Ancient Origins and Global Diversity of Plague: Genomic Evidence for Deep Eurasian Reservoirs and Recurrent Emergence
Abstract
1. Introduction
- How did major Y. pestis lineages diverge and persist across millennia?
- What evolutionary patterns differentiate ancient pandemic strains from modern environmental lineages?
- What do the geographic origins and host associations of these strains reveal about the nature of plague reservoirs and outbreak dynamics?
2. Methods
2.1. Genome Selection and Data Acquisition
2.2. Sequence Alignment and Variant Calling
2.3. SNP-Based Maximum Likelihood (ML) Phylogenetic Analysis
2.4. Interactive Visualization of Estimated Divergence Tree Using Nextstrain
3. Results
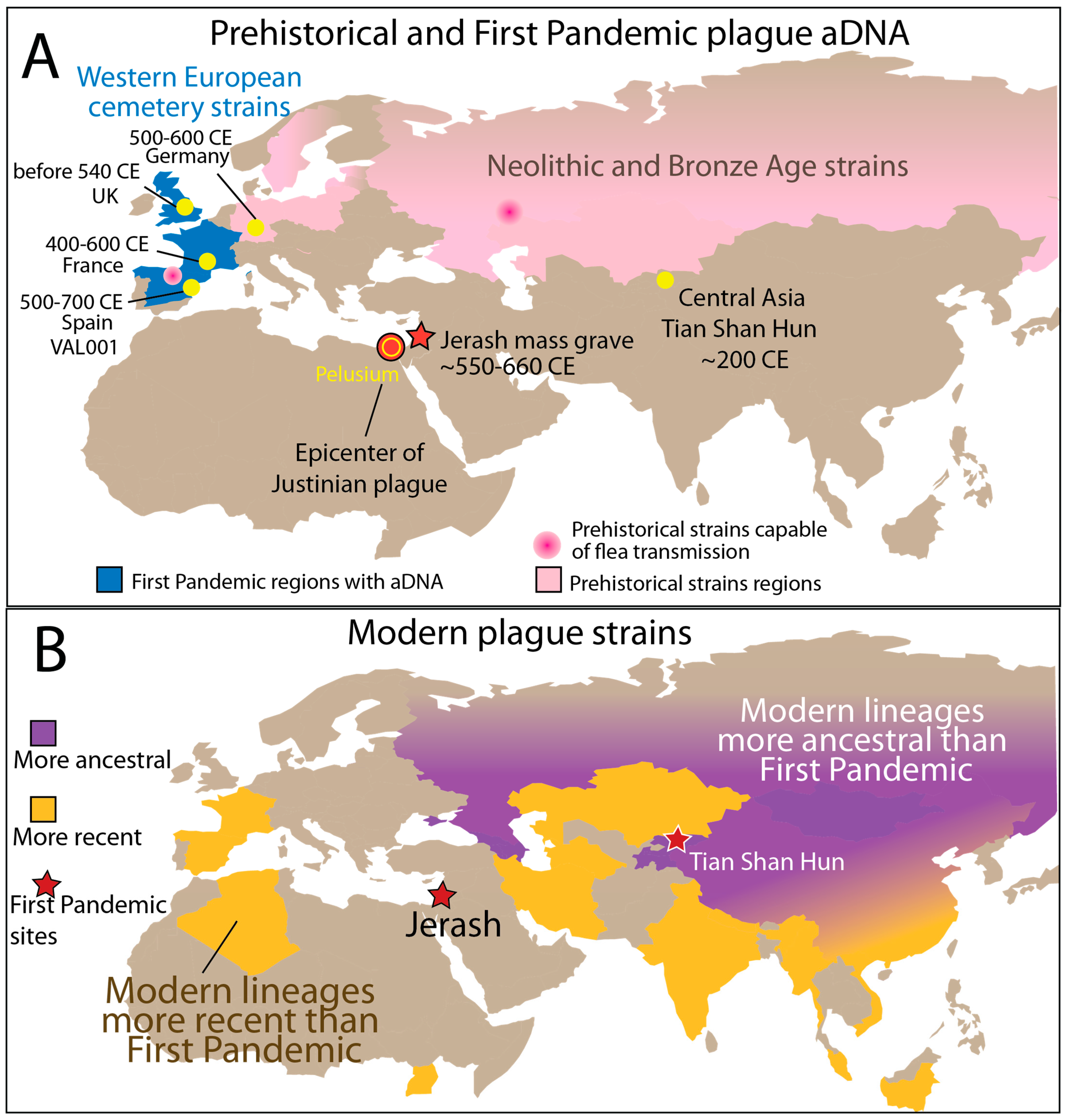
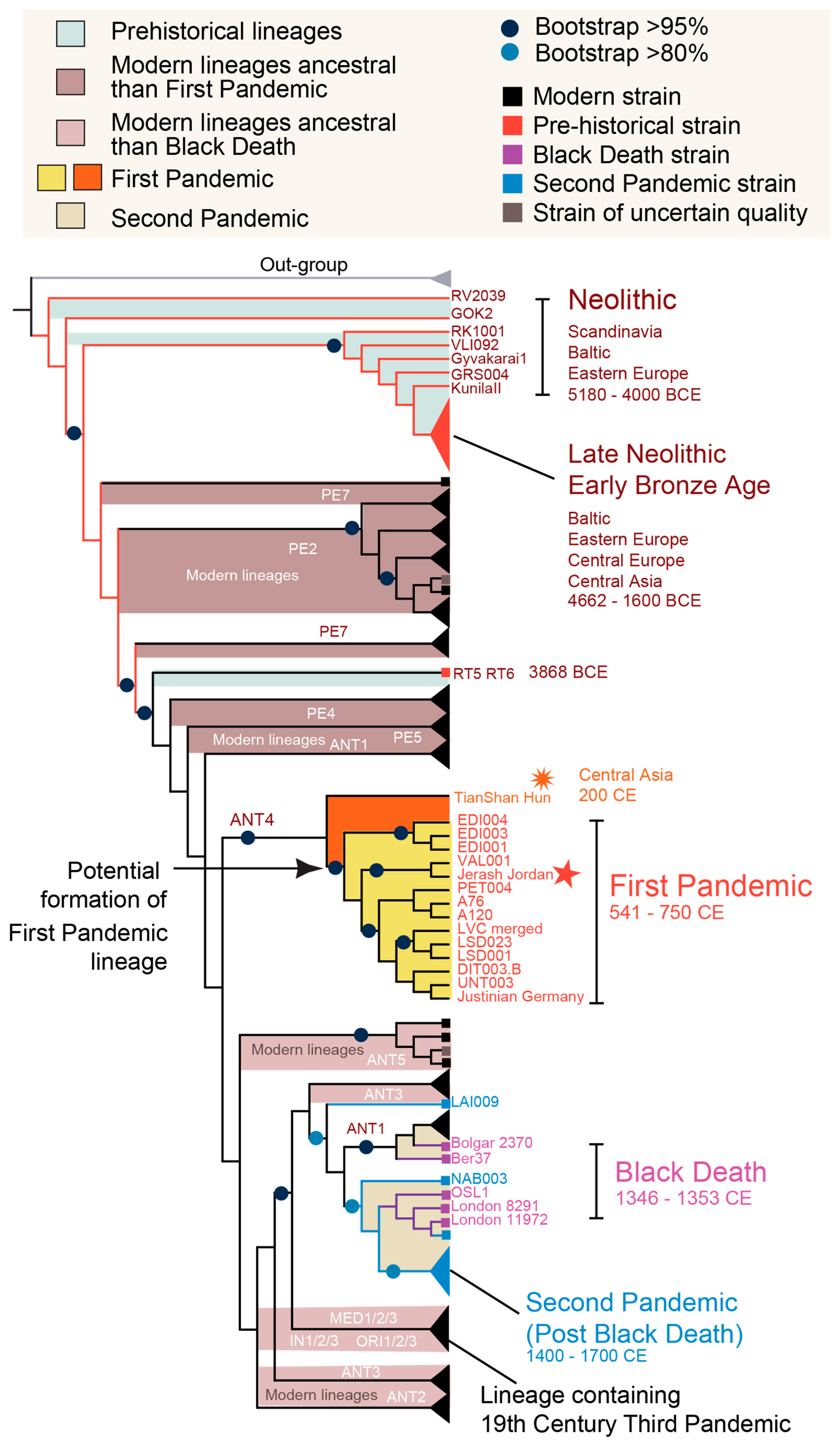
3.1. Spatiotemporal Patterns of Y. pestis Genomes Reveal Deep Eurasian Roots
3.2. Ancient Evolutionary Patterns of Global Plague Lineages
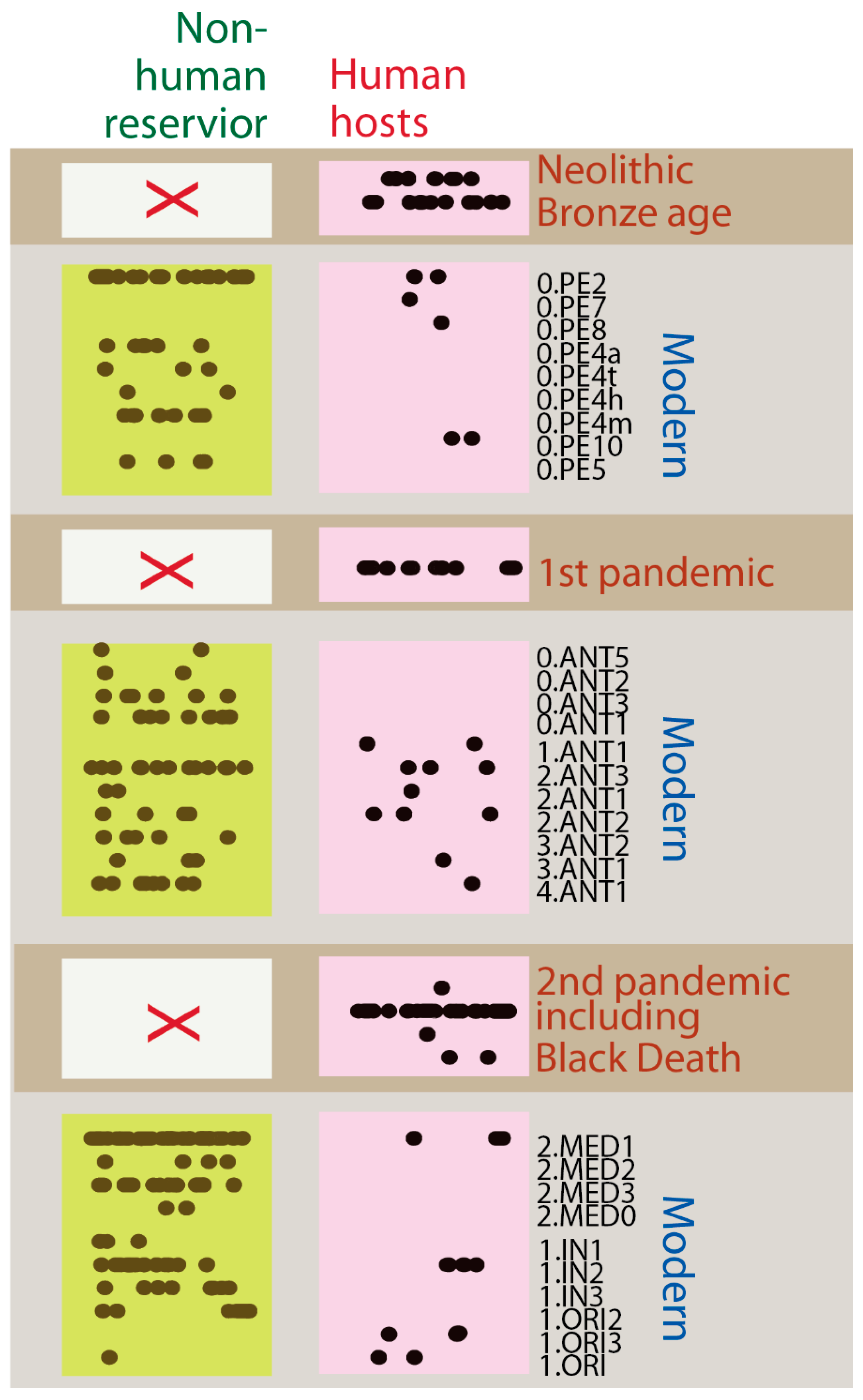
3.3. Human-Only Host Range in Historical Plague Lineages Suggests Pandemic-Specific Transmission Modes
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conrad, L.I. The plague in Bilad al-Sham in pre-Islamic times. In Proceedings of the Fourth International Conference on the History of Bilad al-Sham During the Byzantine Period; University of Jordan: Amman, Jordan, 1986; Volume 2, pp. 143–163. [Google Scholar]
- Sarris, P. The Justinianic plague: Origins and effects. Contin. Change 2002, 17, 169–182. [Google Scholar] [CrossRef]
- Benedictow, O.J. The Complete History of the Black Death; Boydell & Brewer: Woodbridge, UK, 2021. [Google Scholar]
