Twenty Years of Equine Piroplasmosis Research: Global Distribution, Molecular Diagnosis, and Phylogeny
Abstract
:1. Current Knowledge of Equine Piroplasmosis
1.1. Life Cycle, Vectors, and Transmission
1.2. Clinical Disease
1.3. Immunity, Treatment, and Control
1.4. Diagnosis
2. Epidemiology
A Review of EP Epidemiology in the Last 20 Years
3. Genotyping
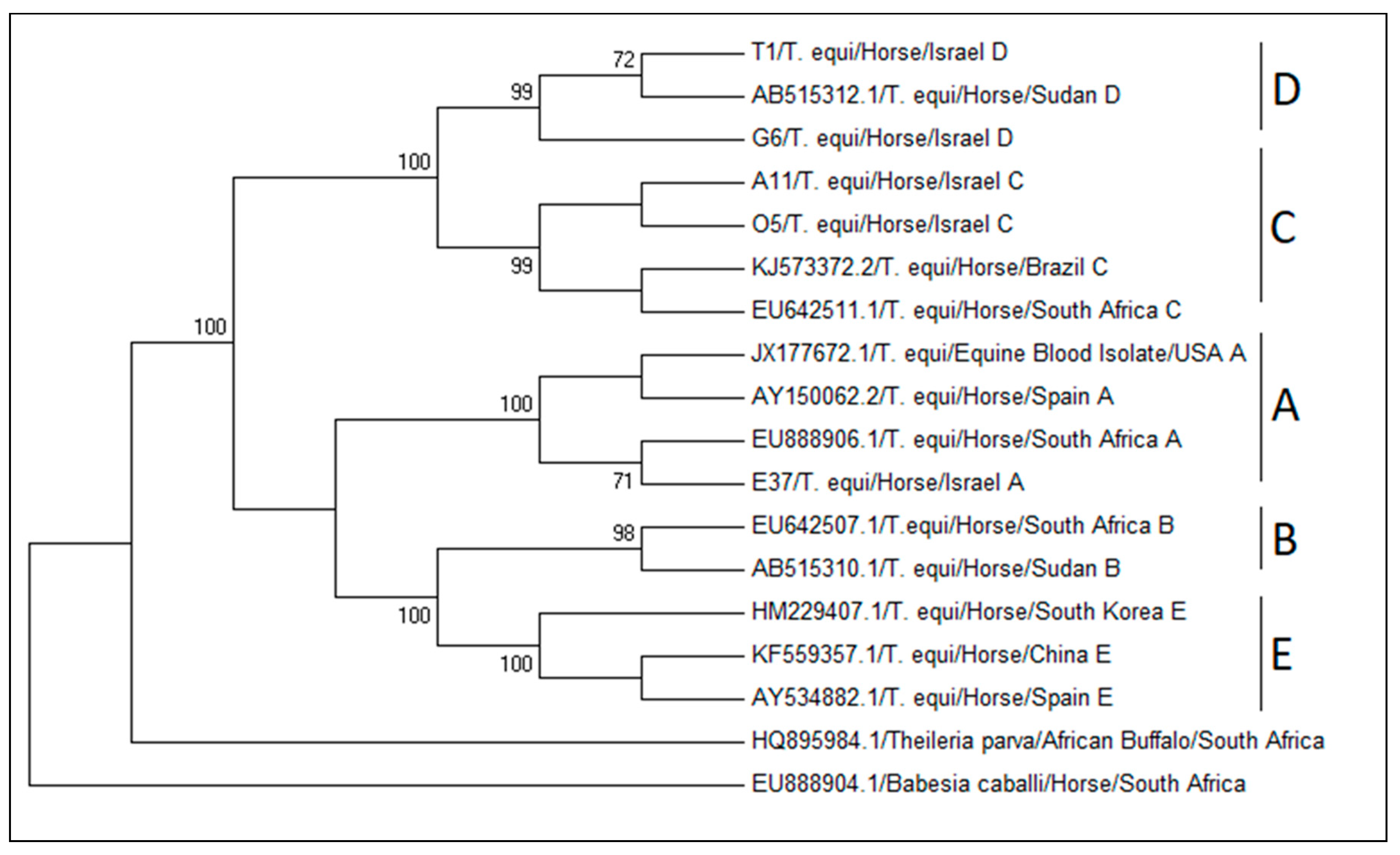
3.1. Theileria equi 18S rRNA Genotypes and Their Global Distribution
3.2. Theileria equi ema Genotypes and Their Global Distribution
3.3. Babesia caballi Genotypes and Their Global Distribution
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Friedhoff, K.T.; Tenter, A.M.; Muller, I. Haemoparasites of equines: Impact on international trade of horses. Rev. Sci. Tech. 1990, 9, 1187–1194. [Google Scholar] [PubMed]
- Rothschild, C.M. Equine piroplasmosis. J. Equine Vet. Sci. 2013, 23, 115–120. [Google Scholar] [CrossRef]
- Wise, L.N.; Kappmeyer, L.S.; Mealey, R.H.; Knowles, D.P. Review of equine piroplasmosis. J. Vet. Intern. Med. 2013, 27, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Knowles, D.P.; Kappmeyer, L.S.; Haney, D.; Herndon, D.R.; Fry, L.M.; Munro, J.B.; Sears, K.; Ueti, M.W.; Wise, L.N.; Silva, M.; et al. Discovery of a novel species, Theileria haneyi n. sp., infective to equids, highlights exceptional genomic diversity within the genus Theileria: Implications for apicomplexan parasite surveillance. Int. J. Parasitol. 2018, 48, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Onyiche, T.E.; Suganuma, K.; Igarashi, I.; Yokoyama, N.; Xuan, X.; Thekisoe, O. A Review on Equine Piroplasmosis: Epidemiology, Vector Ecology, Risk Factors, Host Immunity, Diagnosis and Control. Int. J. Environ. Res. Public Health 2019, 16, 1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allsopp, M.; Cavalier-Smith, T.; de Waal, D.; Allsopp, B. Phylogeny and evolution of the piroplasms. Parasitology 1994, 108, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.D.; Ueti, M.W.; Johnson, W.C.; Scoles, G.A.; Knowles, D.P.; Mealey, R.H. Lymphocytes and macrophages are infected by Theileria equi, but T cells and B cells are not required to establish infection in vivo. PLoS ONE 2013, 8, e76996. [Google Scholar] [CrossRef]
- Scoles, G.A.; Ueti, M.W. Vector ecology of equine piroplasmosis. Annu. Rev. Entomol. 2015, 60, 561–580. [Google Scholar] [CrossRef]
- Allsopp, M.T.; Lewis, B.D.; Penzhorn, B.L. Molecular evidence for transplacental transmission of Theileria equi from carrier mares to their apparently healthy foals. Vet. Parasitol. 2007, 148, 130–136. [Google Scholar] [CrossRef] [Green Version]
- Chhabra, S.; Ranjan, R.; Uppal, S.K.; Singla, L.D. Transplacental transmission of Babesia equi (Theileria equi) from carrier mares to foals. J. Parasit. Dis. 2012, 36, 31–33. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, J.; Phipps, L.P.; Watkins, K.L. Evidence of Maternal Antibodies to Babesia-Equi and Babesia-Caballi in Foals of Seropositive Mares. Equine Vet. J. 1982, 14, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Georges, K.C.; Ezeokoli, C.D.; Sparagano, O.; Pargass, I.; Campbell, M.; D’Abadie, R.; Yabsley, M.J. A case of transplacental transmission of Theileria equi in a foal in Trinidad. Vet. Parasitol. 2011, 175, 363–366. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, R.; Gupta, A.K.; Dwivedi, S.K. Passive transfer of Theileria equi antibodies to neonate foals of immune tolerant mares. Vet. Parasitol. 2008, 151, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Levi, M.M.; Tirosh-Levy, S.; Dahan, R.; Berlin, D.; Steinman, A.; Edery, N.; Savitski, I.; Lebovich, B.; Knowles, D.; Suarez, C.E.; et al. First Detection of Diffuse and Cerebral Theileria equi Infection in Neonatal Filly. J. Equine Vet. Sci. 2018, 60, 23–28. [Google Scholar] [CrossRef]
- Oliveira, A.; Pinheiro, G.; Souza, T.; Flecher, M.; Santos, R. Abortion in association with transplacental Theileria equi infection in a mare from the State of Espírito Santo, southeast Brazil: Case report. Arq. Bras. Med. Vet. Zootec. 2019, 71, 369–373. [Google Scholar] [CrossRef]
- Phipps, L.P.; Otter, A. Transplacental transmission of Theileria equi in two foals born and reared in the United Kingdom. Vet. Rec. 2004, 154, 406–408. [Google Scholar] [CrossRef]
- Sant, C.; Allicock, O.M.; d’Abadie, R.; Charles, R.A.; Georges, K. Phylogenetic analysis of Theileria equi and Babesia caballi sequences from thoroughbred mares and foals in Trinidad. Parasitol. Res. 2019, 118, 1171–1177. [Google Scholar] [CrossRef]
- Sant, C.; d’Abadie, R.; Pargass, I.; Basu, A.K.; Asgarali, Z.; Charles, R.A.; Georges, K.C. Prospective study investigating transplacental transmission of equine piroplasmosis in thoroughbred foals in Trinidad. Vet. Parasitol. 2016, 226, 132–137. [Google Scholar] [CrossRef]
- Sudan, V.; Jaiswal, A.K.; Srivastava, A.; Saxena, A.; Shanker, D. A rare clinical presentation of transplacental transmission and subsequent abortion by Babesia (Theileria) equi in a mare. J. Parasit. Dis. 2015, 39, 336–338. [Google Scholar] [CrossRef] [Green Version]
- De Waal, D. Equine piroplasmosis: A review. Brit. Vet. J. 1992, 148, 6–14. [Google Scholar] [CrossRef]
- Lewis, B.D.; Penzhorn, B.L.; Volkmann, D.H. Could treatment of pregnant mares prevent abortions due to equine piroplasmosis? J. S. Afr. Vet. Assoc. 1999, 70, 90–91. [Google Scholar] [CrossRef] [Green Version]
- Tirosh-Levy, S.; Gottlieb, Y.; Mimoun, L.; Mazuz, M.L.; Steinman, A. Transplacental Transmission of Theileria equi is not a Common Cause of Abortions and Infection of Foals in Israel. Animals 2020, 10, 341. [Google Scholar] [CrossRef] [Green Version]
- Zobba, R.; Ardu, M.; Niccolini, S.; Chessa, B.; Manna, L.; Cocco, R.; Parpaglia, M.L.P. Clinical and laboratory findings in equine piroplasmosis. J. Equine Vet. Sci. 2008, 28, 301–308. [Google Scholar] [CrossRef]
- Prasad, A.; Kumar, V.; Kumar, B. First Report of Acute Bilateral Hyphema in a Theileria equi-Infected Kathiawari Horse. J. Equine Vet. Sci. 2019, 77, 72–74. [Google Scholar] [CrossRef]
- Diana, A.; Guglielmini, C.; Candini, D.; Pietra, M.; Cipone, M. Cardiac arrhythmias associated with piroplasmosis in the horse: A case report. Vet. J. 2007, 174, 193–195. [Google Scholar] [CrossRef]
- Pasolini, M.P.; Pagano, T.B.; Costagliola, A.; Biase, D.; Lamagna, B.; Auletta, L.; Fatone, G.; Greco, M.; Coluccia, P.; Vincenzo, V.; et al. Inflammatory Myopathy in Horses with Chronic Piroplasmosis. Vet. Pathol. 2018, 55, 133–143. [Google Scholar] [CrossRef]
- Padalino, B.; Rosanowski, S.M.; di Bella, C.; Lacinio, R.; Rubino, G.T.R. Piroplasmosis in Italian Standardbred Horses: 15 Years of Surveillance Data. J. Equine Vet. Sci. 2019, 83, 102813. [Google Scholar] [CrossRef]
- Tirosh-Levy, S.; Gottlieb, Y.; Steinman, A. Stress conditions do not affect Theileria equi parasitemia levels in sub-clinically infected horses. Tick Tick-Borne Dis. 2020. [Google Scholar] [CrossRef]
- Sears, K.P.; Kappmeyer, L.S.; Wise, L.N.; Silva, M.; Ueti, M.W.; White, S.; Reif, K.E.; Knowles, D.P. Infection dynamics of Theileria equi and Theileria haneyi, a newly discovered apicomplexan of the horse. Vet. Parasitol. 2019, 271, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Knowles, D.P., Jr.; Kappmeyer, L.S.; Stiller, D.; Hennager, S.G.; Perryman, L.E. Antibody to a recombinant merozoite protein epitope identifies horses infected with Babesia equi. J. Clin. Microbiol. 1992, 30, 3122–3126. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.J.; Wagner, B.; Woof, J.M. The different effector function capabilities of the seven equine IgG subclasses have implications for vaccine strategies. Mol. Immunol. 2008, 45, 818–827. [Google Scholar] [CrossRef]
- Mealey, R.H.; Kappmeyer, L.S.; Ueti, M.W.; Wagner, B.; Knowles, D.P. Protective effects of passively transferred merozoite-specific antibodies against Theileria equi in horses with severe combined immunodeficiency. Clin. Vaccine Immunol. 2012, 19, 100–104. [Google Scholar] [CrossRef] [Green Version]
- Butler, C.M.; Nijhof, A.M.; van der Kolk, J.H.; de Haseth, O.B.; Taoufik, A.; Jongejan, F.; Houwers, D.J. Repeated high dose imidocarb dipropionate treatment did not eliminate Babesia caballi from naturally infected horses as determined by PCR-reverse line blot hybridization. Vet. Parasitol. 2008, 151, 320–322. [Google Scholar] [CrossRef]
- Hines, S.A.; Ramsay, J.D.; Kappmeyer, L.S.; Lau, A.O.; Ojo, K.K.; van Voorhis, W.C.; Knowles, D.P.; Mealey, R.H. Theileria equi isolates vary in susceptibility to imidocarb dipropionate but demonstrate uniform in vitro susceptibility to a bumped kinase inhibitor. Parasites Vectors 2015, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Grause, J.F.; Ueti, M.W.; Nelson, J.T.; Knowles, D.P.; Kappmeyer, L.S.; Bunn, T.O. Efficacy of imidocarb dipropionate in eliminating Theileria equi from experimentally infected horses. Vet. J. 2013, 196, 541–546. [Google Scholar] [CrossRef]
- Schwint, O.N.; Ueti, M.W.; Palmer, G.H.; Kappmeyer, L.S.; Hines, M.T.; Cordes, R.T.; Knowles, D.P.; Scoles, G.A. Imidocarb dipropionate clears persistent Babesia caballi infection with elimination of transmission potential. Antimicrob. Agents Chemother. 2009, 53, 4327–4332. [Google Scholar] [CrossRef] [Green Version]
- Nugraha, A.B.; Tuvshintulga, B.; Guswanto, A.; Tayebwa, D.S.; Rizk, M.A.; Gantuya, S.; El-Saber Batiha, G.; Beshbishy, A.M.; Sivakumar, T.; Yokoyama, N.; et al. Screening the Medicines for Malaria Venture Pathogen Box against piroplasm parasites. Int. J. Parasitol. Drugs Drug Resist. 2019, 10, 84–90. [Google Scholar] [CrossRef]
