Metabolic Changes of Mycobacterium tuberculosis during the Anti-Tuberculosis Therapy
Abstract
:1. Introduction
2. Results and Discussion
2.1. Clinical Isolates Characteristic
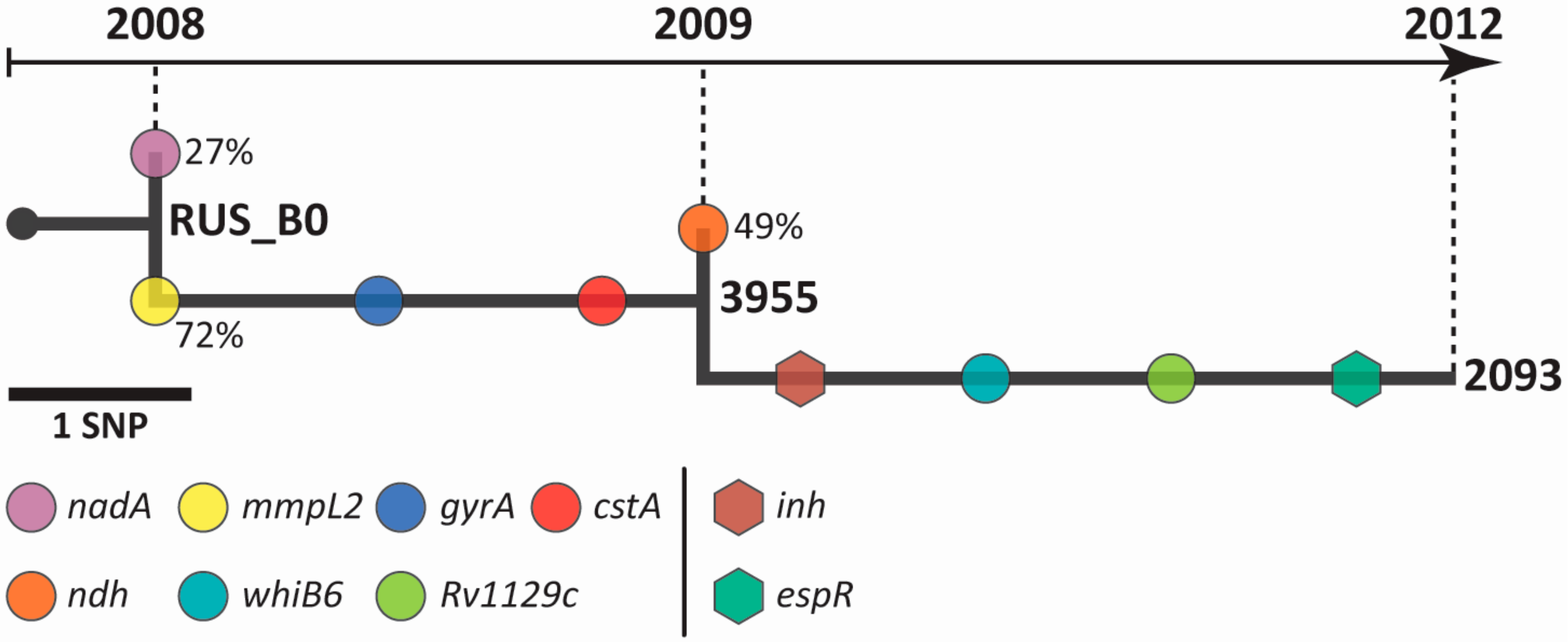
2.2. Intra-host Genome Evolution of M. tuberculosis Serial Isolates
2.3. Analysis of Drug Resistance Genes on Multi-omics Level
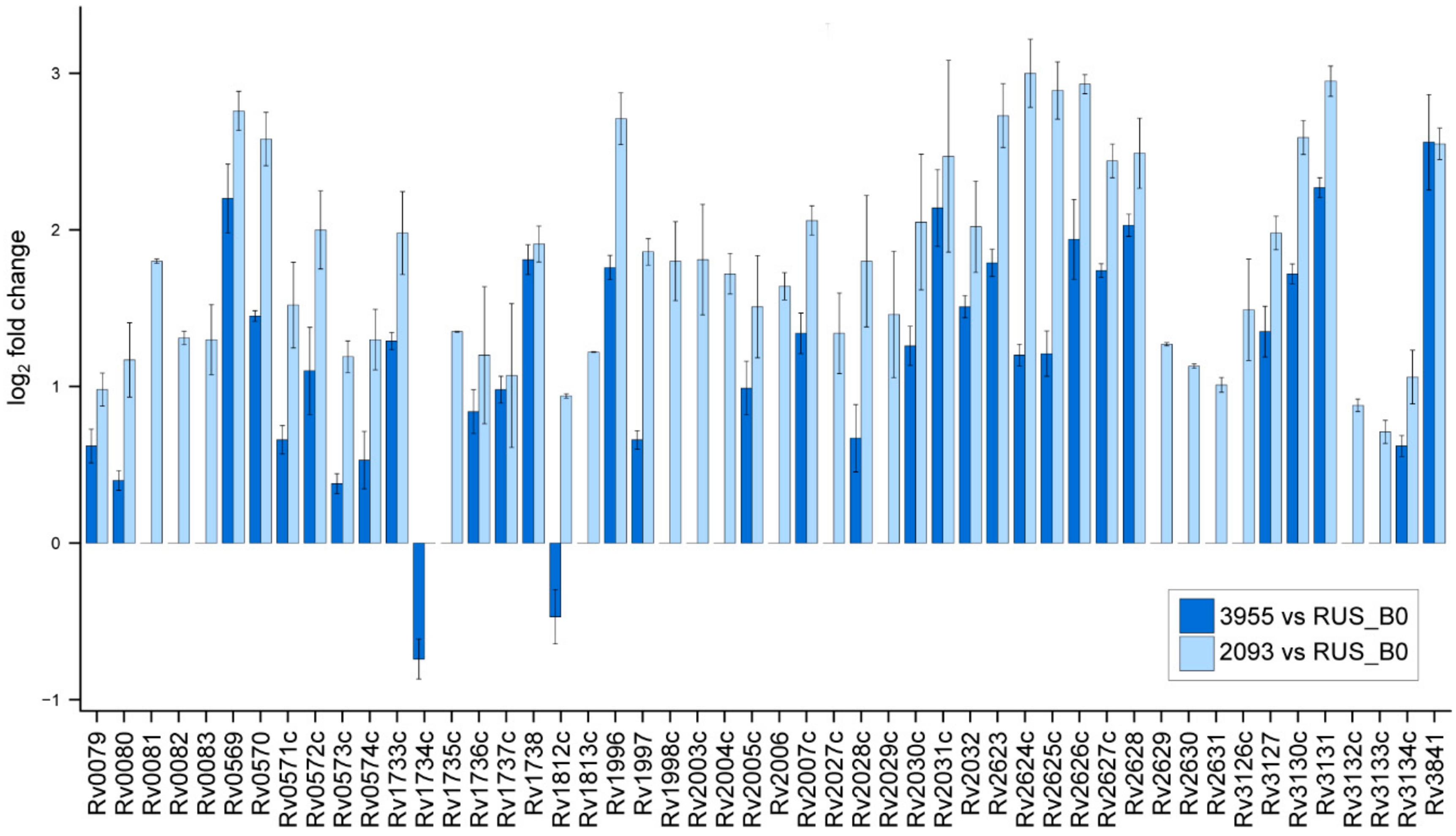
2.4. Non-Specific Bacterial Response to Anti-tuberculosis Therapy
2.5. Variability in the Virulence Factors Representation
3. Materials and Methods
3.1. Mycobacterium Tuberculosis Strains and Growth Conditions
3.2. Genomic Analysis
3.3. Transcriptomic Analysis
3.3.1. RNA Extraction
3.3.2. RNA-seq and Analysis
3.4. Proteomic Analysis
3.4.1. Protein Extraction and Trypsin Digestion
3.4.2. LC-MS/MS Analysis
3.4.3. Protein Identification and Quantitation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Weekly Epidemiological Record; WHO: Geneva, Switzerland, 2020; Volume 95, pp. 1–12. [Google Scholar]
- WHO. Weekly Epidemiological Record No 45; WHO: Geneva, Switzerland, 2006; Volume 81, pp. 425–432. [Google Scholar]
- WHO. Treatment of TB: Guidelines for National Programmes; WHO/TB/97.220; WHO: Geneva, Switzerland, 1997. [Google Scholar]
- Calver, A.D.; Falmer, A.A.; Murray, M.; Strauss, O.J.; Streicher, E.M.; Hanekom, M.; Liversage, T.; Masibi, M.; Van Helden, P.D.; Warren, R.M.; et al. Emergence of increased resistance and extensively drug-resistant tuberculosis despite treatment adherence, South Africa. Emerg. Infect. Dis. 2010, 16, 264–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dye, C. Doomsday postponed? Preventing and reversing epidemics of drug-resistant tuberculosis. Nat. Rev. Microbiol. 2009, 7, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Borrell, S.; Rose, G.; Gagneux, S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 2013, 29, 160–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didelot, X.; Walker, A.S.; Peto, T.E.; Crook, D.W.; Wilson, D.J. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 2016, 14, 150–162. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.; Luo, T.; Yang, C.; Dong, X.; Li, J.; Zhu, Y.; Zheng, H.; Tian, W.; Wang, S.; Barry, C.E.; et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 2012, 206, 1724–1733. [Google Scholar] [CrossRef]