- Dennis, D.T.; Gage, K.L.; Gratz, N.G.; Poland, J.D.; Tikhomirov, E.; World Health Organization. Plague Manual: Epidemiology, Distribution, Surveillance and Control; World Health Organization: Geneva, Switzerland, 1999. [Google Scholar]
- Green, M.H. A New Definition of the Black Death: Genetic Findings and Historical Interpretations. Medio Aevo 2022, 11, 139–155. [Google Scholar] [CrossRef]
- Green, M.H.; Fancy, N. Plague history, Mongol history, and the processes of focalisation leading up to the Black Death: A response to Brack et al. Med. Hist. 2024, 68, 411–435. [Google Scholar] [CrossRef] [PubMed]
- Andrades Valtueña, A.; Neumann, G.U.; Spyrou, M.A.; Musralina, L.; Aron, F.; Beisenov, A.; Belinskiy, A.B.; Bos, K.I.; Buzhilova, A.; Conrad, M.; et al. Stone Age Yersinia pestis genomes shed light on the early evolution, diversity, and ecology of plague. Proc. Natl. Acad. Sci. USA 2022, 119, e2116722119. [Google Scholar] [CrossRef]
- Damgaard, P.d.B.; Marchi, N.; Rasmussen, S.; Peyrot, M.; Renaud, G.; Korneliussen, T.; Moreno-Mayar, J.V.; Pedersen, M.W.; Goldberg, A.; Usmanova, E.; et al. 137 ancient human genomes from across the Eurasian steppes. Nature 2018, 557, 369–374. [Google Scholar] [CrossRef]
- Rascovan, N.; Sjögren, K.-G.; Kristiansen, K.; Nielsen, R.; Willerslev, E.; Desnues, C.; Rasmussen, S. Emergence and Spread of Basal Lineages of Yersinia pestis during the Neolithic Decline. Cell 2019, 176, 295–305.e10. [Google Scholar] [CrossRef]
- Rasmussen, S.; Allentoft, M.E.; Nielsen, K.; Orlando, L.; Sikora, M.; Sjogren, K.G.; Pedersen, A.G.; Schubert, M.; Van Dam, A.; Kapel, C.M.; et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 2015, 163, 571–582. [Google Scholar] [CrossRef]
