Test Performance Study on qPCR Assays for Detection of Phyllosticta citricarpa
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Test Selection
2.3. Real-Time PCR
2.4. TPS Participants
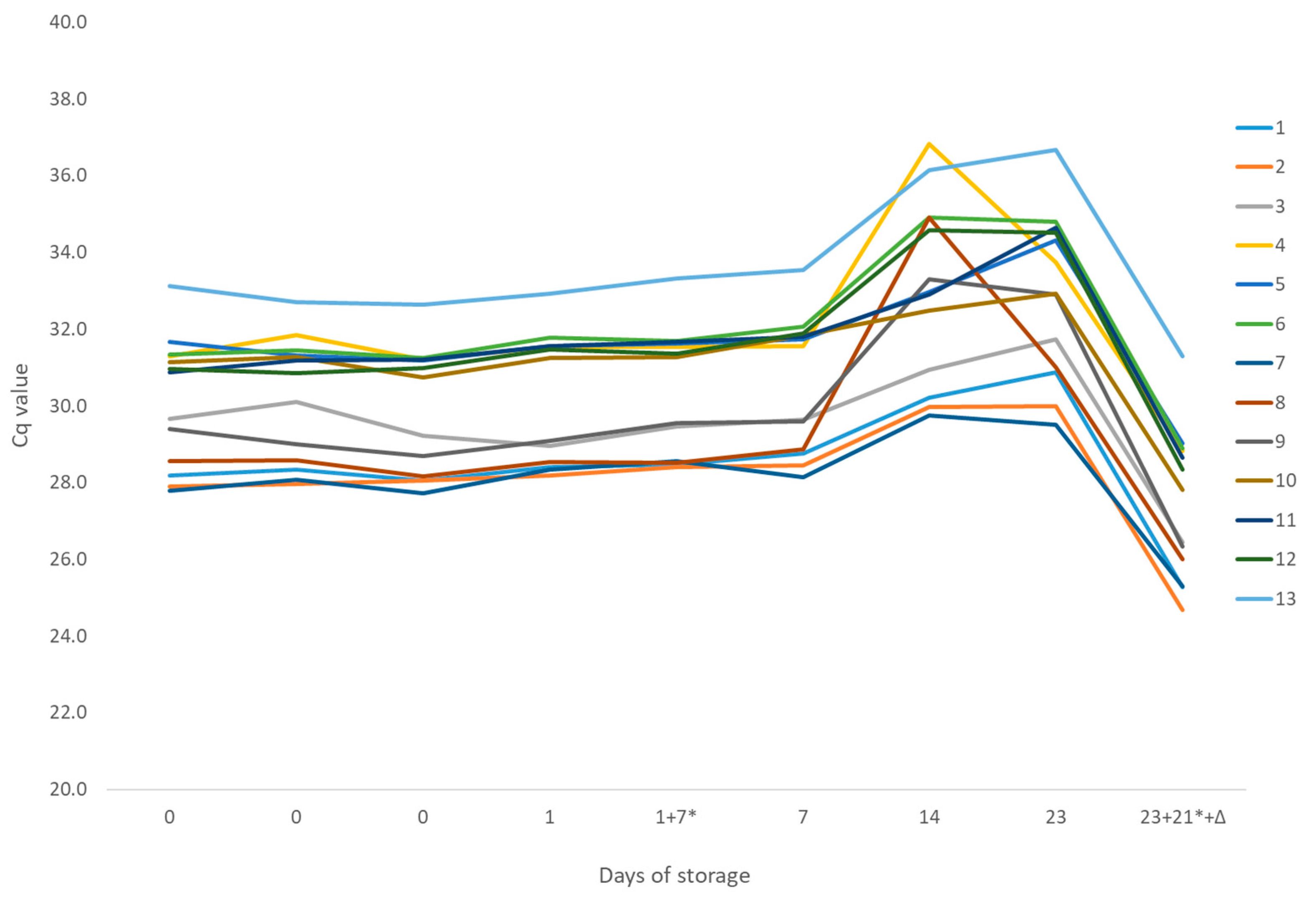
2.5. Homogeneity and Stability
3. Results
3.1. Preliminary Evaluation of Samples and Primers and Probe Mixes
3.2. Data on Test Performance Study Execution by Participants
3.3. Repeatability and Reproducibility
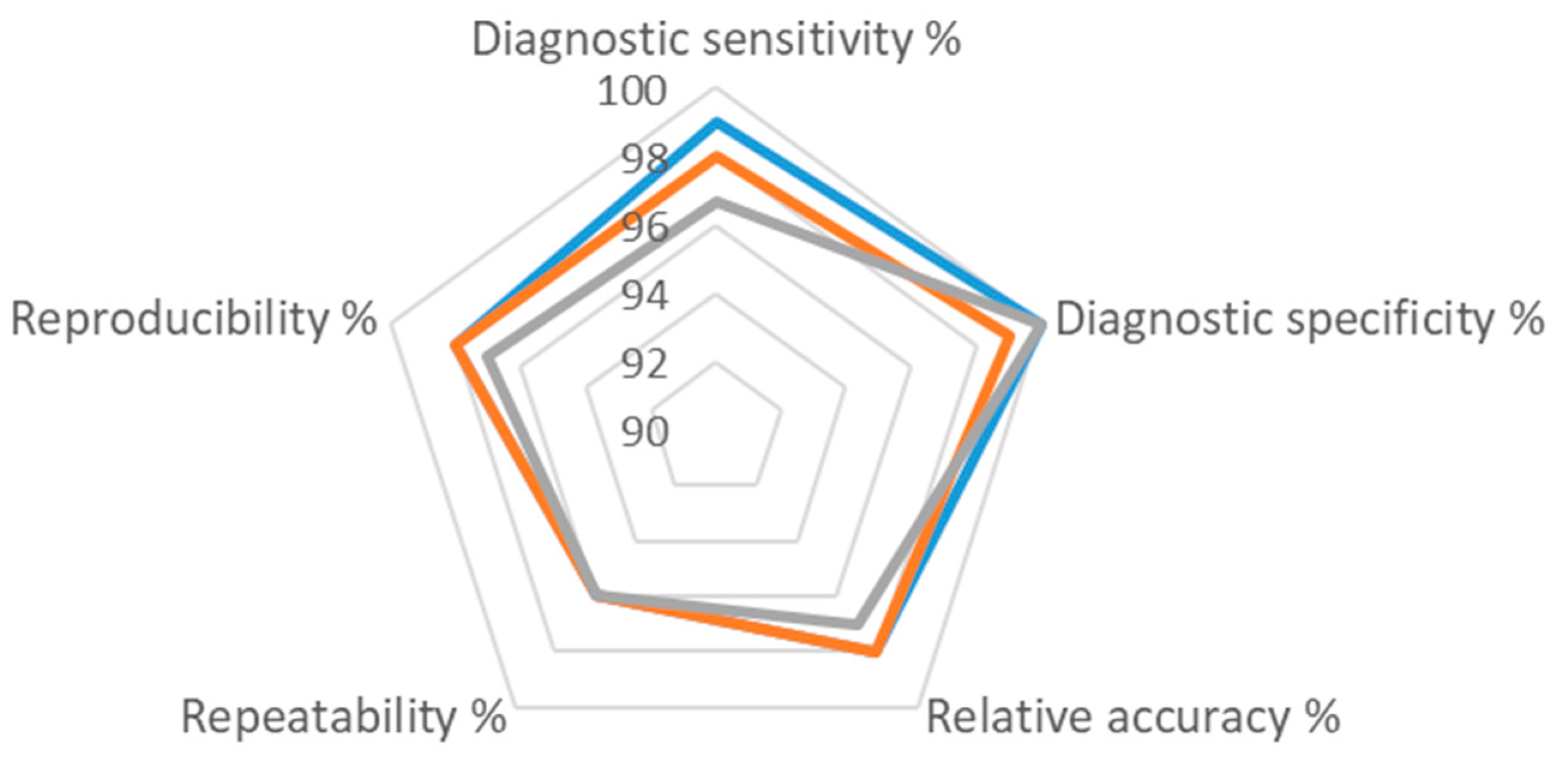
3.4. Evaluation of Other Assay Performance Criteria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parnell, S.; Schenk, M.; Schrader, G.; Vicent, A.; Delbianco, A.; Vos, S. Pest Survey Card on Phyllosticta citricarpa. EFSA Support. Publ. 2020, 17, 1863E. [Google Scholar] [CrossRef]
- Kotzé, J.M. Epidemiology and Control of Citrus Black Spot in South Africa. Plant Dis. 1981, 65, 945. [Google Scholar] [CrossRef]
- Boughalleb-M’Hamdi, N.; Fathallah, A.; Benfradj, N.; Mahmoud, S.B.; Ali, A.B.H.; Medhioub, L.; Jaouadi, I.; Huber, J.; Jeandel, C.; Ioos, R. First Report of Citrus Black Spot Disease Caused by Phyllosticta citricarpa on Citrus limon and C. sinensis in Tunisia. New Dis. Rep. 2020, 41, 8. [Google Scholar] [CrossRef]
- REGULATION (EU) 2019/1702; European Commission Delegated Regulation (EU) 2019/1702 of 1 August 2019 Supplementing Regulation (EU) 2016/2031 of the European Parliament and of the Council by Establishing the List of Priority Pests. Off. J. Eur. Union, Brussels, Bulgium, 2019. Available online: http://data.europa.eu/eli/reg_del/2019/1702/oj/eng (accessed on 18 April 2025).
- EPPO. PM 7/017 (3) Phyllosticta citricarpa (Formerly Guignardia citricarpa). EPPO Bull. 2020, 50, 440–461. [Google Scholar] [CrossRef]
- Van Gent-Pelzer, M.P.E.; Van Brouwershaven, I.R.; Kox, L.F.F.; Bonants, P.J.M. A TaqMan PCR Method for Routine Diagnosis of the Quarantine Fungus Guignardia citricarpa on Citrus Fruit. J. Phytopathol. 2007, 155, 357–363. [Google Scholar] [CrossRef]