- Nagai, A.; Yokoyama, N.; Matsuo, T.; Bork, S.; Hirata, H.; Xuan, X.; Zhu, Y.; Claveria, F.G.; Fujisaki, K.; Igarashi, I. Growth-inhibitory effects of artesunate, pyrimethamine, and pamaquine against Babesia equi and Babesia caballi in in vitro cultures. Antimicrob. Agents Chemother. 2003, 47, 800–803. [Google Scholar] [CrossRef] [Green Version]
- Rizk, M.A.; El-Sayed, S.A.E.; El-Khodery, S.; Yokoyama, N.; Igarashi, I. Discovering the in vitro potent inhibitors against Babesia and Theileria parasites by repurposing the Malaria Box: A review. Vet. Parasitol. 2019, 274, 108895. [Google Scholar] [CrossRef]
- Tuvshintulga, B.; AbouLaila, M.; Davaasuren, B.; Ishiyama, A.; Sivakumar, T.; Yokoyama, N.; Iwatsuki, M.; Otoguro, K.; Omura, S.; Igarashi, I. Clofazimine Inhibits the Growth of Babesia and Theileria Parasites In Vitro and In Vivo. Antimicrob. Agents Chemother. 2016, 60, 2739–2746. [Google Scholar] [CrossRef] [Green Version]
- Silva, M.G.; Villarino, N.F.; Knowles, D.P.; Suarez, C.E. Assessment of Draxxin((R)) (tulathromycin) as an inhibitor of in vitro growth of Babesia bovis, Babesia bigemina and Theileria equi. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Rizk, M.A.; El-Sayed, S.A.; AbouLaila, M.; Yokoyama, N.; Igarashi, I. Evaluation of the inhibitory effect of N-acetyl-L-cysteine on Babesia and Theileria parasites. Exp. Parasitol. 2017, 179, 43–48. [Google Scholar] [CrossRef]
- Omar, M.A.; Salama, A.; Elsify, A.; Rizk, M.A.; Al-Aboody, M.S.; Aboulaila, M.; El-Sayed, S.A.; Igarashi, I. Evaluation of in vitro inhibitory effect of enoxacin on Babesia and Theileria parasites. Exp. Parasitol. 2016, 161, 62–67. [Google Scholar] [CrossRef]
- Salama, A.A.; Aboulaila, M.; Moussa, A.A.; Nayel, M.A.; El-Sify, A.; Terkawi, M.A.; Hassan, H.Y.; Yokoyama, N.; Igarashi, I. Evaluation of in vitro and in vivo inhibitory effects of fusidic acid on Babesia and Theileria parasites. Vet. Parasitol. 2013, 191, 1–10. [Google Scholar] [CrossRef]
- Ikadai, H.; Tanaka, T.; Shibahara, N.; Tanaka, H.; Matsuu, A.; Kudo, N.; Shimazaki, K.; Igarashi, I.; Oyamada, T. Inhibitory effect of lactoferrin on in vitro growth of Babesia caballi. Am. J. Trop. Med. Hyg. 2005, 73, 710–712. [Google Scholar] [CrossRef]
- Maji, C.; Goel, P.; Suthar, A.; Mandal, K.D.; Gopalakrishnan, A.; Kumar, R.; Tripathi, B.N.; Kumar, S. Lumefantrine and o-choline—Parasite metabolism specific drug molecules inhibited in vitro growth of Theileria equi and Babesia caballi in MASP culture system. Tick Tick-Borne Dis. 2019, 10, 568–574. [Google Scholar] [CrossRef]
- Gimenez, F.; Hines, S.A.; Evanoff, R.; Ojo, K.K.; van Voorhis, W.C.; Maly, D.J.; Vidadala, R.S.R.; Mealey, R.H. In vitro growth inhibition of Theileria equi by bumped kinase inhibitors. Vet. Parasitol. 2018, 251, 90–94. [Google Scholar] [CrossRef]
- Silva, M.G.; Knowles, D.P.; Antunes, S.; Domingos, A.; Esteves, M.A.; Suarez, C.E. Inhibition of the in vitro growth of Babesia bigemina, Babesia caballi and Theileria equi parasites by trifluralin analogues. Tick Tick-Borne Dis. 2017, 8, 593–597. [Google Scholar] [CrossRef]
- Gopalakrishnan, A.; Maji, C.; Dahiya, R.K.; Suthar, A.; Kumar, R.; Gupta, A.K.; Dimri, U.; Kumar, S. In vitro growth inhibitory efficacy of some target specific novel drug molecules against Theileria equi. Vet. Parasitol. 2016, 217, 1–6. [Google Scholar] [CrossRef]
- AbouLaila, M.; Batadoj, D.; Salama, A.; Munkhjargal, T.; Ichikawa-Seki, M.; Terkawi, M.A.; Yokoyama, N.; Igarashi, I. Evaluation of the inhibitory effects of miltefosine on the growth of Babesia and Theileria parasites. Vet. Parasitol. 2014, 204, 104–110. [Google Scholar] [CrossRef]
- Aboulaila, M.; Munkhjargal, T.; Sivakumar, T.; Ueno, A.; Nakano, Y.; Yokoyama, M.; Yoshinari, T.; Nagano, D.; Katayama, K.; El-Bahy, N.; et al. Apicoplast-targeting antibacterials inhibit the growth of Babesia parasites. Antimicrob. Agents Chemother. 2012, 56, 3196–3206. [Google Scholar] [CrossRef]
- Wise, L.N.; Ueti, M.W.; Kappmeyer, L.S.; Hines, M.T.; White, S.N.; Davis, W.; Knowles, D.P. In vitro activity of ponazuril against Theileria equi. Vet. Parasitol. 2012, 185, 282–285. [Google Scholar] [CrossRef]
- Aboulaila, M.; Nakamura, K.; Govind, Y.; Yokoyama, N.; Igarashi, I. Evaluation of the in vitro growth-inhibitory effect of epoxomicin on Babesia parasites. Vet. Parasitol. 2010, 167, 19–27. [Google Scholar] [CrossRef]
- Tayebwa, D.S.; Tuvshintulga, B.; Guswanto, A.; Nugraha, A.B.; Batiha, G.E.; Gantuya, S.; Rizk, M.A.; Vudriko, P.; Sivakumar, T.; Yokoyama, N.; et al. The effects of nitidine chloride and camptothecin on the growth of Babesia and Theileria parasites. Tick Tick-Borne Dis. 2018, 9, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Guswanto, A.; Nugraha, A.B.; Tuvshintulga, B.; Tayebwa, D.S.; Rizk, M.A.; Batiha, G.E.; Gantuya, S.; Sivakumar, T.; Yokoyama, N.; Igarashi, I. 17-DMAG inhibits the multiplication of several Babesia species and Theileria equi on in vitro cultures, and Babesia microti in mice. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 104–111. [Google Scholar] [CrossRef]
- Boschi, D.; Pippione, A.C.; Sainas, S.; Lolli, M.L. Dihydroorotate dehydrogenase inhibitors in anti-infective drug research. Eur. J. Med. Chem. 2019, 183, 111681. [Google Scholar] [CrossRef] [PubMed]
- Kamyingkird, K.; Cao, S.; Tuvshintulga, B.; Salama, A.; Mousa, A.A.; Efstratiou, A.; Nishikawa, Y.; Yokoyama, N.; Igarashi, I.; Xuan, X. Effects of dihydroorotate dehydrogenase (DHODH) inhibitors on the growth of Theileria equi and Babesia caballi in vitro. Exp. Parasitol. 2017, 176, 59–65. [Google Scholar] [CrossRef]
- Beshbishy, A.M.; Batiha, G.E.; Yokoyama, N.; Igarashi, I. Ellagic acid microspheres restrict the growth of Babesia and Theileria in vitro and Babesia microti in vivo. Parasites Vectors 2019, 12, 269. [Google Scholar] [CrossRef] [Green Version]
- Batiha, G.E.; Beshbishy, A.M.; Tayebwa, D.S.; Shaheen, H.M.; Yokoyama, N.; Igarashi, I. Inhibitory effects of Syzygium aromaticum and Camellia sinensis methanolic extracts on the growth of Babesia and Theileria parasites. Tick Tick-Borne Dis. 2019, 10, 949–958. [Google Scholar] [CrossRef]
- Ganchimeg, D.; Batbold, B.; Murata, T.; Davaapurev, B.O.; Munkhjargal, T.; Tuvshintulga, B.; Suganuma, K.; Igarashi, I.; Buyankhishig, B.; Sasaki, K.; et al. Flavonoids isolated from the flowers of Pulsatilla flavescens and their anti-piroplasm activity. J. Nat. Med. 2019, 73, 633–640. [Google Scholar] [CrossRef]
- Badral, D.; Odonbayar, B.; Murata, T.; Munkhjargal, T.; Tuvshintulga, B.; Igarashi, I.; Suganuma, K.; Inoue, N.; Brantner, A.H.; Odontuya, G.; et al. Flavonoid and Galloyl Glycosides Isolated from Saxifraga spinulosa and Their Antioxidative and Inhibitory Activities against Species That Cause Piroplasmosis. J. Nat. Prod. 2017, 80, 2416–2423. [Google Scholar] [CrossRef]
- El-Sayed, S.A.E.; Rizk, M.A.; Yokoyama, N.; Igarashi, I. Evaluation of the in vitro and in vivo inhibitory effect of thymoquinone on piroplasm parasites. Parasites Vectors 2019, 12, 37. [Google Scholar] [CrossRef]
- Naidoo, V.; Zweygarth, E.; Eloff, J.N.; Swan, G.E. Identification of anti-babesial activity for four ethnoveterinary plants in vitro. Vet. Parasitol. 2005, 130, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Equine piroplasmosis visits Australia in 2000. Aust. Vet. J. 2000, 78, 380. [CrossRef]
- Martin, R. Equine piroplasmosis: The temporary importation of seropositive horses into Australia. Aust. Vet. J. 1999, 77, 308–309. [Google Scholar] [CrossRef]
- Bruning, A. Equine piroplasmosis an update on diagnosis, treatment and prevention. Br. Vet. J. 1996, 152, 139–151. [Google Scholar] [CrossRef]
- Bonfini, B.; Semproni, G.; Savini, G. Use of an in vitro culture system to detect Theileria equi strains from infected equids and/or reservoirs. Vet. Ital. 2006, 42, 209–215, 217–223. [Google Scholar]
- Mans, B.J.; Pienaar, R.; Latif, A.A. A review of Theileria diagnostics and epidemiology. Int. J. Parasitol. Parasites Wildl. 2015, 4, 104–118. [Google Scholar] [CrossRef] [Green Version]
- Zweygarth, E.; Josemans, A.I. L-cysteine replaces microaerophilous culture conditions for the in vitro initiation of Theileria equi. Parasitol. Res. 2014, 113, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Zweygarth, E.; Lopez-Rebollar, L.M.; Nurton, J.; Guthrie, A.J. Culture, isolation and propagation of Babesia caballi from naturally infected horses. Parasitol. Res. 2002, 88, 460–462. [Google Scholar] [CrossRef] [PubMed]
- Friedhoff, K.; Soule, C. An account on equine babesioses. Rev. Sci. Tech. 1996, 15, 1191. [Google Scholar] [CrossRef]
- Ogunremi, O.; Halbert, G.; Mainar-Jaime, R.; Benjamin, J.; Pfister, K.; Lopez-Rebollar, L.; Georgiadis, M.P. Accuracy of an indirect fluorescent-antibody test and of a complement-fixation test for the diagnosis of Babesia caballi in field samples from horses. Prev. Vet. Med. 2008, 83, 41–51. [Google Scholar] [CrossRef]
- Ogunremi, O.; Georgiadis, M.P.; Halbert, G.; Benjamin, J.; Pfister, K.; Lopez-Rebollar, L. Validation of the indirect fluorescent antibody and the complement fixation tests for the diagnosis of Theileria equi. Vet. Parasitol. 2007, 148, 102–108. [Google Scholar] [CrossRef]
