Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Initial Isolate Screening
2.2. Isolate Selection and Phenotypic Confirmation
2.3. Genotypic Confirmation
2.4. Whole Genome Sequencing
3. Results
3.1. Screening and Phenotypic Characterization
3.2. Genotypic Characterization
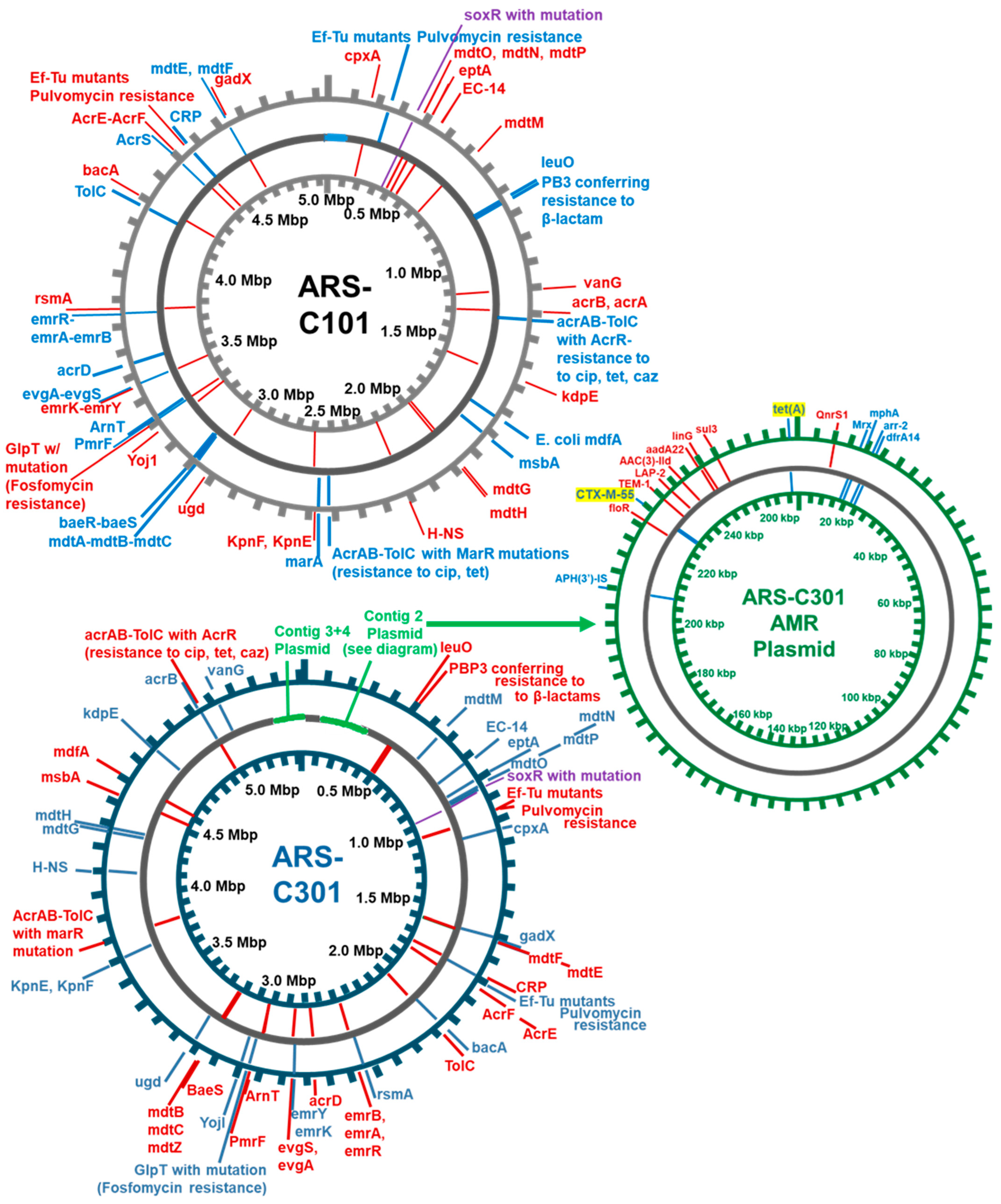
3.3. Assembly and Annotation of Two Agriculturally Sourced E. coli Genomes
3.4. Finding Best Hits for Assembled Genomes Using Mash
3.5. Identification of Antibiotic Resistance Genes Using CARD-RGI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larsson, D.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Abramova, A.; Berendonk, T.U.; Coelho, L.P.; Forslund, S.K.; Gschwind, R.; Heikinheimo, A.; Jarquín-Díaz, V.H.; Khan, A.A.; Klümper, U.; et al. Towards monitoring of antimicrobial resistance in the environment: For what reasons, how to implement it, and what are the data needs? Environ. Int. 2023, 178, 108089. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Miller, D.N.; Wienhold, B.J. Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes. PLoS ONE 2012, 7, e48325. [Google Scholar] [CrossRef]
- Cytryn, E. The soil resistome: The anthropogenic, the native, and the unknown. Soil Biol. Biochem. 2013, 63, 18–23. [Google Scholar] [CrossRef]
- Wang, F.; Fu, Y.H.; Sheng, H.J.; Topp, E.; Jiang, X.; Zhu, Y.G.; Tiedje, J.M. Antibiotic resistance in the soil ecosystem: A One Health perspective. Curr. Opin. Environ. Sci. Health 2021, 20, 100230. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef]
- Williams-Nguyen, J.; Sallach, J.B.; Bartelt-Hunt, S.; Boxall, A.B.; Durso, L.M.; McLain, J.E.; Singer, R.S.; Snow, D.D.; Zilles, J.L. Antibiotics and antibiotic resistance in agroecosystems: State of the science. J. Environ. Qual. 2016, 45, 394–406. [Google Scholar] [CrossRef]