- Ioos, R.; Puertolas, A.; Renault, C.; Ndiaye, A.; Cerf-Wendling, I.; Hubert, J.; Wang, W.; Jiao, C.; Li, H.; Armengol, J.; et al. Harnessing the Power of Comparative Genomics to Support the Distinction of Sister Species within Phyllosticta and Development of Highly Specific Detection of Phyllosticta citricarpa Causing Citrus Black Spot by Real-Time PCR. PeerJ 2023, 11, e16354. [Google Scholar] [CrossRef] [PubMed]
- Schirmacher, A.M.; Tomlinson, J.A.; Barnes, A.V.; Barton, V.C. Species-specific Real-time PCR for Diagnosis of Phyllosticta citricarpa on Citrus Species. EPPO Bull. 2019, 49, 306–313. [Google Scholar] [CrossRef]
- CREA. Validation Process of the Real Time PCR for the Identification of Phyllosticta citricarpa (van Gent-Pelzer et al., 2007). Available online: https://dc.eppo.int/validation_data/dwvalidation?id=140 (accessed on 18 April 2025).
- Ahmed, Y.; Hussein, A.; Hubert, J.; Fourrier-Jeandel, C.; Aguayo, J.; Ioos, R. New Multiplex Conventional PCR and Quadruplex Real-Time PCR Assays for One-Tube Detection of Phyllosticta citricarpa, Elsinoë fawcettii, Elsinoë australis, and Pseudocercospora angolensis in Citrus: Development and Validation. Appl. Microbiol. Biotechnol. 2020, 104, 9363–9385. [Google Scholar] [CrossRef] [PubMed]
- Zajc, J.; Kogej, Z.; Fišer, S.; Gostinčar, C.; Vicent, A.; Galvañ Domenech, A.; Ricconi, L.; Boonham, N.; Ravnikar, M.; Kogovšek, P.; et al. Highly Specific qPCR and Amplicon Sequencing Method for Detection of Quarantine Citrus Pathogen Phyllosticta citricarpa Applicable for Air Samples. Plant Pathol. 2023, 72, 548–563. [Google Scholar] [CrossRef]
- van Ingen-Buijs, V.A.; van Westerhoven, A.C.; Skiadas, P.; Zuijdgeest, X.C.L.; Haridas, S.; Daum, C.; Duffy, K.; Guo, J.; Hundley, H.; LaButti, K.; et al. Phyllosticta paracitricarpa Is Synonymous with the EU Quarantine Fungus P. citricarpa Based on Phylogenomic Analyses. Fungal Genet. Biol. 2024, 175, 103925. [Google Scholar] [CrossRef] [PubMed]
- Vučurović, A.; Mehle, N.; Anthoine, G.; Dreo, T.; Ravnikar, M. (Eds.) Critical Points for the Organisation of Test Performance Studies in Microbiology: Plant Pathogens as a Case Study; Plant Pathology in the 21st Century; Springer International Publishing: Cham, Switzerland, 2022; Volume 12, ISBN 978-3-030-99810-3. [Google Scholar]