- Asenzo, G.; Wilkowsky, S.; Barrandeguy, M.; Mesplet, M.; Benitez, D.; Florin-Christensen, M. Development of an indirect ELISA for the diagnosis of equine piroplasmosis. Ann. N. Y. Acad. Sci. 2008, 1149, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Baldani, C.D.; Nakaghi, A.C.; Machado, R.Z. Occurrence of Theileria equi in horses raised in the Jaboticabal microregion, Sao Paulo State, Brazil. Rev. Bras. Parasitol. Vet. 2010, 19, 228–232. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, R.; Gupta, A.K.; Yadav, S.C.; Goyal, S.K.; Khurana, S.K.; Singh, R.K. Development of EMA-2 recombinant antigen based enzyme-linked immunosorbent assay for seroprevalence studies of Theileria equi infection in Indian equine population. Vet. Parasitol. 2013, 198, 10–17. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, Y.; Malhotra, D.V.; Dhar, S.; Nichani, A.K. Standardisation and comparison of serial dilution and single dilution enzyme linked immunosorbent assay (ELISA) using different antigenic preparations of the Babesia (Theileria) equi parasite. Vet. Res. 2003, 34, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Kappmeyer, L.S.; Perryman, L.E.; Hines, S.A.; Baszler, T.V.; Katz, J.B.; Hennager, S.G.; Knowles, D.P. Detection of equine antibodies to Babesia caballi by recombinant B. caballi rhoptry-associated protein 1 in a competitive-inhibition enzyme-linked immunosorbent assay. J. Clin. Microbiol. 1999, 37, 2285–2290. [Google Scholar] [CrossRef] [Green Version]
- Baldani, C.D.; Hilario, E.; Nakaghi, A.C.; Bertolini, M.C.; Machado, R.Z. Production of recombinant EMA-1 protein and its application for the diagnosis of Theileria equi using an enzyme immunoassay in horses from Sao Paulo State, Brazil. Rev. Bras. Parasitol. Vet. 2011, 20, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, J.; Song, R.; Zhang, M.; Wu, L.; Li, F.; Yan, Y.; Zhou, J.; Chahan, B.; Liao, M. Preparation of monoclonal antibodies against Bc48 and development of a rapid detection assay for infection with Babesia caballi in China. Folia Parasitol. 2019, 66. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.T.; Wang, Z.B.; Bolati; Li, H.; Bayinchahan. A Duplex PCR Method for Detection of Babesia caballi and Theileria equi. Chin. J. Parasitol. Parasit. Dis. 2015, 33, 105–109. [Google Scholar]
- Bhoora, R.; Quan, M.; Zweygarth, E.; Guthrie, A.J.; Prinsloo, S.A.; Collins, N.E. Sequence heterogeneity in the gene encoding the rhoptry-associated protein-1 (RAP-1) of Babesia caballi isolates from South Africa. Vet. Parasitol. 2010, 169, 279–288. [Google Scholar] [CrossRef]
- Mahmoud, M.S.; El-Ezz, N.T.; Abdel-Shafy, S.; Nassar, S.A.; El Namaky, A.H.; Khalil, W.K.; Knowles, D.; Kappmeyer, L.; Silva, M.G.; Suarez, C.E. Assessment of Theileria equi and Babesia caballi infections in equine populations in Egypt by molecular, serological and hematological approaches. Parasites Vectors 2016, 9, 260. [Google Scholar] [CrossRef] [Green Version]
- Rapoport, A.; Aharonson-Raz, K.; Berlin, D.; Tal, S.; Gottlieb, Y.; Klement, E.; Steinman, A. Molecular characterization of the Babesia caballi rap-1 gene and epidemiological survey in horses in Israel. Infect. Genet. Evol. 2014, 23, 115–120. [Google Scholar] [CrossRef]
- Alhassan, A.; Govind, Y.; Tam, N.T.; Thekisoe, O.M.; Yokoyama, N.; Inoue, N.; Igarashi, I. Comparative evaluation of the sensitivity of LAMP, PCR and in vitro culture methods for the diagnosis of equine piroplasmosis. Parasitol. Res. 2007, 100, 1165–1168. [Google Scholar] [CrossRef]
- Alhassan, A.; Iseki, H.; Kim, C.; Yokoyama, N.; Igarashi, I. Comparison of polymerase chain reaction methods for the detection of Theileria equi infection using whole blood compared with pre-extracted DNA samples as PCR templates. Trop. Anim. Health Prod. 2007, 39, 369–374. [Google Scholar] [CrossRef]
- Alhassan, A.; Pumidonming, W.; Okamura, M.; Hirata, H.; Battsetseg, B.; Fujisaki, K.; Yokoyama, N.; Igarashi, I. Development of a single-round and multiplex PCR method for the simultaneous detection of Babesia caballi and Babesia equi in horse blood. Vet. Parasitol. 2005, 129, 43–49. [Google Scholar] [CrossRef]
- Alhassan, A.; Thekisoe, O.M.; Yokoyama, N.; Inoue, N.; Motloang, M.Y.; Mbati, P.A.; Yin, H.; Katayama, Y.; Anzai, T.; Sugimoto, C.; et al. Development of loop-mediated isothermal amplification (LAMP) method for diagnosis of equine piroplasmosis. Vet. Parasitol. 2007, 143, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Bhoora, R.V.; Pienaar, R.; Cornelius, F.; Josemans, A.; Matthee, O.; Marumo, R.; Troskie, C.; Mans, B.J. Multiplex hydrolysis-probe assay for the simultaneous detection of Theileria equi and Babesia caballi infections in equids. Vet. Parasitol. 2018, 255, 61–68. [Google Scholar] [CrossRef]
- Montes Cortes, M.G.; Fernandez-Garcia, J.L.; Habela Martinez-Estellez, M.A. A multinested PCR for detection of the equine piroplasmids Babesia caballi and Theileria equi. Tick Tick-Borne Dis. 2019, 10, 305–313. [Google Scholar] [CrossRef]
- Xie, J.; Liu, G.; Tian, Z.; Luo, J. Development of loop-mediated isothermal amplification (LAMP) for detection of Theileria equi. Acta Trop. 2013, 127, 245–250. [Google Scholar] [CrossRef]
- Salim, B.; Bakheit, M.A.; Sugimoto, C. Rapid detection and identification of Theileria equi and Babesia caballi by high-resolution melting (HRM) analysis. Parasitol. Res. 2013, 112, 3883–3886. [Google Scholar] [CrossRef]
- Nicolaiewsky, T.B.; Richter, M.F.; Lunge, V.R.; Cunha, C.W.; Delagostin, O.; Ikuta, N.; Fonseca, A.S.; da Silva, S.S.; Ozaki, L.S. Detection of Babesia equi (Laveran, 1901) by nested polymerase chain reaction. Vet. Parasitol. 2001, 101, 9–21. [Google Scholar] [CrossRef]
- Bhoora, R.; Quan, M.; Franssen, L.; Butler, C.M.; van der Kolk, J.H.; Guthrie, A.J.; Zweygarth, E.; Jongejan, F.; Collins, N.E. Development and evaluation of real-time PCR assays for the quantitative detection of Babesia caballi and Theileria equi infections in horses from South Africa. Vet. Parasitol. 2010, 168, 201–211. [Google Scholar] [CrossRef]
- Bhoora, R.; Quan, M.; Matjila, P.T.; Zweygarth, E.; Guthrie, A.J.; Collins, N.E. Sequence heterogeneity in the equi merozoite antigen gene (ema-1) of Theileria equi and development of an ema-1-specific TaqMan MGB assay for the detection of T. equi. Vet. Parasitol. 2010, 172, 33–45. [Google Scholar] [CrossRef]
- Ueti, M.W.; Palmer, G.H.; Kappmeyer, L.S.; Scoles, G.A.; Knowles, D.P. Expression of equi merozoite antigen 2 during development of Babesia equi in the midgut and salivary gland of the vector tick Boophilus microplus. J. Clin. Microbiol. 2003, 41, 5803–5809. [Google Scholar] [CrossRef] [Green Version]
- Lobanov, V.A.; Peckle, M.; Massard, C.L.; Brad Scandrett, W.; Gajadhar, A.A. Development and validation of a duplex real-time PCR assay for the diagnosis of equine piroplasmosis. Parasites Vectors 2018, 11, 125. [Google Scholar] [CrossRef] [Green Version]
- Alanazi, A.D.; Said, A.E.; Morin-Adeline, V.; Alyousif, M.S.; Slapeta, J. Quantitative PCR detection of Theileria equi using laboratory workflows to detect asymptomatic persistently infected horses. Vet. Parasitol. 2014, 206, 138–145. [Google Scholar] [CrossRef]
- Kim, C.M.; Blanco, L.B.; Alhassan, A.; Iseki, H.; Yokoyama, N.; Xuan, X.; Igarashi, I. Diagnostic real-time PCR assay for the quantitative detection of Theileria equi from equine blood samples. Vet. Parasitol. 2008, 151, 158–163. [Google Scholar] [CrossRef]
- Bhoora, R.; Franssen, L.; Oosthuizen, M.C.; Guthrie, A.J.; Zweygarth, E.; Penzhorn, B.L.; Jongejan, F.; Collins, N.E. Sequence heterogeneity in the 18S rRNA gene within Theileria equi and Babesia caballi from horses in South Africa. Vet. Parasitol. 2009, 159, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Gubbels, J.M.; de Vos, A.P.; van der Weide, M.; Viseras, J.; Schouls, L.M.; de Vries, E.; Jongejan, F. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 1999, 37, 1782–1789. [Google Scholar] [CrossRef] [Green Version]
- Bhoora, R.V.; Collins, N.E.; Schnittger, L.; Troskie, C.; Marumo, R.; Labuschagne, K.; Smith, R.M.; Dalton, D.L.; Mbizeni, S. Molecular genotyping and epidemiology of equine piroplasmids in South Africa. Tick Tick-Borne Dis. 2020, 11, 101358. [Google Scholar] [CrossRef]
- Al-Obaidi, Q.; Mohd, M.; Al-Sultan, I.; Azlinda, A.; Mohd, A. Equine piroplasmosis in Kelantan, Malaysia: Clinico-hemato-biochemical alterations in subclinically and clinically infected equids. Trop. Biomed. 2016, 33, 619–631. [Google Scholar]
- Jaffer, O.; Abdishakur, F.; Hakimuddin, F.; Riya, A.; Wernery, U.; Schuster, R.K. A comparative study of serological tests and PCR for the diagnosis of equine piroplasmosis. Parasitol. Res. 2010, 106, 709–713. [Google Scholar] [CrossRef]
- Kamyingkird, K.; Yangtara, S.; Desquesnes, M.; Cao, S.; Moumouni, A.; Jittapalapong, S.; Nimsupan, B.; Terkawi, M.; Masatani, T.; Nishikawa, Y. Seroprevalence of Babesia caballi and Theileria equi in horses and mules from Northern Thailand. J. Protozool. Res. 2016, 24, 11–17. [Google Scholar]
- Rhalem, A.; Sahibi, H.; Lasri, S.; Johnson, W.C.; Kappmeyer, L.S.; Hamidouch, A.; Knowles, D.P.; Goff, W.L. Validation of a competitive enzyme-linked immunosorbent assay for diagnosing Babesia equi infections of Moroccan origin and its use in determining the seroprevalence of B. equi in Morocco. J. Vet. Diagn. Investig. 2001, 13, 249–251. [Google Scholar] [CrossRef] [Green Version]
- Manna, G.; Cersini, A.; Nardini, R.; Bartolome Del Pino, L.E.; Antognetti, V.; Zini, M.; Conti, R.; Lorenzetti, R.; Veneziano, V.; Autorino, G.L.; et al. Genetic diversity of Theileria equi and Babesia caballi infecting horses of Central-Southern Italy and preliminary results of its correlation with clinical and serological status. Tick Tick-Borne Dis. 2018, 9, 1212–1220. [Google Scholar] [CrossRef]
- Del Pino, L.E.B.; Nardini, R.; Veneziano, V.; Iacoponi, F.; Cersini, A.; Autorino, G.L.; Buono, F.; Scicluna, M. Babesia caballi and Theileria equi infections in horses in Central-Southern Italy: Sero-molecular survey and associated risk factors. Tick Tick-Borne Dis. 2016, 7, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Sumbria, D.; das Singla, L.; Sharma, A. Theileria equi and Babesia caballi infection of equids in Punjab, India: A serological and molecular survey. Trop. Anim. Health Prod. 2016, 48, 45–52. [Google Scholar] [CrossRef]
