Earthworms Significantly Alter the Composition, Diversity, Abundance and Pathogen Load of Fungal Communities in Sewage Sludge from Different Urban Wastewater Treatment Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sewage Sludge and Sampling of Earthworm Casts
2.2. Amplification, Sequencing and Analysis of the ITS Region of rRNA Genes
2.3. Bioinformatic and Statistical Analysis
3. Results
3.1. Impact of Earthworm Gut Transit on the Composition of Fungal Communities in Sewage Sludge
3.2. Impact of Passage Through the Earthworm Gut on the Diversity of Fungal Communities in Sewage Sludge
3.3. Effect of Earthworm Gut Transit on Fungal Guilds and Pathogens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bolesta, W.; Głodniok, M.; Styszko, K. From sewage sludge to the soil-transfer of pharmaceuticals: A review. Int. J. Environ. Res. Public Health 2022, 19, 10246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, X.; Liu, H.; Dong, S.; Zhang, Z.; Wang, Z.; Li, J.; Nghiem, L.D.; Khan, S.J.; Wang, Q. Occurrence, fate, and remediation for per-and polyfluoroalkyl substances (PFAS) in sewage sludge: A comprehensive review. J. Hazard. Mater. 2024, 466, 133637. [Google Scholar] [CrossRef]
- Li, M.; Song, G.; Liu, R.; Huang, X.; Liu, H. Inactivation and risk control of pathogenic microorganisms in municipal sludge treatment: A review. Front. Environ. Sci. Eng. 2022, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 93, 1–10. [Google Scholar] [CrossRef]
- Corrin, T.; Rabeenthira, P.; Young, K.M.; Mathiyalagan, G.; Baumeister, A.; Pussegoda, K.; Waddell, L.A. A scoping review of human pathogens detected in untreated human wastewater and sludge. J. Water Health 2024, 22, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Hudcová, H.; Vymazal, J.; Rozkošný, M. Present restrictions of sewage sludge application in agriculture within the European Union. Soil Water Res. 2019, 14, 104–120. [Google Scholar] [CrossRef]
- World Health Organization. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. 2022. Available online: https://www.who.int/publications/i/item/9789240060241 (accessed on 12 January 2025).
- Zhang, H.; Feng, J.; Chen, S.; Li, B.; Sekar, R.; Zhao, Z.; Jia, J.; Wang, Y.; Kang, P. Disentangling the drivers of diversity and distribution of fungal community composition in wastewater treatment plants across spatial scales. Front. Microbiol. 2018, 9, 1291. [Google Scholar] [CrossRef]
- Yang, W.; Cai, C.; Guo, Y.; Wu, H.; Guo, Y.; Dai, X. Diversity and fate of human pathogenic bacteria, fungi, protozoa, and viruses in full-scale sludge treatment plants. J. Clean. Prod. 2022, 380, 134990. [Google Scholar] [CrossRef]
- Domínguez, J.; Aira, M.; Crandall, K.A.; Pérez-Losada, M. Earthworms drastically change fungal and bacterial communities during vermicomposting of sewage sludge. Sci. Rep. 2021, 11, 15556. [Google Scholar] [CrossRef]
- Dume, B.; Hanc, A.; Svehla, P.; Michal, P.; Chane, A.D.; Nigussie, A. Composting and vermicomposting of sewage sludge at various C/N ratios: Technological feasibility and end-product quality. Ecotoxicol. Environ. Saf. 2023, 263, 115255. [Google Scholar] [CrossRef]
- Swati, A.; Hait, S. A comprehensive review of the fate of pathogens during vermicomposting of organic wastes. J. Environ. Qual. 2018, 47, 16–29. [Google Scholar] [CrossRef]
- Lei, X.; Cui, G.; Sun, H.; Hou, S.; Deng, H.; Li, B.; Yang, Z.; Xu, Q.; Huo, X.; Cai, J. How do earthworms affect the pathway of sludge bio-stabilization via vermicomposting? Sci. Total Environ. 2024, 916, 170411. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhang, T.; Tian, G.; Zhang, L.; Bian, B. Pilot-scale vermicomposting of sewage sludge mixed with mature vermicompost using earthworm reactor of frame composite structure. Sci. Total Environ. 2021, 767, 144217. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, H.; Peng, S.; Wang, Z.; Cai, S.; Chen, Z.; Yang, B.; Yang, P.; Wang, D.; Guo, J.; et al. Vermicomposting preferably alters fungal communities in wasted activated sludge and promotes the production of plant growth-promoting biostimulants in the vermicompost. Chem. Eng. J. 2024, 495, 153232. [Google Scholar] [CrossRef]
- Gómez-Roel, A.; Aira, M.; Domínguez, J. Vermicomposting enhances microbial detoxification of sewage sludge, enabling potential application of the treated product in agroecosystems. Appl. Sci. 2024, 14, 7894. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Pedersen, T.L. Patchwork: The Composer of Plots, Version 1.3.0; R Core Team: Vienna, Austria, 2024. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, Version 2.6.8; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests, version 0.7.2; R Core Team: Vienna, Austria, 2023. [Google Scholar]
- van den Brand, T. ggh4x: Hacks for ‘ggplot2’. R Package Version 0.2.8.9000. 2024. Available online: https://teunbrand.github.io/ggh4x/ (accessed on 12 January 2025).