- EPPO. PM 7/129 (2) DNA Barcoding as an Identification Tool for a Number of Regulated Pests. EPPO Bull. 2021, 51, 100–143. [Google Scholar] [CrossRef]
- Hu, J.; Johnson, E.G.; Wang, N.-Y.Y.; Davoglio, T.; Dewdney, M.M. qPCR Quantification of Pathogenic Guignardia citricarpa and Nonpathogenic G. mangiferae in Citrus. Plant Dis. 2014, 98, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Kachur, S.; Dwan, M.G.; Abraham, A.G.; Aziz, M.; Hsueh, P.R.; Huang, Y.T.; Busch, J.D.; Lamit, L.J.; Gehring, C.A.; et al. FungiQuant: A Broad-Coverage Fungal Quantitative Real-Time PCR Assay. BMC Microbiol. 2012, 12, 255. [Google Scholar] [CrossRef] [PubMed]
- EPPO. PM 7/122 (2) Guidelines for the Organization of Interlaboratory Comparisons by Plant Pest Diagnostic Laboratories. EPPO Bull. 2022, 52, 604–618. [Google Scholar] [CrossRef]
- EPPO. PM 7/98 (4) Specific Requirements for Laboratories Preparing Accreditation for a Plant Pest Diagnostic Activity. EPPO Bull. 2019, 49, 530–563. [Google Scholar] [CrossRef]
- Langton, S.D.; Chevennement, R.; Nagelkerke, N.; Lombard, B. Analysing Collaborative Trials for Qualitative Microbiological Methods: Accordance and Concordance. Int. J. Food Microbiol. 2002, 79, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Lebas, B.; Chabirand, A.; Chappé, A.-M.; Dreo, T.; Faggioli, F.; Harrison, C.; Macarthur, R.; Mehle, N.; Mezzalama, M.; et al. Guidelines for Improving Statistical Analyses of Validation Datasets for Plant Pest Diagnostic Tests. EPPO Bull. 2022, 52, 419–433. [Google Scholar] [CrossRef]
- Ioos, R.; Fourrier, C.; Iancu, G.; Gordon, T.R. Sensitive Detection of Fusarium circinatum in Pine Seed by Combining an Enrichment Procedure with a Real-Time Polymerase Chain Reaction Using Dual-Labeled Probe Chemistry. Phytopathology 2009, 99, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.L.D.; Crichton, E.M.; Allison, M.J.; Dema, A.H.; Bonderud, M.T.; Helbing, C.C. Effects of Storage Conditions on the Stability of qPCR Reagents: Implications for Environmental DNA Detection. BMC Res. Notes 2024, 17, 199. [Google Scholar] [CrossRef] [PubMed]