- Posada-Guzman, M.F.; Dolz, G.; Romero-Zuniga, J.J.; Jimenez-Rocha, A.E. Detection of Babesia caballi and Theileria equi in Blood from Equines from Four Indigenous Communities in Costa Rica. Vet. Med. Int. 2015, 2015, 236278. [Google Scholar] [CrossRef]
- Butler, C.M.; Sloet van Oldruitenborgh-Oosterbaan, M.M.; Stout, T.A.; van der Kolk, J.H.; Wollenberg, L.; Nielen, M.; Jongejan, F.; Werners, A.H.; Houwers, D.J. Prevalence of the causative agents of equine piroplasmosis in the South West of The Netherlands and the identification of two autochthonous clinical Theileria equi infections. Vet. J. 2012, 193, 381–385. [Google Scholar] [CrossRef] [Green Version]
- Munkhjargal, T.; Sivakumar, T.; Battsetseg, B.; Nyamjargal, T.; Aboulaila, M.; Purevtseren, B.; Bayarsaikhan, D.; Byambaa, B.; Terkawi, M.A.; Yokoyama, N.; et al. Prevalence and genetic diversity of equine piroplasms in Tov province, Mongolia. Infect. Genet. Evol. 2013, 16, 178–185. [Google Scholar] [CrossRef]
- Ruegg, S.R.; Torgerson, P.; Deplazes, P.; Mathis, A. Age-dependent dynamics of Theileria equi and Babesia caballi infections in southwest Mongolia based on IFAT and/or PCR prevalence data from domestic horses and ticks. Parasitology 2007, 134, 939–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, M.G.; Yun, S.H.; Choi, S.K.; Cho, G.J.; Park, Y.S.; Kwon, O.D.; Cho, K.H.; Kim, T.H.; Jeong, K.S.; Park, S.J.; et al. Seroprevalence of equine piroplasms in the Republic of Korea. Vet. Parasitol. 2011, 179, 224–226. [Google Scholar] [CrossRef]
- Abutarbush, S.M.; Alqawasmeh, D.M.; Mukbel, R.M.; Al-Majali, A.M. Equine babesiosis: Seroprevalence, risk factors and comparison of different diagnostic methods in Jordan. Transbound. Emerg. Dis. 2012, 59, 72–78. [Google Scholar] [CrossRef]
- Grandi, G.; Molinari, G.; Tittarelli, M.; Sassera, D.; Kramer, L.H. Prevalence of Theileria equi and Babesia caballi infection in horses from northern Italy. Vector Borne Zoonotic Dis. 2011, 11, 955–956. [Google Scholar] [CrossRef]
- Moretti, A.; Mangili, V.; Salvatori, R.; Maresca, C.; Scoccia, E.; Torina, A.; Moretta, I.; Gabrielli, S.; Tampieri, M.P.; Pietrobelli, M. Prevalence and diagnosis of Babesia and Theileria infections in horses in Italy: A preliminary study. Vet. J. 2010, 184, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Abedi, V.; Razmi, G.; Seifi, H.; Naghibi, A. Molecular and serological detection of Theileria equi and Babesia caballi infection in horses and ixodid ticks in Iran. Ticks Tick-Borne Dis. 2014, 5, 239–244. [Google Scholar] [CrossRef]
- Laus, F.; Veronesi, F.; Passamonti, F.; Paggi, E.; Cerquetella, M.; Hyatt, D.; Tesei, B.; Fioretti, D.P. Prevalence of tick borne pathogens in horses from Italy. J. Vet. Med. Sci. 2013, 75, 715–720. [Google Scholar] [CrossRef] [Green Version]
- Kizilarslan, F.; Yildirim, A.; Duzlu, O.; Inci, A.; Onder, Z.; Ciloglu, A. Molecular detection and characterization of Theileria equi and Babesia caballi in horses (Equus ferus caballus) in Turkey. J. Equine Vet. Sci. 2015, 35, 830–835. [Google Scholar] [CrossRef]
- Sgorbini, M.; Bonelli, F.; Nardoni, S.; Rocchigiani, G.; Corazza, M.; Mancianti, F. Seroprevalence and molecular analysis of Babesia caballi and Theileria equi in horses from central Italy during a 10-year period. J. Equine Vet. Sci. 2015, 35, 865–868. [Google Scholar] [CrossRef]
- Wise, L.N.; Kappmeyer, L.S.; Silva, M.G.; White, S.N.; Grause, J.F.; Knowles, D.P. Verification of post-chemotherapeutic clearance of Theileria equi through concordance of nested PCR and immunoblot. Tick Tick-Borne Dis. 2018, 9, 135–140. [Google Scholar] [CrossRef]
- Guidi, E.; Pradier, S.; Lebert, I.; Leblond, A. Piroplasmosis in an endemic area: Analysis of the risk factors and their implications in the control of Theileriosis and Babesiosis in horses. Parasitol. Res. 2015, 114, 71–83. [Google Scholar] [CrossRef]
- Ruegg, S.R.; Heinzmann, D.; Barbour, A.D.; Torgerson, P.R. Estimation of the transmission dynamics of Theileria equi and Babesia caballi in horses. Parasitology 2008, 135, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Tirosh-Levy, S.; Gottlieb, Y.; Mazuz, M.L.; Savisky, I.; Steinman, A. Infection dynamics of Theileria equi in carrier horses is associated with management and tick exposure. Tick Tick-Borne Dis. 2020, 101508. [Google Scholar] [CrossRef]
- Coultous, R.M.; Phipps, P.; Dalley, C.; Lewis, J.; Hammond, T.A.; Shiels, B.R.; Weir, W.; Sutton, D.G.M. Equine piroplasmosis status in the UK: An assessment of laboratory diagnostic submissions and techniques. Vet. Rec. 2019, 184, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baptista, C.; Lopes, M.S.; Tavares, A.C.; Rojer, H.; Kappmeyer, L.; Mendonca, D.; da Camara Machado, A. Diagnosis of Theileria equi infections in horses in the Azores using cELISA and nested PCR. Tick Tick-Borne Dis. 2013, 4, 242–245. [Google Scholar] [CrossRef]
- Davitkov, D.; Vucicevic, M.; Stevanovic, J.; Krstic, V.; Slijepcevic, D.; Glavinic, U.; Stanimirovic, Z. Molecular detection and prevalence of Theileria equi and Babesia caballi in horses of central Balkan. Acta Parasitol. 2016, 61, 337–342. [Google Scholar] [CrossRef]
- Xuan, X.; Nagai, A.; Battsetseg, B.; Fukumoto, S.; Makala, L.H.; Inoue, N.; Igarashi, I.; Mikami, T.; Fujisaki, K. Diagnosis of equine piroplasmosis in Brazil by serodiagnostic methods with recombinant antigens. J. Vet. Med. Sci. 2001, 63, 1159–1160. [Google Scholar] [CrossRef] [Green Version]
- Heim, A.; Passos, L.M.; Ribeiro, M.F.; Costa-Junior, L.M.; Bastos, C.V.; Cabral, D.D.; Hirzmann, J.; Pfister, K. Detection and molecular characterization of Babesia caballi and Theileria equi isolates from endemic areas of Brazil. Parasitol. Res. 2007, 102, 63–68. [Google Scholar] [CrossRef]
- Kerber, C.E.; Labruna, M.B.; Ferreira, F.; de Waal, D.T.; Knowles, D.P.; Gennari, S.M. Prevalence of equine Piroplasmosis and its association with tick infestation in the State of Sao Paulo, Brazil. Rev. Bras. Parasitol. Vet. 2009, 18, 1–8. [Google Scholar] [CrossRef]
- Dos Santos, T.M.; Roier, E.C.; Santos, H.A.; Pires, M.S.; Vilela, J.A.; Moraes, L.M.; Almeida, F.Q.; Baldani, C.D.; Machado, R.Z.; Massard, C.L. Factors associated to Theileria equi in equids of two microregions from Rio de Janeiro, Brazil. Rev. Bras. Parasitol. Vet. 2011, 20, 235–241. [Google Scholar] [CrossRef]
- Peckle, M.; Pires, M.S.; Dos Santos, T.M.; Roier, E.C.; da Silva, C.B.; Vilela, J.A.; Santos, H.A.; Massard, C.L. Molecular epidemiology of Theileria equi in horses and their association with possible tick vectors in the state of Rio de Janeiro, Brazil. Parasitol. Res. 2013, 112, 2017–2025. [Google Scholar] [CrossRef] [Green Version]
- Vieira, T.S.; Vieira, R.F.; Finger, M.A.; Nascimento, D.A.; Sicupira, P.M.; Dutra, L.H.; Deconto, I.; Barros-Filho, I.R.; Dornbusch, P.T.; Biondo, A.W.; et al. Seroepidemiological survey of Theileria equi and Babesia caballi in horses from a rural and from urban areas of Parana State, southern Brazil. Tick Tick-Borne Dis. 2013, 4, 537–541. [Google Scholar] [CrossRef]
- Prochno, H.C.; Scorsin, L.M.; de Melo, F.R.; Baldani, C.D.; Falbo, M.K.; de Aquino, L.C.; Lemos, K.R. Seroprevalence rates of antibodies against Theileria equi in team roping horses from central-western region of Parana. Rev. Bras. Parasitol. Vet. 2014, 23, 85–89. [Google Scholar] [CrossRef]
- Braga, M.; Costa, F.N.; Gomes, D.R.M.; Xavier, D.R.; Andre, M.R.; Goncalves, L.R.; Freschi, C.R.; Machado, R.Z. Genetic diversity of piroplasmids species in equids from island of Sao Luis, northeastern Brazil. Rev. Bras. Parasitol. Vet. 2017, 26, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Costa, S.C.L.; Freitas, J.S.; Silva, A.N.D.; Lacerda, L.C.; Cruz, R.D.S.; Carvalho, F.S.; Pereira, M.J.S.; Munhoz, A.D. Frequency and factors associated with Theileria equi, Babesia caballi and Trypanosoma evansi in equids from Bahia (Northeast Brazil). Rev. Bras. Parasitol. Vet. 2019, 28, 47–58. [Google Scholar] [CrossRef]
- Minervino, A.H.H.; Torres, A.C.; Moreira, T.R.; Vinholte, B.P.; Sampaio, B.M.; Bianchi, D.; Portela, J.M.; Sarturi, C.; Marcili, A.; Barreto Junior, R.A.; et al. Factors associated with the prevalence of antibodies against Theileria equi in equids of Western Para, Brazil. Transbound. Emerg. Dis. 2020, 67, 100–105. [Google Scholar] [CrossRef]
- Campos, J.B.V.; Andre, M.R.; Goncalves, L.R.; Freschi, C.R.; Santos, F.M.; de Oliveira, C.E.; Piranda, E.M.; de Andrade, G.B.; Macedo, G.C.; Machado, R.Z.; et al. Assessment of equine piroplasmids in the Nhecolandia sub-region of Brazilian Pantanal wetland using serological, parasitological, molecular, and hematological approaches. Tick Tick-Borne Dis. 2019, 10, 714–721. [Google Scholar] [CrossRef]
- Dahmana, H.; Amanzougaghene, N.; Davoust, B.; Normand, T.; Carette, O.; Demoncheaux, J.P.; Mulot, B.; Fabrizy, B.; Scandola, P.; Chik, M.; et al. Great diversity of Piroplasmida in Equidae in Africa and Europe, including potential new species. Vet. Parasitol. Reg. Stud. Rep. 2019, 18, 100332. [Google Scholar] [CrossRef]
- Xuan, X.; Chahan, B.; Huang, X.; Yokoyama, N.; Makala, L.H.; Igarashi, I.; Fujisaki, K.; Maruyama, S.; Sakai, T.; Mikami, T. Diagnosis of equine piroplasmosis in Xinjiang province of China by the enzyme-linked immunosorbent assays using recombinant antigens. Vet. Parasitol. 2002, 108, 179–182. [Google Scholar] [CrossRef]