- Seersholm, F.V.; Sjögren, K.-G.; Koelman, J.; Blank, M.; Svensson, E.M.; Staring, J.; Fraser, M.; Pinotti, T.; McColl, H.; Gaunitz, C.; et al. Repeated plague infections across six generations of Neolithic Farmers. Nature 2024, 632, 114–121. [Google Scholar] [CrossRef]
- Spyrou, M.A.; Tukhbatova, R.I.; Wang, C.C.; Valtuena, A.A.; Lankapalli, A.K.; Kondrashin, V.V.; Tsybin, V.A.; Khokhlov, A.; Kuhnert, D.; Herbig, A.; et al. Analysis of 3800-year-old Yersinia pestis genomes suggests Bronze Age origin for bubonic plague. Nat. Commun. 2018, 9, 2234. [Google Scholar] [CrossRef] [PubMed]
- Chouikha, I.; Hinnebusch, B.J. Silencing urease: A key evolutionary step that facilitated the adaptation of Yersinia pestis to the flea-borne transmission route. Proc. Natl. Acad. Sci. USA 2014, 111, 18709–18714. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yu, C.; Yan, Y.; Li, D.; Li, Y.; Jombart, T.; Weinert, L.A.; Wang, Z.; Guo, Z.; Xu, L.; et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl. Acad. Sci. USA 2013, 110, 577–582. [Google Scholar] [CrossRef]
- Zhou, D.; Yang, R. Molecular Darwinian evolution of virulence in Yersinia pestis. Infect. Immun. 2009, 77, 2242–2250. [Google Scholar] [CrossRef]
- Keller, M.; Spyrou, M.A.; Scheib, C.L.; Neumann, G.U.; Kröpelin, A.; Haas-Gebhard, B.; Päffgen, B.; Haberstroh, J.; Ribera, I.L.A.; Raynaud, C.; et al. Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541–750). Proc. Natl. Acad. Sci. USA 2019, 116, 12363–12372. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, H. Justinianic Plague in Syria and the Archaeological Evidence. In Plague and the End of Antiquity: The Pandemic of 541–750; Little, L., Ed.; Cambridge University Press: Cambridge, UK, 2007; pp. 87–98. [Google Scholar]
- Wagner, D.M.; Klunk, J.; Harbeck, M.; Devault, A.; Waglechner, N.; Sahl, J.W.; Enk, J.; Birdsell, D.N.; Kuch, M.; Lumibao, C.; et al. Yersinia pestis and the plague of Justinian 541–543 AD: A genomic analysis. Lancet Infect. Dis. 2014, 14, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Kehrberg, I. Pottery and glass sherd-tools from Roman and Byzantine workshops at the Gerasa hippodrome and other sites. A reappraisal of earlier findings. Stud. Hist. Archaeol. Jordan (SHAJ) 2016, 12, 411–441. [Google Scholar]
- Kehrberg, I.; Ostrasz, A.A. Ancient Burials at the Hippodrome of Gerasa/Jarash. Annu. Dep. Antiq. Jordan 2017, 58, 181–213. [Google Scholar]
- Bourke, S.H.K. Twenty Years of Australian Physical Anthropology in Jordan: Retrospects and Prospects; Australians Uncovering Ancient Jordan, The University of Sydney and Department of Antiquities Jordan: Sydney, Australia, 2001; pp. 27–88. [Google Scholar]
- Hendrix, K. Preliminary Investigation of the Human Skeletal Remains from the Hippodrome Jarash. Annu. Dep. Antiq. Jordan/Hashemite Kingd. Jordan 1995, 39, 560–562. [Google Scholar]
- Hendrix, K. Further investigations of the Human Skeletal Remains from the Hippodrome Jarash. Annu. Dep. Antiq. Jordan/Hashemite Kingd. Jordan 1998, 42, 639–640. [Google Scholar]