- Merker, M.; Kohl, T.A.; Roetzer, A.; Truebe, L.; Richter, E.; Rüsch-Gerdes, S.; Fattorini, L.; Oggioni, M.R.; Cox, H.; Varaine, F.; et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS ONE 2013, 8, e82551. [Google Scholar] [CrossRef] [Green Version]
- Eldholm, V.; Norheim, G.; von der Lippe, B.; Kinander, W.; Dahle, U.R.; Caugant, D.A.; Mannsåker, T.; Mengshoel, A.T.; Dyrhol-Riise, A.M.; Balloux, F. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 2014, 15, 490. [Google Scholar] [CrossRef] [Green Version]
- Ley, S.D.; de Vos, M.; Van Rie, A.; Warren, R.M. Deciphering Within-Host Microevolution of Mycobacterium tuberculosis through Whole-Genome Sequencing: The Phenotypic Impact and Way Forward. Microbiol. Mol. Biol. Rev. 2019, 83, e00062-18. [Google Scholar] [CrossRef] [Green Version]
- Davies, A.P.; Billington, O.J.; McHugh, T.D.; Mitchison, D.A.; Gillespie, S.H. Comparison of phenotypic and genotypic methods for pyrazinamide susceptibility testing with Mycobacterium tuberculosis. J. Clin. Microbiol. 2000, 38, 3686–3688. [Google Scholar] [CrossRef] [Green Version]
- María Nieto, L.R.; Mehaffy, C.; Dobos, K.M. Comparing isogenic strains of Beijing genotype Mycobacterium tuberculosis after acquisition of Isoniazid resistance: A proteomics approach. Proteomics 2016, 16, 1376–1380. [Google Scholar] [CrossRef]
- Danelishvili, L.; Shulzhenko, N.; Chinison, J.J.J.; Babrak, L.; Hu, J.; Morgun, A.; Burrows, G.; Bermudeza, L.E. Mycobacterium tuberculosis proteome response to antituberculosis compounds reveals metabolic “escape” pathways that prolong bacterial survival. Antimicrob. Agents Chemother. 2017, 61, e00430-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.G.; Teo, A.S.M.; Wang, S.Y. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2001, 45, 2157–2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finken, M.; Kirschner, P.; Meier, A.; Wrede, A.; Böttger, E.C. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 1993, 9, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Bespyatykh, J.; Shitikov, E.; Butenko, I.; Altukhov, I.; Alexeev, D.; Mokrousov, I.; Dogonadze, M.; Zhuravlev, V.; Yablonsky, P.; Ilina, E.; et al. Proteome analysis of the Mycobacterium tuberculosis Beijing B0/W148 cluster. Sci. Rep. 2016, 6, 28985. [Google Scholar] [CrossRef] [Green Version]