- Wang, M.; Guo, W.; Igarashi, I.; Xuan, X.; Wang, X.; Xiang, W.; Jia, H. Epidemiological investigation of equine piroplasmosis in China by enzyme-linked immunosorbent assays. J. Vet. Med. Sci. 2014, 76, 549–552. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chahan, B.; Liu, S.; Song, R.; Li, Y.; Huercha; Guo, Q.; Wu, H.; Zhu, Y. Epidemiologic studies on Theileria equi infections for grazing horses in Ili of Xinjiang province. Vet. Parasitol. 2017, 244, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, J.; Yang, J.; Wang, X.; Li, Z.; Jianlin, X.; Li, X.; Xiang, Q.; Li, Y.; Liu, Z.; et al. The first molecular detection and genetic diversity of Babesia caballi and Theileria equi in horses of Gansu province, China. Tick Tick-Borne Dis. 2019, 10, 528–532. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Moumouni, P.F.A.; Lee, S.H.; Galon, E.M.; Tumwebaze, M.A.; Yang, H.; Huercha; Liu, M.; Guo, H.; et al. First description of Coxiella burnetii and Rickettsia spp. infection and molecular detection of piroplasma co-infecting horses in Xinjiang Uygur Autonomous Region, China. Parasitol. Int. 2020, 76, 102028. [Google Scholar] [CrossRef]
- Diaz-Sanchez, A.A.; Pires, M.S.; Estrada, C.Y.; Canizares, E.V.; Del Castillo Dominguez, S.L.; Cabezas-Cruz, A.; Rivero, E.L.; da Fonseca, A.H.; Massard, C.L.; Corona-Gonzalez, B. First molecular evidence of Babesia caballi and Theileria equi infections in horses in Cuba. Parasitol. Res. 2018, 117, 3109–3118. [Google Scholar] [CrossRef] [PubMed]
- Fritz, D. A PCR study of piroplasms in 166 dogs and 111 horses in France (March 2006 to March 2008). Parasitol. Res. 2010, 106, 1339–1342. [Google Scholar] [CrossRef] [PubMed]
- Kouam, M.K.; Kantzoura, V.; Gajadhar, A.A.; Theis, J.H.; Papadopoulos, E.; Theodoropoulos, G. Seroprevalence of equine piroplasms and host-related factors associated with infection in Greece. Vet. Parasitol. 2010, 169, 273–278. [Google Scholar] [CrossRef]
- Kouam, M.K.; Kantzoura, V.; Masuoka, P.M.; Gajadhar, A.A.; Theodoropoulos, G. Genetic diversity of equine piroplasms in Greece with a note on speciation within Theileria genotypes (T. equi and T. equi-like). Infect. Genet. Evol. 2010, 10, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Teglas, M.; Matern, E.; Lein, S.; Foley, P.; Mahan, S.M.; Foley, J. Ticks and tick-borne disease in Guatemalan cattle and horses. Vet. Parasitol. 2005, 131, 119–127. [Google Scholar] [CrossRef]
- Farkas, R.; Tanczos, B.; Gyurkovszky, M.; Foldvari, G.; Solymosi, N.; Edelhofer, R.; Hornok, S. Serological and molecular detection of Theileria equi infection in horses in Hungary. Vet. Parasitol. 2013, 192, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Nugraha, A.B.; Cahyaningsih, U.; Amrozi, A.; Ridwan, Y.; Agungpriyono, S.; Taher, D.M.; Guswanto, A.; Gantuya, S.; Tayebwa, D.S.; Tuvshintulga, B.; et al. Serological and molecular prevalence of equine piroplasmosis in Western Java, Indonesia. Vet. Parasitol. Reg. Stud. Rep. 2018, 14, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Malekifard, F.; Tavassoli, M.; Yakhchali, M.; Darvishzadeh, R. Detection of Theileria equi and Babesia caballi using microscopic and molecular methods in horses in suburb of Urmia, Iran. Vet. Res. Forum 2014, 5, 129–133. [Google Scholar] [PubMed]
- Bahrami, S.; Ghadrdan, A.; Mirabdollahi, S.; Fayed, M. Diagnosis of subclinical equine theileriosis in center of Iran using parasitological and molecular methods. Trop. Biomed. 2014, 31, 110–117. [Google Scholar]
- Habibi, G.; Esmaeilnia, K.; Hablolvarid, M.H.; Afshari, A.; Zamen, M.; Bozorgi, S. Microscopic and Molecular Detection of Theileria (Babesia) Equi Infection in Equids of Kurdistan Province, Iran. Iran. J. Parasitol. 2016, 11, 86–90. [Google Scholar]
- Ebrahimi, M.; Adinehbeigi, K.; Hamidinejat, H.; Tabandeh, M.R. Molecular characterization of Theileria equi infection in horse populations belonging to West Azerbaijan, Iran: Insights into the importance of Equine Merozoite Antigen (EMA)-1 in its diagnosis. Ann. Parasitol. 2018, 64, 21–27. [Google Scholar] [CrossRef]
- Aharonson-Raz, K.; Rapoport, A.; Hawari, I.M.; Lensky, I.M.; Berlin, D.; Zivotofsky, D.; Klement, E.; Steinman, A. Novel description of force of infection and risk factors associated with Theileria equi in horses in Israel and in The Palestinian Authority. Tick Tick-Borne Dis. 2014, 5, 366–372. [Google Scholar] [CrossRef]
- Steinman, A.; Zimmerman, T.; Klement, E.; Lensky, I.M.; Berlin, D.; Gottlieb, Y.; Baneth, G. Demographic and environmental risk factors for infection by Theileria equi in 590 horses in Israel. Vet. Parasitol. 2012, 187, 558–562. [Google Scholar] [CrossRef]
- Ebani, V.V.; Nardoni, S.; Bertelloni, F.; Rocchigiani, G.; Mancianti, F. Tick-borne infections in horses from Tuscany, Italy. J. Equine Vet. Sci. 2015, 35, 290–294. [Google Scholar] [CrossRef]
- Zanet, S.; Bassano, M.; Trisciuoglio, A.; Taricco, I.; Ferroglio, E. Horses infected by Piroplasms different from Babesia caballi and Theileria equi: Species identification and risk factors analysis in Italy. Vet. Parasitol. 2017, 236, 38–41. [Google Scholar] [CrossRef]
- Ikadai, H.; Nagai, A.; Xuan, X.; Igarashi, I.; Tsugihiko, K.; Tsuji, N.; Oyamada, T.; Suzuki, N.; Fujisaki, K. Seroepidemiologic studies on Babesia caballi and Babesia equi infections in Japan. J. Vet. Med. Sci. 2002, 64, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Qablan, M.A.; Obornik, M.; Petrzelkova, K.J.; Sloboda, M.; Shudiefat, M.F.; Horin, P.; Lukes, J.; Modry, D. Infections by Babesia caballi and Theileria equi in Jordanian equids: Epidemiology and genetic diversity. Parasitology 2013, 140, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.G.; Yun, S.H.; Choi, S.K.; Cho, G.J.; Park, Y.S.; Cho, K.H.; Kwon, O.D.; Kwak, D. Molecular and phylogenetic analysis of equine piroplasms in the Republic of Korea. Res. Vet. Sci. 2013, 94, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Al-Obaidi, Q.; Arshad, M.; Al-Sultan, I.; Azlinda, A.; Mohd-Azam, K. Comparison between microscopic examination and competitive ELISA for diagnosis of equine piroplasmosis in Kelantan, Malaysia. Malays. J. Vet. Res. 2016, 7, 23–29. [Google Scholar]
- Cantu-Martinez, M.A.; Segura-Correa, J.C.; Silva-Paez, M.L.; Avalos-Ramirez, R.; Wagner, G.G. Prevalence of antibodies to Theileria equi and Babesia caballi in horses from northeastern Mexico. J. Parasitol. 2012, 98, 869–870. [Google Scholar] [CrossRef]
- Ayala-Valdovinos, M.A.; Lemus-Flores, C.; Galindo-Garcia, J.; Banuelos-Pineda, J.; Rodriguez-Carpena, J.G.; Sanchez-Chipres, D.; Duifhuis-Rivera, T. Diagnosis and prevalence of Theileria equi horses in western Mexico by nested PCR. Parasitol. Int. 2017, 66, 821–824. [Google Scholar] [CrossRef]
- Boldbaatar, D.; Xuan, X.; Battsetseg, B.; Igarashi, I.; Battur, B.; Batsukh, Z.; Bayambaa, B.; Fujisaki, K. Epidemiological study of equine piroplasmosis in Mongolia. Vet. Parasitol. 2005, 127, 29–32. [Google Scholar] [CrossRef]
- Sloboda, M.; Jirku, M.; Lukesova, D.; Qablan, M.; Batsukh, Z.; Fiala, I.; Horin, P.; Modry, D.; Lukes, J. A survey for piroplasmids in horses and Bactrian camels in North-Eastern Mongolia. Vet. Parasitol. 2011, 179, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Myagmarsuren, P.; Sivakumar, T.; Enkhtaivan, B.; Davaasuren, B.; Zoljargal, M.; Narantsatsral, S.; Davkharbayar, B.; Mungun-Ochir, B.; Battur, B.; Inoue, N.; et al. A Seroepidemiological Survey of Theileria equi and Babesia caballi in Horses in Mongolia. J. Parasitol. 2019, 105, 580–586. [Google Scholar] [CrossRef]
- Tyrrell, J.D.; Qurollo, B.A.; Tornquist, S.J.; Schlaich, K.G.; Kelsey, J.; Chandrashekar, R.; Breitschwerdt, E.B. Molecular identification of vector-borne organisms in Ehrlichia seropositive Nicaraguan horses and first report of Rickettsia felis infection in the horse. Acta Trop. 2019, 200, 105170. [Google Scholar] [CrossRef]
- Mshelia, P.W.; Kappmeyer, L.; Johnson, W.C.; Kudi, C.A.; Oluyinka, O.O.; Balogun, E.O.; Richard, E.E.; Onoja, E.; Sears, K.P.; Ueti, M.W. Molecular detection of Theileria species and Babesia caballi from horses in Nigeria. Parasitol. Res. 2020, 119, 2955–2963. [Google Scholar] [CrossRef]
- Hussain, M.H.; Saqib, M.; Raza, F.; Muhammad, G.; Asi, M.N.; Mansoor, M.K.; Saleem, M.; Jabbar, A. Seroprevalence of Babesia caballi and Theileria equi in five draught equine populated metropolises of Punjab, Pakistan. Vet. Parasitol. 2014, 202, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Ybanez, A.P.; Ybanez, R.H.D.; Talle, M.G.; Arreglo, R.M.T.; Geens, M.J.C.; Villas, J.G.I., III; Villar, S.R.; Laruga, C.L.; Cao, S.; Moumouni, F.P.A.; et al. Serological and molecular detection of Theileria equi and Babesia caballi in Philippine horses. Tick Tick-Borne Dis. 2018, 9, 1125–1128. [Google Scholar] [CrossRef]
- Slivinska, K.; Vichova, B.; Werszko, J.; Szewczyk, T.; Wroblewski, Z.; Petko, B.; Ragac, O.; Demeshkant, V.; Karbowiak, G. Molecular surveillance of Theileria equi and Anaplasma phagocytophilum infections in horses from Ukraine, Poland and Slovakia. Vet. Parasitol. 2016, 215, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.J.; Cardoso, L.; Maia, J.M.; Coutinho, T.; Cotovio, M. Prevalence of Theileria equi, Babesia caballi, and Anaplasma phagocytophilum in horses from the north of Portugal. Parasitol. Res. 2013, 112, 2611–2617. [Google Scholar] [CrossRef]
- Gallusova, M.; Qablan, M.A.; D’Amico, G.; Obornik, M.; Petrzelkova, K.J.; Mihalca, A.D.; Modry, D. Piroplasms in feral and domestic equines in rural areas of the Danube Delta, Romania, with survey of dogs as a possible reservoir. Vet. Parasitol. 2014, 206, 287–292. [Google Scholar] [CrossRef]
- Alanazi, A.D.; Alyousif, M.S.; Hassieb, M.M. Seroprevalence study on Theileria equi and Babesia caballi antibodies in horses from central province of Saudi Arabia. J. Parasitol. 2012, 98, 1015–1017. [Google Scholar] [CrossRef]
- Motloang, M.Y.; Thekisoe, O.M.; Alhassan, A.; Bakheit, M.; Motheo, M.P.; Masangane, F.E.; Thibedi, M.L.; Inoue, N.; Igarashi, I.; Sugimoto, C.; et al. Prevalence of Theileria equi and Babesia caballi infections in horses belonging to resource-poor farmers in the north-eastern Free State Province, South Africa. Onderstepoort J. Vet. Res. 2008, 75, 141–146. [Google Scholar] [CrossRef]