- EPA. Factsheet on Water Quality Parameters, Escherichia coli. 2021. Available online: https://www.epa.gov/system/files/documents/2021-07/parameter-factsheet_e.-coli.pdf (accessed on 1 February 2025).
- Savageau, M.A. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 1983, 122, 732–744. [Google Scholar] [CrossRef]
- Lang, N.L.; Smith, S.R. Influence of soil type, moisture content and biosolids application on the fate of Escherichia coli in agricultural soil under controlled laboratory conditions. J. Appl. Microbiol. 2007, 103, 2122–2131. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Hur, H.G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Navarro-González, N.; Porrero, M.C.; Mentaberre, G.; Serrano, E.; Mateos, A.; Domínguez, L.; Lavín, S. Antimicrobial resistance in indicator Escherichia coli isolates from free-ranging livestock and sympatric wild ungulates in a natural environment (Northeastern Spain). Appl. Environ. Microbiol. 2013, 79, 6184–6186. [Google Scholar] [CrossRef]
- Agunos, A.; Gow, S.P.; Léger, D.F.; Carson, C.A.; Deckert, A.E.; Bosman, A.L.; Loest, D.; Irwin, R.J.; Reid-Smith, R.J. Antimicrobial use and antimicrobial resistance Indicators—Integration of Farm-Level surveillance data from broiler chickens and turkeys in British Columbia, Canada. Front. Vet. Sci. 2019, 6, 131. [Google Scholar] [CrossRef]
- World Health Organization. WHO Integrated Global Surveillance on ESBL-Producing E. coli Using a “One Health” Approach: Implementation and Opportunities; World Health Organization: Geneva, Switzerland, 2021; ISBN 978-92-4-002140-2. Available online: https://www.who.int/publications-detail-redirect/9789240021402 (accessed on 1 February 2025).