- Ostrasz, A.A. The Excavation and Restoration of the Hippodrome Jerash. A Synopsis. Annu. Dep. Antiq. Jordan/Hashemite Kingd. Jordan 1991, 35, 237–250. [Google Scholar]
- Adapa, S.R.; Hendrix, K.; Upadhyay, A.; Dutta, S.; Vianello, A.; O’Corry-Crowe, G.; Ferrer, T.; Monroy, J.; Wood, E.; Ferreira, G.; et al. Genetic Evidence of Yersinia pestis from the First Pandemic. Genes 2025, 16, 926. [Google Scholar] [CrossRef]
- Yates, J.A.F.; Lamnidis, T.C.; Borry, M.; Valtueña, A.A.; Fagernäs, Z.; Clayton, S.; Garcia, M.U.; Neukamm, J.; Peltzer, A. Reproducible, portable, and efficient ancient genome reconstruction with nf-core/eager. PeerJ 2021, 9, e10947. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- To, T.-H.; Jung, M.; Lycett, S.; Gascuel, O. Fast dating using least-squares criteria and algorithms. Syst. Biol. 2016, 65, 82–97. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.; Hadfield, J.; Sibley, T.R.; Lee, J.; Fay, K.; Ilcisin, M.; Harkins, E.; Bedford, T.; Neher, R.A.; Hodcroft, E.B. Augur: A bioinformatics toolkit for phylogenetic analyses of human pathogens. J. Open Source Softw. 2021, 6, 2906. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Eaton, K.; Featherstone, L.; Duchene, S.; Carmichael, A.G.; Varlık, N.; Golding, G.B.; Holmes, E.C.; Poinar, H.N. Plagued by a cryptic clock: Insight and issues from the global phylogeny of Yersinia pestis. Commun. Biol. 2023, 6, 23. [Google Scholar] [CrossRef]
- Barbieri, R.; Signoli, M.; Chevé, D.; Costedoat, C.; Tzortzis, S.; Aboudharam, G.; Raoult, D.; Drancourt, M. Yersinia pestis: The Natural History of Plague. Clin. Microbiol. Rev. 2020, 34, e00044-19. [Google Scholar] [CrossRef]