- Ioos, R.; Aloi, F.; Piškur, B.; Guinet, C.; Mullett, M.; Berbegal, M.; Bragança, H.; Cacciola, S.O.; Oskay, F.; Cornejo, C.; et al. Transferability of PCR-Based Diagnostic Protocols: An International Collaborative Case Study Assessing Protocols Targeting the Quarantine Pine Pathogen Fusarium circinatum. Sci. Rep. 2019, 9, 8195. [Google Scholar] [CrossRef] [PubMed]
| Fungi | Isolate | Collection/Source | Origin | Collection Year | Original Host Plant | Accession Numbers |
|---|---|---|---|---|---|---|
| P. citricarpa | IVIA115 | IVIA | Argentina | 2014 | C. limon | PV495870 a, PV527763 c |
| P. citricarpa | IIA-GC003NA | IVIA | Angola | 2016 | C. sinensis | MF693405 a |
| P. citricarpa | CBS 127455 | CBS/CREA | Australia | 1973 | C. sinensis | KF206305 a |
| P. citriasiana | IVIA120 | IVIA | China | 2017 | C. maxima | PV495871 a, PV527764 c |
| P. paracitricarpa | CBS 141357 | CBS/CREA | Greece | 2017 | C. limon | JBBWUC000000000 b |
| P. capitalensis | CBS 120490 | CBS/CREA | USA | 1968 | C. paradisi | FJ538338 a, FJ538396 c |
| Plenodomus tracheiphilus | ER 2221 | CREA | Italy | Na | C. sinensis | Na |
| Colletotrichum sp. | ER 2100 | CREA | Italy | Na | C. maxima | Na |
| Assay/PPS Mix | Primer and Probe Sequence | Final Concentration in Reaction | Reference | |
|---|---|---|---|---|
| PC | FP | GGTGATGGAAGGGAGGCCT | 0.3 µM | [5,6] |
| RP | GCAACATGGTAGATACACAAGGGT | 0.3 µM | ||
| Probe | FAM-AAAAAGCCGCCCGACCTACCTTCA-TAMRA | 0.2 µM | ||
| Pc-TEF1 | FP | GAAGGTCAGTTGCCTCACACTTT | 0.3 µM | [11] |
| RP | GTCATATAACCGAGCGCCAAA | 0.3 µM | ||
| Probe | HEX-TTGCGCCTCCACTTG-MGBNFQ | 0.2 µM | ||
| FQ | FP | GGRAAACTCACCAGGTCCAG | 0.9 µM | [16] |
| RP | GSWCTATCCCCAKCACGA | 0.9 µM | ||
| Probe | FAM-TGGTGCATGGCCGTT-MGBNFQ | 0.25 µM |
| Participant | Country |
|---|---|
| Agricultural Institute of Slovenia, Ljubljana | Slovenia |
| Benaki Phytopathological Institute, Laboratory of Mycology, Kifissia | Greece |
| Centro Attività Vivaistiche, Faenza | Italy |
| Centro de Citricultura Sylvio Moreira, Instituto Agronômico, Cordeiropolis, Sao Paulo | Brasil |
| Council for Agricultural Research and Economics (CREA-DC), Plant Protection and Certification, Rome | Italy |
| Directorate of plant protection central research institute, Mycology Laboratory, Ankara | Turkey |