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2023. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Furneaux, B.; Song, Z.; FUNGuildR: Look Up Guild Information for Fungi. R Package Version 0.2.0.9000. 2025. Available online: https://github.com/brendanf/FUNGuildR (accessed on 12 January 2025).
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Assress, H.A.; Selvarajan, R.; Nyoni, H.; Ntushelo, K.; Mamba, B.B.; Msagati, T.A.M. Diversity, co-occurrence and implications of fungal communities in wastewater treatment plants. Sci. Rep. 2019, 9, 14056. [Google Scholar] [CrossRef]
- Niu, L.; Li, Y.; Xu, L.; Wang, P.; Zhang, W.; Wang, C.; Cai, W.; Wang, L. Ignored fungal community in activated sludge wastewater treatment plants: Diversity and altitudinal characteristics. Environ. Sci. Pollut. Res. 2017, 24, 4185–4193. [Google Scholar] [CrossRef]
- Lejoly, J.D.M.; Quideau, S.A.; Laganière, J.; Karst, J.; Martineau, C.; Samad, A. Earthworm cast microbiomes differ across soil types in northern forests. Appl. Soil Ecol. 2024, 200, 105466. [Google Scholar] [CrossRef]
- Yan, Z.Z.; Hu, H.W.; Xiong, C.; Peleg, A.Y.; Chen, Q.L.; Sáez-Sandino, T.; Maestre, F.; Delgado-Baquerizo, M.; Singh, B.K. Environmental microbiome, human fungal pathogens, and antimicrobial resistance. Trends Microbiol. 2025, 33, 112–129. [Google Scholar] [CrossRef]
- Goberna, M.; Simón, P.; Hernández, M.T.; García, C. Prokaryotic communities and potential pathogens in sewage sludge: Response to wastewaster origin, loading rate and treatment technology. Sci. Total Environ. 2018, 615, 360–368. [Google Scholar] [CrossRef]
- Byzov, B.A.; Khomyakov, N.V.; Kharin, S.A.; Kurakov, A.V. Fate of soil bacteria and fungi in the gut of earthworms. Eur. J. Soil Biol. 2007, 43, S149–S156. [Google Scholar] [CrossRef]
- Sampedro, L.; Jeannotte, R.; Whalen, J.K. Trophic transfer of fatty acids from gut microbiota to the earthworm Lumbricus terrestris L. Soil Biol. Biochem. 2006, 38, 2188–2198. [Google Scholar] [CrossRef]