- Shi, L.; Yang, G.; Zhang, Z.; Xia, L.; Liang, Y.; Tan, H.; He, J.; Xu, J.; Song, Z.; Li, W.; et al. Reemergence of human plague in Yunnan, China in 2016. PLoS ONE 2018, 13, e0198067. [Google Scholar] [CrossRef]
- Bos, K.I.; Schuenemann, V.J.; Golding, G.B.; Burbano, H.A.; Waglechner, N.; Coombes, B.K.; McPhee, J.B.; DeWitte, S.N.; Meyer, M.; Schmedes, S.; et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature 2011, 478, 506–510. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutta, S.; Upadhyay, A.; Adapa, S.R.; O’Corry-Crowe, G.; Tripathy, S.; Jiang, R.H.Y. Ancient Origins and Global Diversity of Plague: Genomic Evidence for Deep Eurasian Reservoirs and Recurrent Emergence. Pathogens 2025, 14, 797. https://doi.org/10.3390/pathogens14080797
Dutta S, Upadhyay A, Adapa SR, O’Corry-Crowe G, Tripathy S, Jiang RHY. Ancient Origins and Global Diversity of Plague: Genomic Evidence for Deep Eurasian Reservoirs and Recurrent Emergence. Pathogens. 2025; 14(8):797. https://doi.org/10.3390/pathogens14080797
Chicago/Turabian StyleDutta, Subhajeet, Aditya Upadhyay, Swamy R. Adapa, Gregory O’Corry-Crowe, Sucheta Tripathy, and Rays H. Y. Jiang. 2025. "Ancient Origins and Global Diversity of Plague: Genomic Evidence for Deep Eurasian Reservoirs and Recurrent Emergence" Pathogens 14, no. 8: 797. https://doi.org/10.3390/pathogens14080797
APA StyleDutta, S., Upadhyay, A., Adapa, S. R., O’Corry-Crowe, G., Tripathy, S., & Jiang, R. H. Y. (2025). Ancient Origins and Global Diversity of Plague: Genomic Evidence for Deep Eurasian Reservoirs and Recurrent Emergence. Pathogens, 14(8), 797. https://doi.org/10.3390/pathogens14080797