- Anjum, M.F.; Schmitt, H.; Börjesson, S.; Berendonk, T.U.; Donner, E.; Stehling, E.G.; Boerlin, P.; Topp, E.; Jardine, C.; Li, X.; et al. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Microbiol. 2021, 64, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Franklin, A.M.; Weller, D.L.; Durso, L.M.; Bagley, M.; Davis, B.C.; Frye, J.G.; Grim, C.J.; Ibekwe, A.M.; Jahne, M.A.; Keely, S.P.; et al. A one health approach for monitoring antimicrobial resistance: Developing a national freshwater pilot effort. Front. Water 2024, 6, 1359109. [Google Scholar] [CrossRef]
- Ochman, H.; Selander, R.K. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 1984, 157, 690–693. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- Picard, B.; Garcia, J.S.; Gouriou, S.; Duriez, P.; Brahimi, N.; Bingen, E.; Elion, J.; Denamur, E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef]
- Kim, J.; Nietfeldt, J.; Benson, A.K. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157: H7 strains in cattle. Proc. Natl. Acad. Sci. USA 1999, 96, 13288–13293. [Google Scholar] [CrossRef] [PubMed]
- Franz, E.; Rotariu, O.; Lopes, B.S.; MacRae, M.; Bono, J.L.; Laing, C.; Gannon, V.; Söderlund, R.; Van Hoek, A.H.; Friesema, I.; et al. Phylogeographic analysis reveals multiple international transmission events have driven the global emergence of Escherichia coli O157: H7. Clin. Infect. Dis. 2019, 69, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Ketkhao, P.; Utrarachkij, F.; Parikumsil, N.; Poonchareon, K.; Kerdsin, A.; Ekchariyawat, P.; Narongpun, P.; Nakajima, C.; Suzuki, Y.; Suthienkul, O. Phylogenetic diversity and virulence gene characteristics of Escherichia coli from pork and patients with urinary tract infections in Thailand. PLoS ONE 2024, 19, 0307544. [Google Scholar] [CrossRef]
- Liguori, K.; Keenum, I.; Davis, B.C.; Calarco, J.; Milligan, E.; Harwood, V.J.; Pruden, A. Antimicrobial resistance monitoring of water environments: A framework for standardized methods and quality control. Environ. Sci. Technol. 2022, 56, 9149–9160. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Cook, K.L. Impacts of antibiotic use in agriculture: What are the benefits and risks? Curr. Opin. Microbiol. 2014, 19, 37–44. [Google Scholar] [CrossRef]
- Stanton, I.C.; Bethel, A.; Leonard, A.F.C.; Gaze, W.H.; Garside, R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: A systematic map. Environ. Evid. 2022, 11, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.; Warren, J.; Wilkinson, H.; Schmidt, W. Environmental surveillance of antimicrobial resistance (AMR), perspectives from a national environmental regulator in 2023. Eurosurveillance 2023, 28, 2200367. [Google Scholar] [CrossRef]
- Hua, M.; Huang, W.; Chen, A.; Rehmet, M.; Jin, C.; Huang, Z. Comparison of antimicrobial resistance detected in environmental and clinical isolates from historical data for the US. BioMed Res. Int. 2020, 2020, 4254530. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association: Washington, DC, USA; American Water Works Association: Denver, CO, USA; Water Environment Federation: Alexandria, VA, USA, 1998. [Google Scholar]
- Turano, A.; Pirali, F. Quantification Methods in Microbiology. In Laboratory Diagnosis of Infectious Diseases; Balows, A., Hausler, W.J., Ohashi, M., Turano, A., Lennete, E.H., Eds.; Springer: New York, NY, USA, 1988. [Google Scholar] [CrossRef]
- Campoccia, D.; Montanaro, L.; Moriarty, T.F.; Richards, R.G.; Ravaioli, S.; Arciola, C.R. The selection of appropriate bacterial strains in preclinical evaluation of infection-resistant biomaterials. Int. J. Artif. Organs 2008, 31, 841–847. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. Available online: https://www.cdc.gov/DrugResistance/Biggest-Threats.html (accessed on 1 February 2025).
- Durso, L.M.; Schmidt, A.M. Antimicrobial resistance related to agricultural wastewater and biosolids. In Antimicrobial Resistance in Wastewater Treatment Processes; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 219–240. [Google Scholar]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLOS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef]
- Long, N.S.; Hales, K.E.; Berry, E.D.; Legako, J.F.; Woerner, D.R.; Broadway, P.R.; Carroll, J.A.; Burdick Sanchez, N.C.; Fernando, S.C.; Wells, J.E. Antimicrobial Susceptibility of Trimethoprim–Sulfamethoxazole and 3rd-Generation Cephalosporin-Resistant Escherichia coli Isolates Enumerated Longitudinally from Feedlot Arrival to Harvest in High-Risk Beef Cattle Administered Common Metaphylactic Antimicrobials. Foodborne Pathog. Dis. 2023, 20, 252–260. [Google Scholar] [PubMed]
- Agga, G.E.; Schmidt, J.W.; Arthur, T.M. Effects of in-feed chlortetracycline prophylaxis in beef cattle on animal health and antimicrobial-resistant Escherichia coli. Appl. Environ. Microbiol. 2016, 82, 7197–7204. [Google Scholar] [CrossRef] [PubMed]
- M100-S25; Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. The β-Lactamase Disk Test: A Modified Method to Detect Extended-Spectrum-β-Lactamases in Multidrug-Resistant Escherichia coli Isolates. Clinical and Laboratory Standards Institute: Tucson, AZ, USA, 2015; Volume 35.
- EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance, Version 2; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2017. Available online: https://www.eucast.org/resistance_mechanisms (accessed on 1 February 2025).
- Condon, J.C.; Lisa, M. Durso 2025a. ESBL Antibiotic Susceptibility Disc Diffusion Test. protocols.io. Available online: https://www.protocols.io/view/esbl-antibiotic-susceptibility-disc-diffusion-test-eq2lyxpqrgx9/v1 (accessed on 1 February 2025).
- Sanders, E.R. Aseptic laboratory techniques: Plating methods. J. Vis. Exp. 2012, e3064. [Google Scholar] [CrossRef]
- SOP No: MDP-MTH-11; Real-Time PCR Detection of Shiga-Toxin Producing Escherichia coli (STEC), Serotype O157 and non-O157 in Fresh Produce and Food with non-O157 Isolation and Identification. United States Department of Agriculture Agricultural Marketing Service, Science & Technology Microbiological Data Program: Washington, DC, USA, 2011. Available online: https://www.ams.usda.gov/sites/default/files/media/MDP-MTH-11.pdf (accessed on 1 February 2025).
- Condon, J.C.; Lisa, M. Durso 2025b. Standard Uniplex PCR SOP for Escherichia coli and Salmonella. protocols.io. Available online: https://www.protocols.io/view/standard-uniplex-pcr-sop-for-escherichia-coli-and-dm6gp9km1vzp/v1 (accessed on 1 February 2025).
- Ducey, T.F.; Durso, L.M.; Ibekwe, A.M.; Dungan, R.S.; Jackson, C.R.; Frye, J.G.; Castleberry, B.L.; Rashash, D.M.; Rothrock, M.J.; Boykin, D.; et al. A newly developed Escherichia coli isolate panel from a cross section of US animal production systems reveals geographic and commodity-based differences in antibiotic resistance gene carriage. J. Hazard. Mater. 2020, 382, 120991. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Day, M.J.; Hope, R.; Wain, J.; Woodford, N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J. Clin. Microbiol. 2012, 50, 3108–3110. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Escobar-Páramo, P.; Grenet, K.; Le Menac’h, A.; Rode, L.; Salgado, E.; Amorin, C.; Gouriou, S.; Picard, B.; Rahimy, M.C.; Andremont, A.; et al. Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 2004, 70, 5698–5700. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P. Assembly of long error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2014, 25, 1043–1055. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- OIE World organization for Animal Health. OIE List of Antimicrobial Agents of Veterinary Importance. 2021. Available online: https://www.woah.org/app/uploads/2021/06/a-oie-list-antimicrobials-june2021.pdf (accessed on 1 February 2025).
- United States Food and Drug Administration. 2023 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. 2023. Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/2023-summary-report-antimicrobials-sold-or-distributed-use-food-producing-animals (accessed on 1 February 2025).