- Li, N.; Wang, C.; Li, X.; Liu, M. Effects of earthworms and arbuscular mycorrhizal fungi on preventing Fusarium oxysporum infection in the strawberry plant. Plant Soil 2019, 443, 139–153. [Google Scholar] [CrossRef]
- Gudeta, K.; Bhagat, A.; Julka, J.M.; Sinha, R.; Verma, R.; Kumar, A.; Kumari, S.; Ameen, F.; Bhat, S.A.; Amarowicz, R.; et al. Vermicompost and Its Derivatives against Phytopathogenic Fungi in the Soil: A Review. Horticulturae 2022, 8, 311. [Google Scholar] [CrossRef]
- Parthasarathi, K.; Ranganathan, L.S.; Anandi, V.; Zeyer, J. Diversity of microflora in the gut and casts of tropical composting earthworms reared on different substrates. J. Environ. Biol. 2007, 28, 87–97. [Google Scholar]
- Cao, B.; Lv, H.; Nie, T.; Ma, Y.; Jiang, Z.; Hu, Y.; Yang, C.; Zhang, Y. Combined toxicity of acetochlor and metribuzin on earthworm Eisenia fetida: Survival, oxidative stress responses and joint effect. Appl. Soil Ecol. 2022, 178, 104583. [Google Scholar] [CrossRef]
- Junior, S.F.S.; da Silva, E.O.; de Farias Araujo, G.; Soares, L.O.S.; Parente, C.E.T.; Malm, O.; Saggioro, E.M.; Correia, F.V. Antioxidant system alterations and biological health status of earthworms following long-term exposure to antibiotic-contaminated poultry litter. Environ. Sci. Pollut. Res. Int. 2022, 29, 23607–23618. [Google Scholar] [CrossRef]
- Forsell, V.; Saartama, V.; Turja, R.; Haimi, J.; Selonen, S. Reproduction, growth and oxidative stress in earthworm Eisenia andrei exposed to conventional and biodegradable mulching film microplastics. Sci. Total Environ. 2024, 948, 174667. [Google Scholar] [CrossRef]
- Muri, J.; Kopf, M. Redox regulation of immunometabolism. Nat. Rev. Immunol. 2021, 21, 363–381. [Google Scholar] [CrossRef]
- Seyedmousavi, S.; Bosco, S.M.G.; de Hoog, S.; Ebel, F.; Elad, D.; Gomes, R.R.; Jacobsen, I.D.; Jensen, H.E.; Martel, A.; Mignon, B.; et al. Fungal infections in animals: A patchwork of different situations. Med. Mycol. 2018, 56 (Suppl. 1), 165–187. [Google Scholar] [CrossRef] [PubMed]

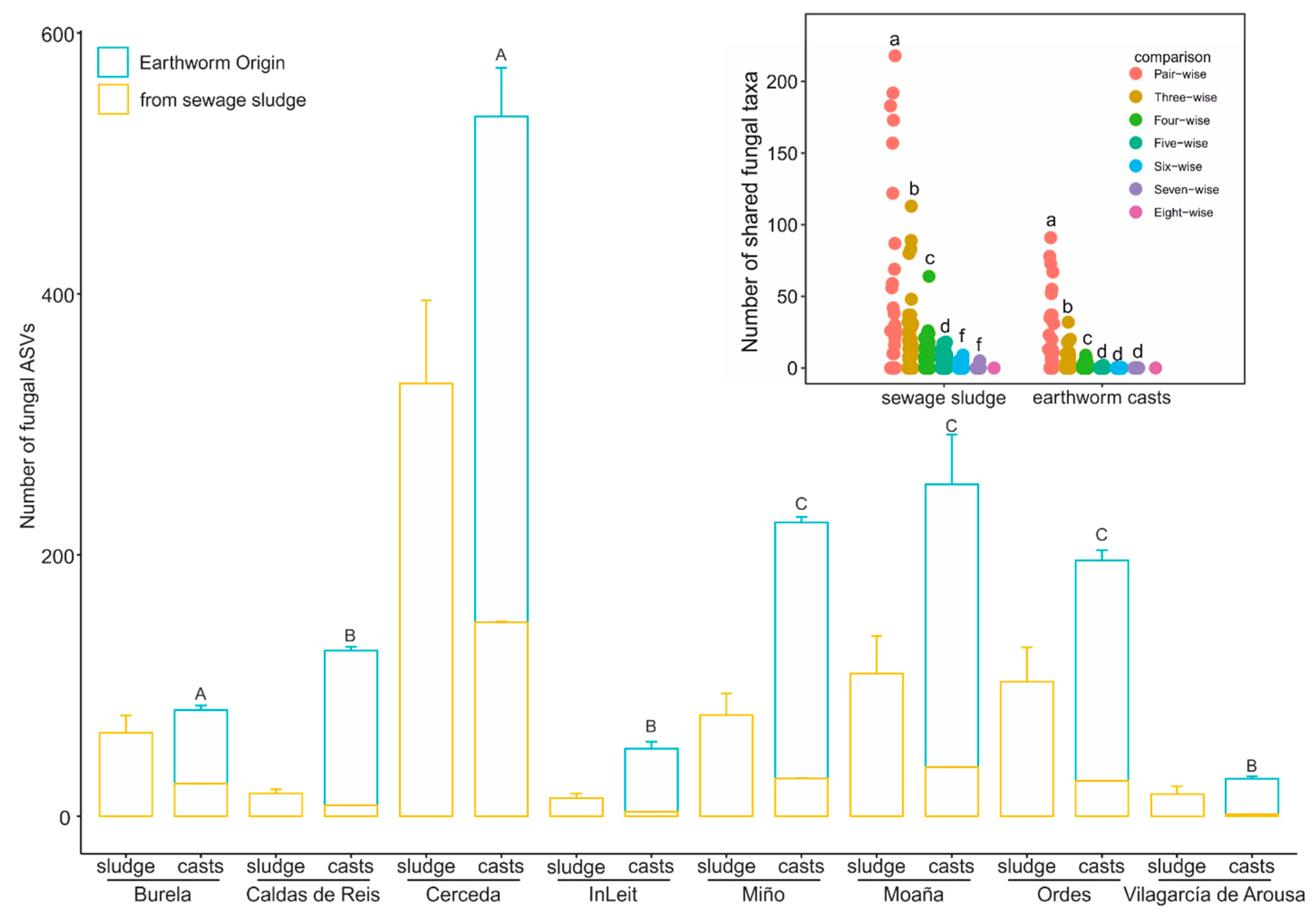