| Fera Science Ltd., York | United Kingdom |
| French Agency for Food, Environmental and Occupational Health & Safety (ANSES), Plant Health Laboratory, Nancy | France |
| General directorate of plant Health and input control (DGSVCIA), Quarantine Laboratory, Tunis | Tunisie |
| Laboratory of Plant Protection Service Lombardy Region, Vertemate con Minoprio | Italy |
| Main Inspectorate of Plant Health and Seed Inspection, Central Laboratory, Phytosanitary Reference Laboratory, Torun | Poland |
| National Institute of Biology, Ljubljana | Slovenia |
| Plant Health Laboratory, DAFM, Celbridge | Ireland |
| State Plant Protection Service of the Republic of Latvia/The National Phytosanitary Laboratory, Riga | Latvia |
| L-01 | L-02 | L-03 | L-04 | L-05 | L-06 | L-07 | L-08-A | L-08-B | L-09 | L-10 | L-11 | L-12 | L-13 | L-14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FQ | 89% | 67% | 100% | 100% | 100% | 94% | 94% | 94% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| PC | 100% | 100% | 100% | 100% | 89% | 83% | 94% | 94% | 100% | 100% | 100% | 94% | 94% | 100% | 94% |
| Pc-TEF1 | 94% | 100% | 100% | 100% | 94% | 94% | NA | 72% | 94% | 100% | 100% | 100% | 100% | 100% | 100% |
| Sample No. | Isolate | Host Background DNA | Fungi | FQ | PC | Pc-TEF1 |
|---|---|---|---|---|---|---|
| 1 | IVIA115 | Citrus limon | P. citricarpa | 100% | 100% | 100% |
| 2 | IVIA115 | Citrus sinensis | P. citricarpa | 100% | 100% | 100% |
| 3 | IVIA115 | Citrus maxima | P. citricarpa | 93% | 100% | 100% |
| 4 | IVIA115 | C. limon | P. citricarpa | 100% | 100% | 93% |
| 5 | IVIA115 | C. sinensis | P. citricarpa | 100% | 100% | 100% |
| 6 | IVIA115 | C. maxima | P. citricarpa | 93% | 100% | 100% |
| 7 | IIA-GC003NA | C. limon | P. citricarpa | 100% | 100% | 100% |
| 8 | IIA-GC003NA | C. sinensis | P. citricarpa | 100% | 100% | 100% |
| 9 | IIA-GC003NA | C. maxima | P. citricarpa | 93% | 100% | 100% |
| 10 | IIA-GC003NA | C. limon | P. citricarpa | 100% | 100% | 93% |
| 11 | IIA-GC003NA | C. sinensis | P. citricarpa | 100% | 100% | 100% |
| 12 | IIA-GC003NA | C. maxima | P. citricarpa | 100% | 100% | 93% |
| 13 | CBS 127455 | / | P. citricarpa | 100% | 100% | 100% |