- Nagore, D.; Garcia-Sanmartin, J.; Garcia-Perez, A.L.; Juste, R.A.; Hurtado, A. Detection and identification of equine Theileria and Babesia species by reverse line blotting: Epidemiological survey and phylogenetic analysis. Vet. Parasitol. 2004, 123, 41–54. [Google Scholar] [CrossRef]
- Camacho, A.T.; Guitian, F.J.; Pallas, E.; Gestal, J.J.; Olmeda, A.S.; Habela, M.A.; Telford, S.R., III; Spielman, A. Theileria (Babesia) equi and Babesia caballi infections in horses in Galicia, Spain. Trop. Anim. Health Prod. 2005, 37, 293–302. [Google Scholar] [CrossRef]
- Adaszek, L.; Garcia-Bocanegra, I.; Arenas-Montes, A.; Carbonero, A.; Arenas, A.; Winiarczyk, S. Identification of piroplasms isolated from asymptomatic equine species from southern Spain. Berliner Und Munchener Tierarztliche Wochenschrift 2012, 125, 509–512. [Google Scholar]
- Garcia-Bocanegra, I.; Arenas-Montes, A.; Hernandez, E.; Adaszek, L.; Carbonero, A.; Almeria, S.; Jaen-Tellez, J.A.; Gutierrez-Palomino, P.; Arenas, A. Seroprevalence and risk factors associated with Babesia caballi and Theileria equi infection in equids. Vet. J. 2013, 195, 172–178. [Google Scholar] [CrossRef]
- Montes Cortes, M.G.; Fernandez-Garcia, J.L.; Habela Martinez-Estellez, M.A. Seroprevalence of Theileria equi and Babesia caballi in horses in Spain. Parasite 2017, 24, 14. [Google Scholar] [CrossRef] [Green Version]
- Camino, E.; Pozo, P.; Dorrego, A.; Carvajal, K.A.; Buendia, A.; Gonzalez, S.; de Juan, L.; Dominguez, L.; Cruz-Lopez, F. Importance of equine piroplasmosis antibody presence in Spanish horses prior to export. Tick Tick-Borne Dis. 2020, 11, 101329. [Google Scholar] [CrossRef]
- Salim, B.O.; Hassan, S.M.; Bakheit, M.A.; Alhassan, A.; Igarashi, I.; Karanis, P.; Abdelrahman, M.B. Diagnosis of Babesia caballi and Theileria equi infections in horses in Sudan using ELISA and PCR. Parasitol. Res. 2008, 103, 1145–1150. [Google Scholar] [CrossRef]
- Sigg, L.; Gerber, V.; Gottstein, B.; Doherr, M.G.; Frey, C.F. Seroprevalence of Babesia caballi and Theileria equi in the Swiss horse population. Parasitol. Int. 2010, 59, 313–317. [Google Scholar] [CrossRef]
- Asgarali, Z.; Coombs, D.K.; Mohammed, F.; Campbell, M.D.; Caesar, E. A serological study of Babesia caballi and Theileria equi in Thoroughbreds in Trinidad. Vet. Parasitol. 2007, 144, 167–171. [Google Scholar] [CrossRef]
- Ros-Garcia, A.; M’Ghirbi, Y.; Hurtado, A.; Bouattour, A. Prevalence and genetic diversity of piroplasm species in horses and ticks from Tunisia. Infect. Genet. Evol. 2013, 17, 33–37. [Google Scholar] [CrossRef]
- Oncel, T.; Vural, G.; Gicik, Y.; Arslan, M.O. Detection of Babesia (Theileria) equi (Laveran, 1901) in horses in the Kars province of Turkey. Turk. Parazitol Derg 2007, 31, 170–172. [Google Scholar]
- Sevinc, F.; Maden, M.; Kumas, C.; Sevinc, M.; Ekici, O.D. A comparative study on the prevalence of Theileria equi and Babesia caballi infections in horse sub-populations in Turkey. Vet. Parasitol. 2008, 156, 173–177. [Google Scholar] [CrossRef]
- Acici, M.; Umur, S.; Guvenc, T.; Arslan, H.H.; Kurt, M. Seroprevalence of equine babesiosis in the Black Sea region of Turkey. Parasitol. Int. 2008, 57, 198–200. [Google Scholar] [CrossRef]
- Karatepe, B.; Karatepe, M.; Cakmak, A.; Karaer, Z.; Ergun, G. Investigation of seroprevalence of Theileria equi and Babesia caballi in horses in Nigde province, Turkey. Trop. Anim. Health Prod. 2009, 41, 109–113. [Google Scholar] [CrossRef]
- Kurt, C.; Yaman, M. The investigation of the prevalence of Babesia equi and Babesia caballi in horses by microscopic and serologic (cELISA) methods in Adana province. Yüzüncü Yıl Üniversitesi Veteriner Fakültesi Dergisi 2012, 23, 1–4. [Google Scholar]
- Guven, E.; Avcioglu, H.; Deniz, A.; Balkaya, I.; Abay, U.; Yavuz, S.; Akyuz, M. Prevalence and molecular characterization of Theileria equi and Babesia caballi in jereed horses in Erzurum, Turkey. Acta Parasitol. 2017, 62, 207–213. [Google Scholar] [CrossRef]
- Mujica, F.F.; Perrone, T.; Forlano, M.; Coronado, A.; Melendez, R.D.; Barrios, N.; Alvarez, R.; Granda, F. Serological prevalence of Babesia caballi and Theileria equi in horses of Lara State, Venezuela. Vet. Parasitol. 2011, 178, 180–183. [Google Scholar] [CrossRef]
- Rosales, R.; Rangel-Rivas, A.; Escalona, A.; Jordan, L.S.; Gonzatti, M.I.; Aso, P.M.; Perrone, T.; Silva-Iturriza, A.; Mijares, A. Detection of Theileria equi and Babesia caballi infections in Venezuelan horses using Competitive-Inhibition ELISA and PCR. Vet. Parasitol. 2013, 196, 37–43. [Google Scholar] [CrossRef]
- Mantran, A.; Votion, D.; Amory, H. Piroplasmosis: A problem in Belgium. In Proceedings of the Annual Congress Belgian Equine Practitioners Society, Brussels, Belgium, 6 November 2004; p. 2013. Available online: http://www.ivis.org/proceedings/BEPS/2004/Amory_nl/ivis.pdf (accessed on 20 April 2020).
- Anon. Equine piroplasmosis confirmed in Ireland. Vet. Rec. 2009, 165, 333. [Google Scholar] [CrossRef]
- Chitimia-Dobler, L.; Nava, S.; Bestehorn, M.; Dobler, G.; Wolfel, S. First detection of Hyalomma rufipes in Germany. Tick Tick-Borne Dis. 2016, 7, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Pena, A. Climate, niche, ticks, and models: What they are and how we should interpret them. Parasitol. Res. 2008, 103, S87–S95. [Google Scholar] [CrossRef]
- Estrada-Pena, A.; Ayllon, N.; de la Fuente, J. Impact of climate trends on tick-borne pathogen transmission. Front. Physiol. 2012, 3, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-Pena, A.; Venzal, J.M. Climate niches of tick species in the Mediterranean region: Modeling of occurrence data, distributional constraints, and impact of climate change. J. Med. Entomol. 2007, 44, 1130–1138. [Google Scholar] [CrossRef]
- Tamzali, Y. Equine piroplasmosis: An updated review. Equine Vet. Educ. 2013, 25, 590–598. [Google Scholar] [CrossRef]
- Adaszek, L.; Gorna, M.; Krzysiak, M.; Adaszek, M.; Garbal, M.; Winiarczyk, S. Identification of the piroplasms isolated from horses with clinical piroplasmosis in Poland. Wiad. Parazytol. 2011, 57, 21–26. [Google Scholar]
- Short, M.A.; Clark, C.K.; Harvey, J.W.; Wenzlow, N.; Hawkins, I.K.; Allred, D.R.; Knowles, D.P.; Corn, J.L.; Grause, J.F.; Hennager, S.G.; et al. Outbreak of equine piroplasmosis in Florida. J. Am. Vet. Med. Assoc. 2012, 240, 588–595. [Google Scholar] [CrossRef]
- Beard, L.A.; Pelzel, A.M.; Rush, B.R.; Wright, A.M.; Galgut, B.I.; Hennager, S.G.; King, A.O.; Traub-Dargatz, J.L. Babesia equi-induced anemia in a Quarter Horse and subsequent regulatory response. J. Am. Vet. Med. Assoc. 2013, 242, 992–996. [Google Scholar] [CrossRef]
- Butler, C.M.; van Gils, J.A.; van der Kolk, J.H. A literature review of equine piroplasmosis after an episode of acute babesiosis in a Dutch Standardbred foal after a stay in Normandy. Tijdschr. Voor Diergeneeskd. 2005, 130, 726–731. [Google Scholar]
- Ionita, M.; Nicorescu, I.M.; Pfister, K.; Mitrea, I.L. Parasitological and molecular diagnostic of a clinical Babesia caballi outbreak in Southern Romania. Parasitol. Res. 2018, 117, 2333–2339. [Google Scholar] [CrossRef]
- Tirosh-Levy, S.; Steinman, A.; Levy, H.; Katz, Y.; Shtilman, M.; Gottlieb, Y. Parasite load and genotype are associated with clinical outcome of piroplasm-infected equines in Israel. Parasites Vectors 2020, 13, 267. [Google Scholar] [CrossRef]
- Camino, E.; Dorrego, A.; Carvajal, K.A.; Buendia-Andres, A.; de Juan, L.; Dominguez, L.; Cruz-Lopez, F. Serological, molecular and hematological diagnosis in horses with clinical suspicion of equine piroplasmosis: Pooling strengths. Vet. Parasitol. 2019, 275, 108928. [Google Scholar] [CrossRef]
- Mehlhorn, H.; Schein, E. Redescription of Babesia equi Laveran, 1901 as Theileria equi Mehlhorn, Schein 1998. Parasitol. Res. 1998, 84, 467–475. [Google Scholar] [CrossRef]
- Aziz, K.J.; Al-Barwary, L.T.O. Epidemiological Study of Equine Piroplasmosis (Theileria equi and Babesia caballi) by Microscopic Examination and Competitive-ELISA in Erbil Province North-Iraq. Iran. J. Parasitol. 2019, 14, 404–412. [Google Scholar]
- Piantedosi, D.; D’Alessio, N.; Di Loria, A.; Di Prisco, F.; Mariani, U.; Neola, B.; Santoro, M.; Montagnaro, S.; Capelli, G.; Veneziano, V. Seroprevalence and risk factors associated with Babesia caballi and Theileria equi infections in donkeys from Southern Italy. Vet. J. 2014, 202, 578–582. [Google Scholar] [CrossRef]
- Tirosh-Levy, S.; Gottlieb, Y.; Arieli, O.; Mazuz, M.L.; King, R.; Horowitz, I.; Steinman, A. Genetic characteristics of Theileria equi in zebras, wild and domestic donkeys in Israel and the Palestinian Authority. Tick Tick-Borne Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Oduori, D.O.; Onyango, S.C.; Kimari, J.N.; MacLeod, E.T. A field survey for the seroprevalence of Theileria equi and Babesia caballi in donkeys from Nuu Division, Kenya. Tick Tick-Borne Dis. 2015, 6, 683–688. [Google Scholar] [CrossRef] [Green Version]
- Afridi, M.J.K.; Mian, A.H.; Saqib, M.; Abbas, G.; Ali, J.; Mansoor, M.K.; Sial, A.U.R.; Rasheed, I.; Hussain, M.H. Seroprevalence and Risk Factors for Theileria equi Infection in Equines from Khyber Pakhtunkhwa Province, Pakistan. Iran. J. Parasitol. 2017, 12, 597–605. [Google Scholar] [PubMed]
- Kumar, S.; Kumar, R.; Sugimoto, C. A perspective on Theileria equi infections in donkeys. Jpn. J. Vet. Res. 2009, 56, 171–180. [Google Scholar]
- Abedi, V.; Razmi, G.; Seifi, H.; Naghibi, A. Molecular detection of equine piroplasms in donkeys (Equus asinus) in North Khorasan province, Iran. Iran. J. Vet. Res. 2015, 16, 202–204. [Google Scholar]