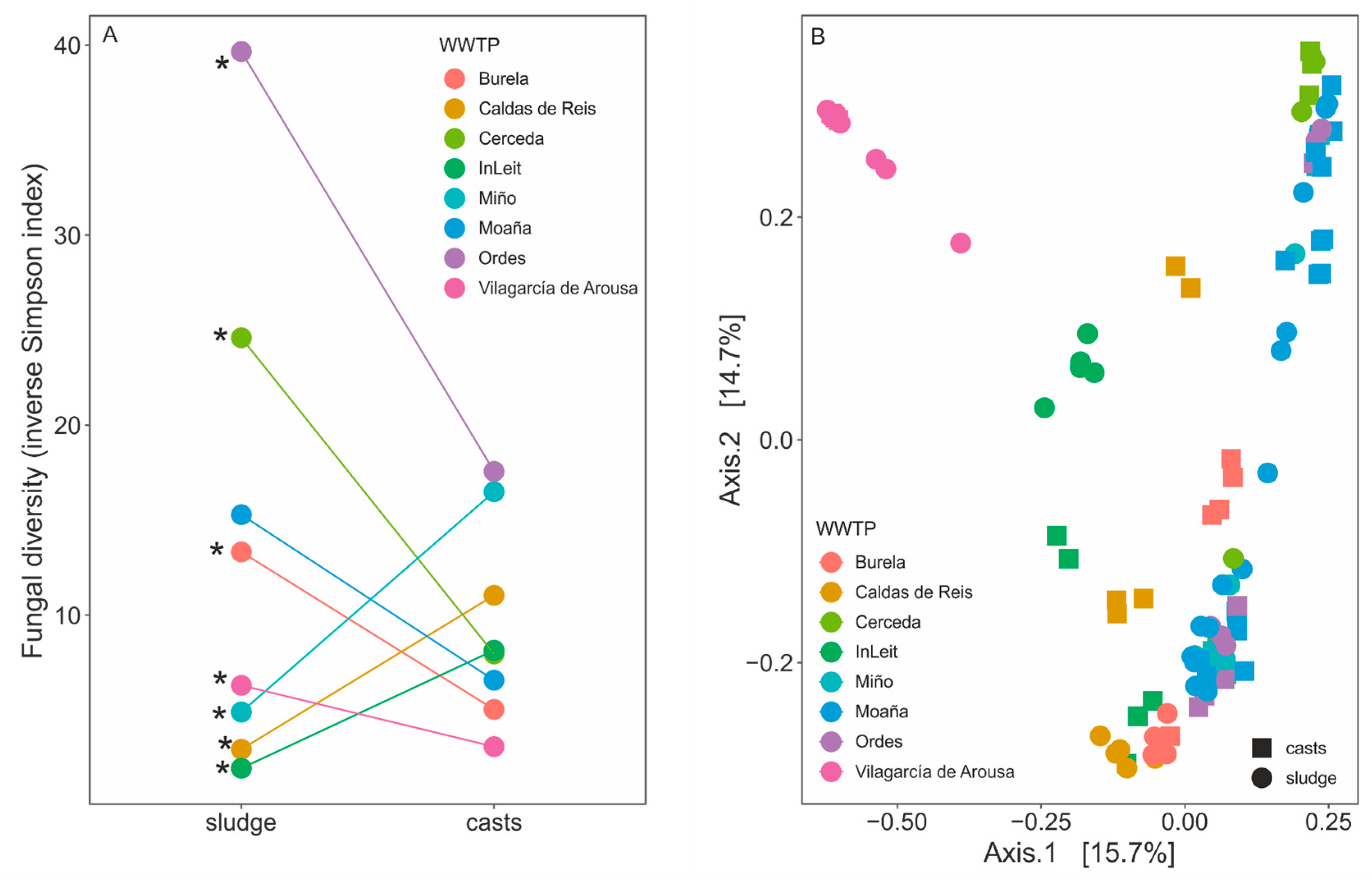

| Burela | Caldas de Reis | Cerceda | InLeit | Miño | Moaña | Ordes | Vilagarcía de Arousa | ||
|---|---|---|---|---|---|---|---|---|---|
| Aphelidiomycota | sludge | 0 ± 0 | 0 ± 0 | 0.004 ± 0.002 | 0 ± 0 | 0 ± 0 | 0.002 ± 0.002 | 0.014 ± 0.014 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Ascomycota | sludge | 48.08 ± 6.85 | 34.68 ± 3.75 | 56.62 ± 1.91 | 95.72 ± 0.60 | 20.12 ± 1.99 | 26.08 ± 6.08 | 52.80 ± 1.34 | 7.69 ± 2.79 |
| casts | 6.64 ± 0.17 | 92.454 ± 1.42 | 36.89 ± 1.36 | 88.11 ± 4.60 | 49.56 ± 5.29 | 23.6 ± 2.13 | 62.01 ± 2.26 | 2.64 ± 0.22 | |
| Basidiobolomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.01 ± 0.01 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Basidiomycota | sludge | 51.91 ± 6.85 | 65.31 ± 3.75 | 43.16 ± 1.93 | 3.93 ± 0.55 | 79.70 ± 2.01 | 73.63 ± 6.08 | 46.97 ± 1.30 | 85.68 ± 2.95 |
| casts | 91.97 ± 0.11 | 6.83 ± 1.4 | 62.94 ± 1.36 | 11.40 ± 4.58 | 49.45 ± 5.32 | 76.12 ± 2.16 | 36.63 ± 2.21 | 70.247 ± 1.69 | |
| Blastocladiomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.33 ± 0.15 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0.002 ± 0.002 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Chytridiomycota | sludge | 0 ± 0 | 0 ± 0 | 0.012 ± 0.002 | 0 ± 0 | 0 ± 0 | 0.004 ± 0.003 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0.002 ± 0.002 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Entorrhizomycota | sludge | 0 ± 0 | 0 ± 0 | 0.001 ± 0.001 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Kickxellomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.024 ± 0.024 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Monoblepharomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.001 ± 0.001 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Mortierellomycota | sludge | 0.002 ± 0.002 | 0 ± 0 | 0.067 ± 0.01 | 0 ± 0 | 0.14 ± 0.02 | 0.25 ± 0.10 | 0.12 ± 0.03 | 6.52 ± 3.79 |
| casts | 1.38 ± 0.07 | 0.705 ± 0.09 | 0.05 ± 0.01 | 0.47 ± 0.14 | 0.83 ± 0.10 | 0.20 ± 0.03 | 1.29 ± 0.06 | 27.10 ± 1.55 | |