- Jacob, M.E.; Keelara, S.; Aidara-Kane, A.; Matheu Alvarez, J.R.; Fedorka-Cray, P.J. Optimizing a screening protocol for potential extended-spectrum β-lactamase Escherichia coli on MacConkey agar for use in a global surveillance program. J. Clin. Microbiol. 2020, 58, e01039–19. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.E.; Laing, G.; McMahon, B.J.; Fanning, S.; Stekel, D.J.; Pahl, O.; Coyne, L.; Latham, S.M.; McIntyre, K.M. The need for One Health systems-thinking approaches to understand multiscale dissemination of antimicrobial resistance. Lancet Planet. Health 2024, 8, e124–e133. [Google Scholar] [CrossRef]
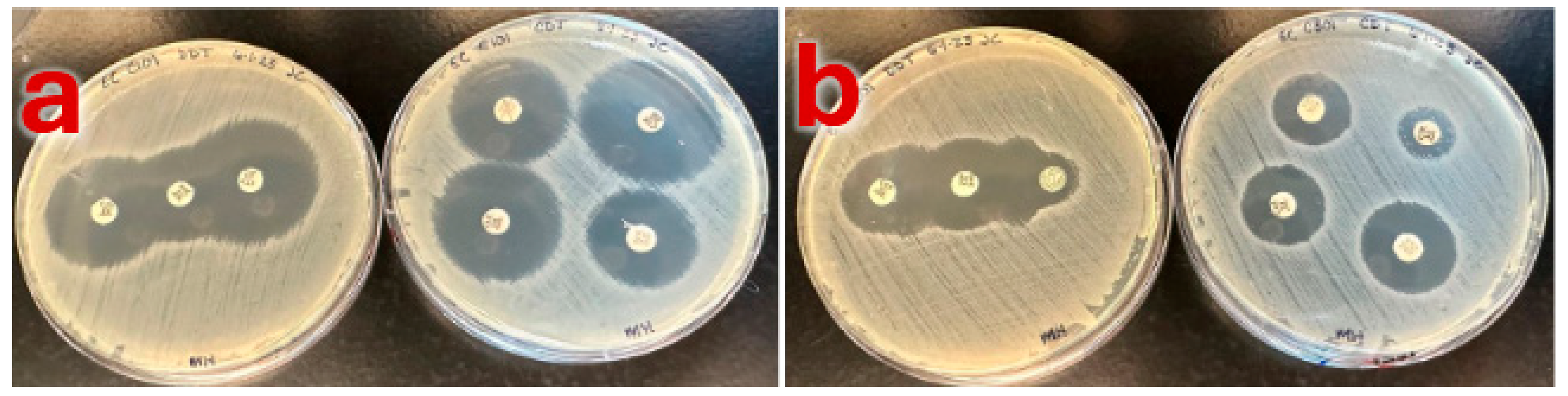

| ARS-C101 (Neg) | ARS-C301 (Pos) | ||||
|---|---|---|---|---|---|
| Drug | MIC | Interpretation | MIC | Interpretation | |
| 1 | Amoxicillin/Clavulanic Acid | 4 | S | 8 | S |
| 2 | Ampicillin | 2 | S | 32 | R |
| 3 | Azithromycin | 8 | S | 32 | R |
| 4 | Cefoxitin | 4 | S | 8 | S |
| 5 | Ceftriaxone | 0.25 | S | 64 | R |
| 6 | Chloramphenicol | 8 | S | 32 | R |
| 7 | Ciprofloxacin | 0.015 | S | 0.25 | S |
| 8 | Gentamicin | 0.5 | S | 16 | R |
| 9 | Meropenem | 0.06 | S | 0.06 | S |
| 10 | Nalidixic Acid | 4 | S | 4 | S |
| 11 | Streptomycin | 8 | S | 64 | R |
| 12 | Sulfisoxazole | 16 | S | 512 | R |
| 13 | Tetracycline | 4 | S | 32 | R |
| 14 | Trimethoprim/Sulfamethoxazole | 0.12 | S | 4 | R |
| Isolate ID | ARS-C101 | ARS-C301 |
|---|---|---|
| Description | E. coli, NEGATIVE CONTROL ESBL−, CTXS, TETS | E. coli, POSITIVE CONTROL ESBL+, CTXR, TETR |
| Date Collected | 7 February 2021 | 28 March 2021 |
| Source | Cattle Fecal Grab | Cattle Fecal Grab |
| Location | Texas | Texas |
| E. coli phenotypic confirmation | Blue-green colonies on TBX agar, blue colonies on CHROMagar ECC, pink colonies on CHROMagar Orientation. Indole positive. | Blue-green colonies on TBX agar, blue colonies on CHROMagar ECC, pink colonies on CHROMagar Orientation. Indole positive. |
| CDT Zone Sizes | (CTX) 31.2 mm, (CTX/CA) 24.7 mm (CAZ) 27.1 mm, (CAZ/CA) 21.8 mm | (CTX) 6.6 mm, (CTX/CA) 16.8 mm (CAZ) 16 mm, (CAZ/CA) 21 mm |
| Combination Disk Diffusion Results |  |  |
| E. coli uidA PCR | uidA positive | uidA positive |
| Phylotyping via PCR | B1 | B1 |
| Target Gene Genotype | Not applicable | CTX-M-55, tet(A) |
| 16S GenBank assignment (V1–V2) * | E. coli O136: H-(GenBank: AB604195), Japan. 528 bp | E. coli JCLys7 (GenBank GQ273520.1), from temperate gley soil, Ireland. 528 bp |
| 16S sequence Accession # | OR269615 | OR269616 |
| NCBI WGS Accession # | Accession number: PRJNA1003888 BioSample: SAMN36909132 | Accession number: PRJNA1003888 BioSample: SAMN36910816 |
| ARS Culture Collection ID | https://nrrl.ncaur.usda.gov/ Accession number: B-65681 Accessed on 23 April 2025 | https://nrrl.ncaur.usda.gov/ Accession number: B-65682 Accessed on 23 April 2025 |
