Prapasarakul, N. Selection and evaluation of lactic acid bacteria from chicken feces in Thailand as potential probiotics. PeerJ 2023, 11, e16637. [Google Scholar] [CrossRef]
- Li, C.; Peng, K.; Xiao, S.; Long, Y.; Yu, Q. The role of Lactobacillus in inflammatory bowel disease: From actualities to prospects. Cell Death Discov. 2023, 9, 361. [Google Scholar] [CrossRef]
- Shan, Y.; Lee, M.; Chang, E.B. The gut microbiome and inflammatory bowel diseases. Annu. Rev. Med. 2022, 73, 455–468. [Google Scholar] [CrossRef]
- Mann, E.R.; Lam, Y.K.; Uhlig, H.H. Short-chain fatty acids: Linking diet, the microbiome and immunity. Nat. Rev. Immunol. 2024, 24, 577–595. [Google Scholar] [CrossRef]
- Shamsuzzaman, M.; Dahal, R.H.; Kim, S.; Kim, J. Genome insight and probiotic potential of three novel species of the genus Corynebacterium. Front. Microbiol. 2023, 14, 1225282. [Google Scholar] [CrossRef]
- Vrakas, S.; Mountzouris, K.C.; Michalopoulos, G.; Karamanolis, G.; Papatheodoridis, G.; Tzathas, C.; Gazouli, M. Intestinal bacteria composition and translocation of bacteria in inflammatory bowel disease. PLoS ONE 2017, 12, e0170034. [Google Scholar] [CrossRef]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Emami, N.K.; Dalloul, R.A. Centennial Review: Recent developments in host-pathogen interactions during necrotic enteritis in poultry. Poult. Sci. 2021, 100, 101330. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, M.; Shojadoost, B.; Boodhoo, N.; Astill, J.; Taha-Abdelaziz, K.; Hodgins, D.C.; Kulkarni, R.R.; Sharif, S. Necrotic enteritis in chickens: A review of pathogenesis, immune responses and prevention, focusing on probiotics and vaccination. Anim. Health Res. Rev. 2021, 22, 147–162. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Zhao, Z.; Broadwater, C.; Tobin, I.; Liu, J.; Whitmore, M.; Zhang, G. Is Intestinal Microbiota Fully Restored After Chickens Have Recovered from Coccidiosis? Pathogens 2025, 14, 81. https://doi.org/10.3390/pathogens14010081
Guo J, Zhao Z, Broadwater C, Tobin I, Liu J, Whitmore M, Zhang G. Is Intestinal Microbiota Fully Restored After Chickens Have Recovered from Coccidiosis? Pathogens. 2025; 14(1):81. https://doi.org/10.3390/pathogens14010081
Chicago/Turabian StyleGuo, Jiaqing, Zijun Zhao, Chace Broadwater, Isabel Tobin, Jing Liu, Melanie Whitmore, and Guolong Zhang. 2025. "Is Intestinal Microbiota Fully Restored After Chickens Have Recovered from Coccidiosis?" Pathogens 14, no. 1: 81. https://doi.org/10.3390/pathogens14010081
APA StyleGuo, J., Zhao, Z., Broadwater, C., Tobin, I., Liu, J., Whitmore, M., & Zhang, G. (2025). Is Intestinal Microbiota Fully Restored After Chickens Have Recovered from Coccidiosis? Pathogens, 14(1), 81. https://doi.org/10.3390/pathogens14010081