| Mucoromycota | sludge | 0 ± 0 | 0 ± 0 | 0.003 ± 0.002 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.08 ± 0.04 |
| casts | 0 ± 0 | 0.002 ± 0.001 | 0.005 ± 0.004 | 0 ± 0 | 0 ± 0 | 0.001 ± 0.001 | 0 ± 0 | 0.002 ± 0.002 | |
| Neocallimastigomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0.003 ± 0.003 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Olpidiomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.006 ± 0.006 | 0.002 ± 0.002 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Rozellomycota | sludge | 0.001 ± 0.001 | 0 ± 0 | 0.12 ± 0.02 | 0 ± 0 | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.08 ± 0.03 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0.10 ± 0.02 | 0 ± 0 | 0.14 ± 0.03 | 0.02 ± 0.01 | 0.06 ± 0.01 | 0 ± 0 | |
| Zoopagomycota | sludge | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| casts | 0 ± 0 | 0.002 ± 0.002 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Burela | Caldas de Reis | Cerceda | InLeit | Miño | Moaña | Ordes | Vilagarcía de Arousa | ||
|---|---|---|---|---|---|---|---|---|---|
| Aspergillus | sludge | 13 ± 4 | 0 ± 0 * | 1225 ± 148 | 1.2 ± 1.2 | 71 ± 8 * | 304 ± 112 * | 157 ± 23 * | 0 ± 0 |
| casts | 28 ± 13 | 44 ± 9 | 1813 ± 359 | 7 ± 3 | 242 ± 14 | 2100 ± 751 | 845 ± 137 | 0 ± 0 | |
| Candida | sludge | 545 ± 186 | 1.4 ± 1.4 | 77 ± 17 | 3 ± 3 | 74 ± 6 * | 290 ± 110 | 176 ± 31 | 54 ± 28 |
| casts | 18 ± 9 | 0 ± 0 | 107 ± 30 | 2 ± 1 | 242 ± 23 | 389 ± 160 | 181 ± 50 | 0.6 ± 0.6 | |
| Cryptococcus | sludge | 1 ± 1 | 0 ± 0 | 208 ± 16 * | 0 ± 0 | 10 ± 3 | 62 ± 24 | 12 ± 5 | 2 ± 1 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 1 ± 1 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Fusarium | sludge | 0 ± 0 | 0 ± 0 | 113 ± 18 * | 0 ± 0 | 3 ± 2 * | 3 ± 2 | 8 ± 2 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0.4 ± 0.4 | 0 ± 0 | 12 ± 4 | 10 ± 5 | 5 ± 3 | 0 ± 0 | |
| Penicillium | sludge | 5 ± 2 | 0 ± 0 | 365 ± 66 | 0 ± 0 | 53 ± 11 * | 123 ± 44 | 148 ± 40 | 0 ± 0 |