| 14 | IVIA120 | C. maxima | P. citriasiana | 73% | 73% | 93% |
| 15 | CBS141357 | C. sinensis | P. paracitricarpa | 100% | 100% | 100% |
| 16 | CBS 120490 | C. limon | P. capitalensis | 100% | 93% | 100% |
| 17 | ER2221 + ER2100 | C. limon | Plenodomus tracheiphilus + Colletotrichum sp. | 100% | 93% | 100% |
| 18 | C. limon + C. sinensis + C. maxima | / | 87% | 100% | 100% | |
| Average reproducibility | 97% | 98% | 98% | |||
| Diagnostic Parameter | FQ | PC | Pc-TEF1 |
|---|---|---|---|
| Total data sets | 15 | 15 | 14 |
| Expected positives | 270 | 225 | 182 |
| Total data points | 270 | 270 | 252 |
| INC | 0 | 3 | 0 |
| TN | 0 | 43 | 69 |
| FP | 0 | 0 | 1 |
| FN | 9 | 3 | 3 |
| TP | 261 | 221 | 179 |
| INC% | 0 | 1 | 0 |
| TN% | 0 | 16 | 27 |
| FP% | 0 | 0 | 0.4 |
| FN% | 3 | 1 | 1 |
| TP% | 97 | 82 | 71 |
| Concordant | 261 | 264 | 248 |
| Non-concordant | 9 | 6 | 4 |
| Concordant% | 97 | 98 | 98 |
| Non-concordant% | 3 | 2 | 2 |
| Diagnostic sensitivity% | 97 | 99 | 98 |
| Diagnostic specificity% | na | 100 | 99 |
| False positive rate% | na | 0 | 1 |
| False negative rate% | 3 | 1 | 2 |
| Relative accuracy% | 97 | 98 | 98 |
| Power% | 97 | 98 | 98 |
| Positive predictive value% | 100 | 100 | 99 |
| Negative predictive value% | na | 96 | 96 |
| Diagnostic odds ratio | na | na | 4117 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakomin, T.; Žunič, J.Z.; Kogovšek, P. Test Performance Study on qPCR Assays for Detection of Phyllosticta citricarpa. Pathogens 2025, 14, 413. https://doi.org/10.3390/pathogens14050413
Jakomin T, Žunič JZ, Kogovšek P. Test Performance Study on qPCR Assays for Detection of Phyllosticta citricarpa. Pathogens. 2025; 14(5):413. https://doi.org/10.3390/pathogens14050413
Chicago/Turabian StyleJakomin, Tjaša, Janja Zajc Žunič, and Polona Kogovšek. 2025. "Test Performance Study on qPCR Assays for Detection of Phyllosticta citricarpa" Pathogens 14, no. 5: 413. https://doi.org/10.3390/pathogens14050413
APA StyleJakomin, T., Žunič, J. Z., & Kogovšek, P. (2025). Test Performance Study on qPCR Assays for Detection of Phyllosticta citricarpa. Pathogens, 14(5), 413. https://doi.org/10.3390/pathogens14050413