- Veronesi, F.; Morganti, G.; Ravagnan, S.; Laus, F.; Spaterna, A.; Diaferia, M.; Moretti, A.; Fioretti, D.P.; Capelli, G. Molecular and serological detection of tick-borne pathogens in donkeys (Equus asinus) in Italy. Vet. Microbiol. 2014, 173, 348–354. [Google Scholar] [CrossRef]
- Hawkins, E.; Kock, R.; McKeever, D.; Gakuya, F.; Musyoki, C.; Chege, S.M.; Mutinda, M.; Kariuki, E.; Davidson, Z.; Low, B.; et al. Prevalence of Theileria equi and Babesia caballi as well as the identification of associated ticks in sympatric Grevy’s zebras (Equus grevyi) and donkeys (Equus africanus asinus) in northern Kenya. J. Wildl. Dis. 2015, 51, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.M.; Bhoora, R.V.; Kotze, A.; Grobler, J.P.; Lee Dalton, D. Translocation a potential corridor for equine piroplasms in Cape mountain zebra (Equus zebra zebra). Int. J. Parasitol. Parasites Wildl. 2019, 9, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Bhoora, R.; Buss, P.; Guthrie, A.J.; Penzhorn, B.L.; Collins, N.E. Genetic diversity of piroplasms in plains zebra (Equus quagga burchellii) and Cape mountain zebra (Equus zebra zebra) in South Africa. Vet. Parasitol. 2010, 174, 145–149. [Google Scholar] [CrossRef]
- Lampen, F.; Bhoora, R.; Collins, N.E.; Penzhorn, B.L. Putative clinical piroplasmosis in a Burchell’s zebra (Equus quagga burchelli). J. S. Afr. Vet. Assoc. 2009, 80, 257–260. [Google Scholar] [CrossRef] [Green Version]
- King’ori, E.M.; Obanda, V.; Ndambiri, E.M.; Runo, S.M.; Chiyo, P.I. Adding injury to infection: The relationship between injury status and genetic diversity of Theileria infecting plains zebra, Equus quagga. Infect. Genet. Evol. 2018, 58, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Zweygarth, E.; Lopez-Rebollar, L.M.; Meyer, P. In vitro isolation of equine piroplasms derived from Cape Mountain zebra (Equus zebra zebra) in South Africa. Onderstepoort J. Vet. Res. 2002, 69, 197–200. [Google Scholar] [PubMed]
- Beck, R.; Vojta, L.; Mrljak, V.; Marinculic, A.; Beck, A.; Zivicnjak, T.; Caccio, S.M. Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int. J. Parasitol. 2009, 39, 843–848. [Google Scholar] [CrossRef]
- Inacio, E.L.; Perez-Macchi, S.; Alabi, A.; Bittencourt, P.; Muller, A. Prevalence and molecular characterization of piroplasmids in domestic dogs from Paraguay. Tick Tick-Borne Dis. 2019, 10, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Adamu, M.; Troskie, M.; Oshadu, D.O.; Malatji, D.P.; Penzhorn, B.L.; Matjila, P.T. Occurrence of tick-transmitted pathogens in dogs in Jos, Plateau State, Nigeria. Parasites Vectors 2014, 7, 119. [Google Scholar] [CrossRef] [Green Version]
- Qablan, M.A.; Kubelova, M.; Siroky, P.; Modry, D.; Amr, Z.S. Stray dogs of northern Jordan as reservoirs of ticks and tick-borne hemopathogens. Parasitol. Res. 2012, 111, 301–307. [Google Scholar] [CrossRef]
- Salim, B.; Alanazi, A.D.; Omori, R.; Alyousif, M.S.; Alanazi, I.O.; Katakura, K.; Nakao, R. Potential role of dogs as sentinels and reservoirs for piroplasms infecting equine and cattle in Riyadh City, Saudi Arabia. Acta Trop. 2019, 193, 78–83. [Google Scholar] [CrossRef]
- Qablan, M.A.; Sloboda, M.; Jirku, M.; Obornik, M.; Dwairi, S.; Amr, Z.S.; Horin, P.; Lukes, J.; Modry, D. Quest for the piroplasms in camels: Identification of Theileria equi and Babesia caballi in Jordanian dromedaries by PCR. Vet. Parasitol. 2012, 186, 456–460. [Google Scholar] [CrossRef]
- Bahrami, S.; Tabandeh, M.R.; Tafreshi, A.R.G. Prevalence and Molecular Identification of Piroplasmids in Iranian Dromedaries (Camelus dromedarius). J. Zoo Wildl. Med. 2017, 48, 1026–1030. [Google Scholar] [CrossRef]
- Sadeddine, R.; Diarra, A.Z.; Laroche, M.; Mediannikov, O.; Righi, S.; Benakhla, A.; Dahmana, H.; Raoult, D.; Parola, P. Molecular identification of protozoal and bacterial organisms in domestic animals and their infesting ticks from north-eastern Algeria. Tick Tick-Borne Dis. 2020, 11, 101330. [Google Scholar] [CrossRef]
- Da Silveira, A.W.; de Oliveira, G.G.; Menezes Santos, L.; da Silva Azuaga, L.B.; Macedo Coutinho, C.R.; Echeverria, J.T.; Antunes, T.R.; do Nascimento Ramos, C.A.; Izabel de Souza, A. Natural Infection of the South American Tapir (Tapirus terrestris) by Theileria equi. J. Wildl. Dis. 2017, 53, 411–413. [Google Scholar] [CrossRef]
- Laveran, A. Contribution a l’etude de Piroplasma equi. CR Soc. Biol. 1901, 12, 385–388. [Google Scholar]
- Allsopp, M.T.; Allsopp, B.A. Molecular sequence evidence for the reclassification of some Babesia species. Ann. N. Y. Acad. Sci. 2006, 1081, 509–517. [Google Scholar] [CrossRef]
- Hikosaka, K.; Watanabe, Y.; Tsuji, N.; Kita, K.; Kishine, H.; Arisue, N.; Palacpac, N.M.; Kawazu, S.; Sawai, H.; Horii, T.; et al. Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol. Biol. Evol. 2010, 27, 1107–1116. [Google Scholar] [CrossRef] [Green Version]
- Kappmeyer, L.S.; Thiagarajan, M.; Herndon, D.R.; Ramsay, J.D.; Caler, E.; Djikeng, A.; Gillespie, J.J.; Lau, A.O.; Roalson, E.H.; Silva, J.C.; et al. Comparative genomic analysis and phylogenetic position of Theileria equi. BMC Genom. 2012, 13, 603. [Google Scholar] [CrossRef] [Green Version]
- Lack, J.B.; Reichard, M.V.; van Den Bussche, R.A. Phylogeny and evolution of the Piroplasmida as inferred from 18S rRNA sequences. Int. J. Parasitol. 2012, 42, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Githaka, N.; Konnai, S.; Bishop, R.; Odongo, D.; Lekolool, I.; Kariuki, E.; Gakuya, F.; Kamau, L.; Isezaki, M.; Murata, S.; et al. Identification and sequence characterization of novel Theileria genotypes from the waterbuck (Kobus defassa) in a Theileria parva-endemic area in Kenya. Vet. Parasitol. 2014, 202, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Caccio, S.; Camma, C.; Onuma, M.; Severini, C. The beta-tubulin gene of Babesia and Theileria parasites is an informative marker for species discrimination. Int. J. Parasitol. 2000, 30, 1181–1185. [Google Scholar] [CrossRef]
- Wise, L.N.; Kappmeyer, L.S.; Knowles, D.P.; White, S.N. Evolution and diversity of the EMA families of the divergent equid parasites, Theileria equi and T. haneyi. Infect. Genet. Evol. 2019, 68, 153–160. [Google Scholar] [CrossRef]
- Knowles, D.P.; Kappmeyer, L.S.; Perryman, L.E. Genetic and biochemical analysis of erythrocyte-stage surface antigens belonging to a family of highly conserved proteins of Babesia equi and Theileria species. Mol. Biochem. Parasitol. 1997, 90, 69–79. [Google Scholar] [CrossRef]
- Hall, C.M.; Busch, J.D.; Scoles, G.A.; Palma-Cagle, K.A.; Ueti, M.W.; Kappmeyer, L.S.; Wagner, D.M. Genetic characterization of Theileria equi infecting horses in North America: Evidence for a limited source of U.S. introductions. Parasites Vectors 2013, 6, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ketter-Ratzon, D.; Tirosh-Levy, S.; Nachum-Biala, Y.; Saar, T.; Qura’n, L.; Zivotofsky, D.; Abdeen, Z.; Baneth, G.; Steinman, A. Characterization of Theileria equi genotypes in horses in Israel, the Palestinian Authority and Jordan. Tick Tick-Borne Dis. 2017, 8, 499–505. [Google Scholar] [CrossRef]
- Salim, B.; Bakheit, M.A.; Kamau, J.; Nakamura, I.; Sugimoto, C. Nucleotide sequence heterogeneity in the small subunit ribosomal RNA gene within Theileria equi from horses in Sudan. Parasitol. Res. 2010, 106, 493–498. [Google Scholar] [CrossRef]
- Bishop, R.P.; Kappmeyer, L.S.; Onzere, C.K.; Odongo, D.O.; Githaka, N.; Sears, K.P.; Knowles, D.P.; Fry, L.M. Equid infective Theileria cluster in distinct 18S rRNA gene clades comprising multiple taxa with unusually broad mammalian host ranges. Parasites Vectors 2020, 13, 261. [Google Scholar] [CrossRef]
- Peckle, M.; Pires, M.S.; Silva, C.B.D.; Costa, R.L.D.; Vitari, G.L.V.; Senra, M.V.X.; Dias, R.J.P.; Santos, H.A.; Massard, C.L. Molecular characterization of Theileria equi in horses from the state of Rio de Janeiro, Brazil. Tick Tick-Borne Dis. 2018, 9, 349–353. [Google Scholar] [CrossRef]
- Vitari, G.L.V.; Costa, R.L.; Abreu, A.P.M.; Peckle, M.; Silva, C.B.; Paulino, P.G.; Pires, M.S.; Massard, C.L.; Santos, H.A. Genetic Diversity of Theileria equi From Horses in Different Regions of Brazil Based On the 18S rRNA Gene. J. Parasitol. 2019, 105, 186–194. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef] [Green Version]
- Tirosh-Levy, S.; Steinman, A.; Einhorn, A.; Apanaskevich, D.A.; Mumcuoglu, K.Y.; Gottlieb, Y. Potential tick vectors for Theileria equi in Israel. Med. Vet. Entomol. 2020. [Google Scholar] [CrossRef]
- Coultous, R.M.; McDonald, M.; Raftery, A.G.; Shiels, B.R.; Sutton, D.G.M.; Weir, W. Analysis of Theileria equi diversity in The Gambia using a novel genotyping method. Transbound. Emerg. Dis. 2019. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Xuan, X.; Igarashi, I.; Tanaka, T.; Fukumoto, S.; Nagasawa, H.; Fujisaki, K.; Mikami, T. Detection of antibodies to Babesia equi in horses by a latex agglutination test using recombinant EMA-1. Clin. Diagn. Lab. Immunol. 2001, 8, 645–646. [Google Scholar] [CrossRef] [Green Version]





| T. equi | B. caballi | |||||||
|---|---|---|---|---|---|---|---|---|
| Location | N | Sero-Prevalence (%) | Prevalence (%) | Sero-Prevalence (%) | Prevalence (%) | Co-Infection (%) | Method * | Ref. |
| Argentina | 180 | 65 | iELISA | [74] | ||||
| Azores | 143 | 2.8 | 2.8 | cELISA/nPCR | [127] | |||
| Balkan | 142 | 22.5 | 2.1 | 0.7 | mPCR | [128] | ||
| Brazil | 47 | 81 | 90 | 75 | ELISA | [129] | ||
| Brazil | 35 | 85.7 | qPCR/ | [99] | ||||
| Brazil | 487 | 91 | 59.7 | 83 | 12.5 | 8.6 | IFAT/MRT-PCR | [130] |
| Brazil | 582 | 21.6 | 54.1 | CFT/cELISA | [131] | |||
| Brazil | 170 | 100 | 63.5 | IFAT/nPCR | [75] | |||
| Brazil | 170 | 95.9 | ELISA | [79] | ||||
| Brazil | 579 | 81.1 | IFAT | [132] | ||||
| Brazil | 314 | 81 | rtPCR | [133] | ||||
| Brazil | 198 | 78.3 | 69.2 | 50 | cELISA | [134] | ||