| ATTC Culture Collection ID | ATCC® BAA-3340™ | ATCC® BAA-3341™ |
| Microbiologics 10–100 CFU Pellets | Not available | ARS-C301 |
| Isolate | CTX | CTX/CA | Difference | CAZ | CAZ/CA | Difference | ESBL? |
|---|---|---|---|---|---|---|---|
| ARS-C301 | 6.6 | 16.8 | 10.2 | 16.0 | 21.0 | 5.0 | Yes |
| ARS-C101 | 32.5 | 32.6 | 0.1 | 28.7 | 29.1 | 0.3 | No |
| Isolate | CTX | AMC | CAZ | ESBL? |
|---|---|---|---|---|
| ARS-C301 | 12.8 | 21.6 | 18.9 | Yes |
| ARS-C101 | 33.5 | 24.8 | 29.8 | No |
| ARS-C101 | ARS-C301 | |
|---|---|---|
| Total bp sequenced | 703,796,709 bp | 866,611,913 bp |
| Total number of reads | 108,488 reads | 130,182 reads |
| Longest read | 100,534 bp | 107,946 bp |
| Raw coverage | 139× | 160× |
| Assembled coverage | 98× | 106× |
| Genome size (Mb) | 5.0 Mb | 5.4 Mb |
| Number of contigs | 3 contigs | 4 contigs |
| Number of genes annotated | 5041 genes | 5432 genes |
| Contig | Size (bp) | Identity (% Matched) | NCBI Accession | Description |
|---|---|---|---|---|
| Contig_1 | 4,937,799 | 99.8 | NZ_JXVO01000001.1 | Escherichia coli strain OLC-157 Cont0001 |
| Contig_2 | 9686 | 100.0 | NZ_CP009169.1 | Escherichia coli 1303 plasmid p1303_5 |
| Contig_3 | 102,406 | 99.2 | NZ_CP010345.1 | Escherichia coli ECC-1470 plasmid pECC-1470_100 |
| Contig | Size (bp) | Identity (% Matched) | NCBI Accession | Description |
|---|---|---|---|---|
| Contig_1 | 4,894,443 | 99.90 | NZ_MPUD01000003.1 | E. coli strain K30 IMT32646_S7_L001_R1_contig_1 |
| Contig_2 | 267,517 | 99.92 | AP027468.1 | E. coli 15.01CC plasmid p15.01CC_DNA |
| Contig_3 | 171,722 | 100.00 | CP063740.1 | E. coli strain 18SC04V04-Ec plasmid pVPS18EC0467-2 |
| Contig_4 | 81,040 | 98.54 | AP019676.1 | E. coli GSH8M-2 plasmid pGSH8M-2-1DNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wells, J.E.; Durso, L.M.; Ibekwe, A.M.; Frye, J.G.; Sharma, M.; Williams, C.F.; Shamimuzzaman, M. Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains. Pathogens 2025, 14, 417. https://doi.org/10.3390/pathogens14050417
Wells JE, Durso LM, Ibekwe AM, Frye JG, Sharma M, Williams CF, Shamimuzzaman M. Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains. Pathogens. 2025; 14(5):417. https://doi.org/10.3390/pathogens14050417
Chicago/Turabian StyleWells, James E., Lisa M. Durso, Abasiofiok M. Ibekwe, Jonathan G. Frye, Manan Sharma, Clinton F. Williams, and Md Shamimuzzaman. 2025. "Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains" Pathogens 14, no. 5: 417. https://doi.org/10.3390/pathogens14050417
APA StyleWells, J. E., Durso, L. M., Ibekwe, A. M., Frye, J. G., Sharma, M., Williams, C. F., & Shamimuzzaman, M. (2025). Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains. Pathogens, 14(5), 417. https://doi.org/10.3390/pathogens14050417