| casts | 0 ± 0 | 27 ± 9. | 445 ± 95 | 4 ± 2 | 161 ± 23 | 255 ± 104 | 107 ± 19 | 0 ± 0 | |
| Rhodotorula | sludge | 4291 ± 957 * | 1 ± 1 | 759 ± 137 * | 28 ± 6 * | 112 ± 7 * | 507 ± 195 | 177 ± 17 | 179 ± 77 |
| casts | 410 ± 32 | 0 ± 0 | 188 ± 32 | 4 ± 2 | 536 ± 44 | 1307 ± 534 | 214 ± 45 | 0 ± 0 | |
| Scedosporium | sludge | 24 ± 8 * | 5 ± 2 * | 514 ± 111 * | 1.2 ± 1.2 * | 8 ± 3 * | 47 ± 10 * | 52 ± 6 * | 0 ± 0 |
| casts | 1.2 ± 1.2 | 383 ± 55 | 278 ± 56 | 102 ± 15 | 15 ± 5 | 121 ± 16.231 | 68 ± 5 | 0 ± 0 | |
| Talaromyces | sludge | 2 ± 1 | 0 ± 0 | 318 ± 66 * | 0 ± 0 | 22 ± 43 | 71 ± 19 | 53 ± 10 | 0 ± 0 |
| casts | 12 ± 10 | 2 ± 1 | 492 ± 70 | 5 ± 4 | 66 ± 14 | 170 ± 61 | 212 ± 57 | 0 ± 0 | |
| Trichosporon | sludge | 45 ± 8 * | 0 ± 0 | 7± 7 | 21 ± 16 | 2 ± 2 | 7 ± 4 | 1 ± 1 | 0 ± 0 |
| casts | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aira, M.; Gómez-Roel, A.; Domínguez, J. Earthworms Significantly Alter the Composition, Diversity, Abundance and Pathogen Load of Fungal Communities in Sewage Sludge from Different Urban Wastewater Treatment Plants. Pathogens 2025, 14, 409. https://doi.org/10.3390/pathogens14050409
Aira M, Gómez-Roel A, Domínguez J. Earthworms Significantly Alter the Composition, Diversity, Abundance and Pathogen Load of Fungal Communities in Sewage Sludge from Different Urban Wastewater Treatment Plants. Pathogens. 2025; 14(5):409. https://doi.org/10.3390/pathogens14050409
Chicago/Turabian StyleAira, Manuel, Ana Gómez-Roel, and Jorge Domínguez. 2025. "Earthworms Significantly Alter the Composition, Diversity, Abundance and Pathogen Load of Fungal Communities in Sewage Sludge from Different Urban Wastewater Treatment Plants" Pathogens 14, no. 5: 409. https://doi.org/10.3390/pathogens14050409
APA StyleAira, M., Gómez-Roel, A., & Domínguez, J. (2025). Earthworms Significantly Alter the Composition, Diversity, Abundance and Pathogen Load of Fungal Communities in Sewage Sludge from Different Urban Wastewater Treatment Plants. Pathogens, 14(5), 409. https://doi.org/10.3390/pathogens14050409