| Brazil | 400 | 61 | ELISA | [135] | ||||
| Brazil | 39 | 43.5 | 38.5 | 7.7 | 60 | 28.2 | ELISA/PCR | [136] |
| Brazil | 430 | 87.4 | 87.9/90.5 | 58.6 | 9.3/7.9 | 8.8 | cELISA/dqPCR/qPCR | [97] |
| Brazil | 528 | 84.3 | 23.5 | nPCR | [137] | |||
| Brazil | 359 | 33.6 | iELISA | [138] | ||||
| Brazil | 170 | 61.8 | 52.9 | 49.4 | ELISA | [139] | ||
| Chad | 96 | 20.8 | PCR | [140] | ||||
| Chad | 59 | 72.8 | PCR | [140] | ||||
| China | 70 | 40 | 24.3 | 15.7 | ELISA | [141] | ||
| China | 55 | 81.8 | 56.3 | LAMP | [85] | |||
| China | 1990 | 11.5 | 51.2 | 7.6 | cELISA | [142] | ||
| China | 723 | 40.8 | PCR | [143] | ||||
| China | 242 | 30.2 | 2.9 | 2.1 | nPCR | [144] | ||
| China | 56 | 57.1 | ICT | [80] | ||||
| China | 200 | 39.5 | 24.5 | PCR | [145] | |||
| Costa Rica | 130 | 88.5 | 46.2 | 69.2 | 20 | 62.3/7.7 | cELISA/nPCR | [110] |
| Cuba | 100 | 73 | 25 | 20 | nPCR | [146] | ||
| DR Congo | 48 | 43.7 | PCR | [140] | ||||
| Dubai | 105 | 32.4/33.3 | 15.3/10.5 | 12.4 | cELISA/IFAT | [104] | ||
| Egypt | 88 | 23.9 | 36.4 | 17 | 19.3 | IFAT/nPCR | [83] | |
| France | 111 | 80 | 1.2 | PCR | [147] | |||
| France | 443 | 58 | 12.9 | CFT | [123] | |||
| France | 51 | 29.4 | PCR | [140] | ||||
| France | 98 | 39.8 | PCR | [140] | ||||
| Ghana | 30 | 53.3 | qPCR | [99] | ||||
| Ghana | 20 | 60 | PCR | [86] | ||||
| Greece | 544 | 11 | 2.2 | 1.7 | cELISA | [148] | ||
| Greece | 772 | 44 | 0 | RLB-PCR | [149] | |||
| Guatemala | 74 | 92.7 | 17 | 16 | IFAT/PCR | [150] | ||
| Hungary | 324 | 32 | cELISA/IFAT | [151] | ||||
| Hungary | 101 | 49 | PCR | [151] | ||||
| India | 5651 | 32.6 | ELISA | [76] | ||||
| India | 426 | 48.6 | 19.7 | iELISA/nPCR | [109] | |||
| Indonesia | 235 | 2.1 | 0.4 | 6.4 | 1.7 | cELISA/nPCR | [152] | |
| Iran | 100 | 48 | 45 | 2 | 0 | 3 | IFAT/PCR | [118] |
| Iran | 240 | 10.8 | 5.8 | 1.6 | PCR | [153] | ||
| Iran | 104 | 22.8 | PCR | [154] | ||||
| Iran | 31 | 96.7 | 0 | PCR | [155] | |||
| Iran | 126 | 27.7 | PCR | [156] | ||||
| Israel | 216 | 50.9 | ELISA | [157] | ||||
| Israel | 590 | 26.4 | PCR | [158] | ||||
| Israel | 257 | 9.3 | PCR | [84] | ||||
| Italy | 412 | 12.4 | 17.9 | 38.1 | IFAT | [117] | ||
| Italy | 294 | 8.2 | 2.7 | 0.3 | 0 | 0 | IFAT/PCR | [116] |
| Italy | 300 | 41 | 11.7 | 26 | 6 | 14.7 | IFAT/PCR | [119] |
| Italy | 1441 | 31.6 | 1.2 | 0.6 | IFAT | |||
| Italy | 177 | 41 | 32.4 | 0 | 0 | IFAT/PCR | [121] | |
| Italy | 160 | 26.9 | 0 | [159] | ||||
| Italy | 673 | 39.8 | 8.9 | cELISA | [108] | |||
| Italy | 135 | 13.3 | PCR | [160] | ||||
| Japan | 2019 | 2.2 | 5.4 | 0 | ELISA | [161] | ||
| Jordan | 253 | 14.6 | 0 | 0 | 0 | cELISA/PCR | [115] | |
| Jordan | 288 | 18.8 | 7.3 | 0 | mPCR | [162] | ||
| Korea | 184 | 1.1 | 0 | cELISA | [114] | |||
| Korea | 224 | 0.9 | PCR | [163] | ||||
| Malaysia | 306 | 51.3 | 63.1 | 34.3 | cELISA | [164] | ||
| Mexico | 248 | 45.2 | 27.4 | IFAT | [165] | |||
| Mexico | 1000 | 19.7 | nPCR | [166] | ||||
| Mongolia | 254 | 72.8 | 40.1 | 30.7 | ELISA | [167] | ||
| Mongolia | 39 | 25.6 | 17.9 | mPCR | [87] | |||
| Mongolia | 510 | 78.8 | 66.5 | 65.7 | 19.1 | IFAT/PCR | [113] | |
| Mongolia | 250 | 19.6 | 6.4 | 51.6 | 6.1 | 10.4/2.5 | ELISA/nPCR | [112] |
| Mongolia | 192 | 92.7 | 0 | nPCR/mPCR | [168] | |||
| Mongolia | 1282 | 33 | 14.2 | 16.8 | ELISA | [169] | ||
| Morocco | 578 | 67 | cELISA | [106] | ||||
| Netherlands | 300 | 4 | 5 | 0 | 0 | IFAT/RLB-PCR | [111] | |
| Nicaragua | 93 | 96.8 | 26.8 | PCR | [170] | |||
| Nigeria | 342 | 73.1 | 4.4 | cELISA | [171] | |||
| Pakistan | 430 | 41.2 | 21.6 | 10.2 | cELISA | [172] | ||
| Palestine | 108 | 29.6 | ELISA | [157] | ||||
| Philippines | 105 | 11.4 | 24.8 | 10.4 | 1.9 | ICT/PCR | [173] | |
| Poland | 76 | 1.3 | PCR | [174] | ||||
| Portugal | 162 | 17.9 | 11.1 | cELISA | [175] | |||
| Portugal | 162 | 9.3 | 1.9 | cELISA/nPCR | [127] | |||
| Romania | 178 | 38.8 | 4.5 | mPCR | [176] | |||
| Saudi Arabia | 141 | 42 | qPCR | [98] | ||||
| Saudi Arabia | 241 | 10.4 | 7.5 | 3 | IFAT | [177] | ||
| Senegal | 127 | 16.5 | 0.01 | qPCR | [140] | |||
| Slovakia | 39 | 0 | PCR | [174] | ||||
| South Africa | 37 | 91.8 | 45.9 | LAMP | [85] | |||
| South Africa | 99 | 97.9 | 9 | 51.5 | 0 | IFAT/PCR | [178] | |
| South Africa | 488 | 50 | 3 | RLB-PCR | [100] | |||
| South Africa | 41 | 83 | 80 | 70 | 78 | IFAT/qPCR | [94] | |
| Spain | 181 | 50.3 | 0.6 | RLB-PCR | [179] | |||
| Spain | 60 | 40 | 28.3 | 20 | IFAT | [180] | ||
| Spain | 135 | 17 | 3 | PCR | [181] | |||
| Spain | 428 | 50.3 | 11.4 | 8.4 | cELISA | [182] | ||
| Spain | 3100 | 44 | 21 | IFAT | [183] | |||
| Spain | 235 | 61.7 | 66 | 3.8 | 29.4 | cELISA/mnPCR | [90] | |
| Spain | 3368 | 21 | 5.6 | 2.5 | cELISA | [184] | ||
| Sudan | 126 | 63.5 | 4.4 | ELISA | [185] | |||
| Sudan | 131 | 25.2 | 0 | PCR | [185] | |||
| Sudan | 499 | 35.9 | 0 | PCR | [92] | |||
| Switzerland | 689 | 5.9 | 3 | 1.5 | IFAT | [186] | ||
| Thailand | 240 | 5.42/8.75 | 1.25 | 2.5/5 | 0 | ELISA/IFAT/PCR | [105] | |
| Trinidad | 93 | 33.3 | 68.8 | 19.4 | IFAT | [187] | ||
| Trinidad | 111 | 24.3 | 3.6 | PCR | [17] | |||
| Tunisia | 104 | 12.5 | 1.9 | 1.9 | RLB-PCR | [188] | ||
| Turkey | 108 | 25 | IFAT | [189] | ||||
| Turkey | 481 | 17.7 | 2.29 | 1.46 | cELISA | [190] | ||
| Turkey | 84 | 23.8 | 38 | 5.6 | IFAT | [191] | ||
| Turkey | 125 | 12.8 | 9.6 | 4 | IFAT | [192] | ||
| Turkey | 220 | 56.8 | 0 | cELISA | [193] | |||
| Turkey | 203 | 2.96 | 1.97 | qPCR | [120] | |||
| Turkey | 125 | 8.8 | 0 | mPCR | [194] | |||
| UK | 1242 | 5.9 | 0.8 | 4.4 | 0 | 2 | IFAT/cELISA/CFT/nPCR | [126] |
| Ukraine | 100 | 29 | [174] | |||||
| Venezuella | 360 | 50.3 | 70.6 | 35.6 | cELISA | [195] | ||
| Venezuella | 694 | 14 | 23.2 | 13 | cELISA | [196] | ||
| Venezuella | 136 | 61.8 | 4.4 | 4.4 | mPCR | [196] |
| TE Seroprevalence | TE Prevalence | BC Seroprevalence | BC Prevalence | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (%) | N | Ref. | (%) | N | Ref. | (%) | N | Ref. | (%) | N | Ref. | |
| Worldwide | 33.17 | 37,398 | 72 | 34.55 | 15,849 | 70 | 20.45 | 27,582 | 56 | 7.35 | 11,840 | 51 |
| Africa | 68.21 | 1274 | 6 | 38.02 | 1867 | 14 | 16.52 | 696 | 5 | 5.14 | 1614 | 9 |
| Asia | 26.79 | 16,217 | 27 | 29.43 | 5418 | 23 | 24.52 | 9540 | 22 | 8.86 | 3871 | 19 |
| Europe | 27.89 | 14,497 | 20 | 22.26 | 4917 | 19 | 9.42 | 1368 | 17 | 2.48 | 4227 | 13 |
| South America | 58.21 | 5410 | 19 | 56.92 | 3647 | 14 | 54.05 | 3478 | 12 | 15.98 | 2128 | 10 |
| Genotype | Total | Horse | Origin | Donkey | Origin | Zebra | Origin | Tick | Origin | Dog | Origin | Camel | Origin | Cattle | Origin | Tapir | Origin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 148 | 122 | Brazil, Cuba, France, India, Iran, Israel, Jordan, Mongolia, Romania, Saudi Arabia, South Africa, South Korea, Spain, Trinidad and Tobago, Turkey, US | 3 | Italy | 1 | Israel | 13 | Brazil, China, Columbia, France, India, Italy, Portugal, Tunisia | 5 | Jordan, Paraguai, Spain, Saudi Arabia | 4 | Jordan | ||||
| B | 13 | 3 | Jordan, South Africa, Sudan | 6 | Italy | 4 | South Africa | ||||||||||
| C | 76 | 74 | Brazil, China, Cuba, Israel, Kenya, Malezia, Mexico, Romania, South Africa | 1 | Italy | 1 | Algiria | ||||||||||
| D | 62 | 44 | Brazil, Iran, Israel, Jordan, Romania, South Africa, Sudan, Turkey | 7 | Italy, Kenya | 7 | Israel, Nigeria, South Africa | 3 | Iran | 1 | Brazil | ||||||
| E | 61 | 57 | China, Hungary, Iran, Iraq, Jordan, Mongolia, Romania, Russia, Saudi Arabia, South Korea, Spain, Switzerland, Turkey, Ukraine | 4 | China, Mongolia |
| Within Genotype | Between Genotypes | |||||
|---|---|---|---|---|---|---|
| Genotype | N | A | B | C | D | |
| A | 55 | 0.004 | ||||
| B | 5 | 0.006 | 0.037 | |||
| C | 40 | 0.004 | 0.030 | 0.038 | ||
| D | 22 | 0.004 | 0.031 | 0.034 | 0.016 | |
| E | 10 | 0.008 | 0.039 | 0.016 | 0.044 | 0.041 |
| (a) ema-1 | Within Genotype | Between Genotypes | |||
|---|---|---|---|---|---|
| Genotype | N | A | B | C1 | |
| A | 83 | 0.004 | |||
| B | 2 | 0.000 | 0.075 | ||
| C1 | 23 | 0.002 | 0.081 | 0.020 | |
| C2 | 13 | 0.014 | 0.155 | 0.125 | 0.123 |
| (b) ema-2 | Within Genotype | Between Genotypes | |||
| Genotype | N | A | B | ||
| A | 11 | 0.000 | |||
| B | 12 | 0.004 | 0.011 | ||
| C | 6 | 0.001 | 0.059 | 0.055 | |
| (a) 18S rRNA | Within Genotype | Between Genotypes | |||
|---|---|---|---|---|---|
| Genotype | N | A | B1 | ||
| A | 27 | 0.005 | |||
| B1 | 15 | 0.01 | 0.065 | ||
| B2 | 14 | 0.017 | 0.052 | 0.031 | |
| (b) rap-1 | Within Genotype | Between Genotypes | |||
| Genotype | N | A1 | A2 | B1 | |
| A1 | 15 | 0.001 | |||
| A2 | 4 | 0 | 0.120 | ||
| B1 | 87 | 0.015 | 0.310 | 0.277 | |
| B2 | 6 | 0.02 | 0.312 | 0.278 | 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tirosh-Levy, S.; Gottlieb, Y.; Fry, L.M.; Knowles, D.P.; Steinman, A. Twenty Years of Equine Piroplasmosis Research: Global Distribution, Molecular Diagnosis, and Phylogeny. Pathogens 2020, 9, 926. https://doi.org/10.3390/pathogens9110926
Tirosh-Levy S, Gottlieb Y, Fry LM, Knowles DP, Steinman A. Twenty Years of Equine Piroplasmosis Research: Global Distribution, Molecular Diagnosis, and Phylogeny. Pathogens. 2020; 9(11):926. https://doi.org/10.3390/pathogens9110926
Chicago/Turabian StyleTirosh-Levy, Sharon, Yuval Gottlieb, Lindsay M. Fry, Donald P. Knowles, and Amir Steinman. 2020. "Twenty Years of Equine Piroplasmosis Research: Global Distribution, Molecular Diagnosis, and Phylogeny" Pathogens 9, no. 11: 926. https://doi.org/10.3390/pathogens9110926
APA StyleTirosh-Levy, S., Gottlieb, Y., Fry, L. M., Knowles, D. P., & Steinman, A. (2020). Twenty Years of Equine Piroplasmosis Research: Global Distribution, Molecular Diagnosis, and Phylogeny. Pathogens, 9(11), 926. https://doi.org/10.3390/pathogens9110926