- Lempens, P.; Meehan, C.J.; Vandelannoote, K.; Fissette, K.; De Rijk, P.; Van Deun, A.; Rigouts, L.; De Jong, B.C. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by high-confidence resistance-conferring mutations. Sci. Rep. 2018, 8, 3246. [Google Scholar] [CrossRef]
- Banerjee, A.; Dubnau, E.; Quemard, A.; Balasubramanian, V.; Um, K.S.; Wilson, T.; Collins, D.; De Lisle, G.; Jacobs, W.R. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 1994, 263, 227–230. [Google Scholar] [CrossRef] [Green Version]
- Ducasse-Cabanot, S.; Cohen-Gonsaud, M.; Marrakchi, H.; Nguyen, M.; Zerbib, D.; Bernadou, J.; Daffé, M.; Labesse, G.; Quémard, A.A. In Vitro Inhibition of the Mycobacterium tuberculosis-Ketoacyl-Acyl Carrier Protein Reductase MabA by Isoniazid. Antimicrob. Agents Chemother. 2004, 48, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Castro-Garza, J.; García-Jacobo, P.; Rivera-Morales, L.G.; Quinn, F.D.; Barber, J.; Karls, R.; Haas, D.; Helms, S.; Gupta, T.; Blumberg, H.; et al. Detection of anti-HspX antibodies and HspX protein in patient sera for the identification of recent latent infection by Mycobacterium tuberculosis. PLoS ONE 2017, 12, e0181714. [Google Scholar] [CrossRef]
- Dhiman, A.; Haldar, S.; Mishra, S.K.; Sharma, N.; Bansal, A.; Ahmad, Y.; Kumar, A.; Sharma, T.K.; Tyagi, J.S. Generation and application of DNA aptamers against HspX for accurate diagnosis of tuberculous meningitis. Tuberculosis 2018, 112, 27–36. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, W.; Gao, F.; Huang, Y.; Lv, C.; Wang, H. Comparison of the proteome of isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis. Microb. Drug Resist. 2006, 12, 231–238. [Google Scholar] [CrossRef]
- Phong, T.Q.; Ha, D.T.T.; Volker, U.; Hammer, E. Using a Label Free Quantitative Proteomics Approach to Identify Changes in Protein Abundance in Multidrug-Resistant Mycobacterium tuberculosis. Indian J. Microbiol. 2015, 55, 219–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Kumar, B.; Gupta, Y.; Singhal, N.; Katoch, V.M.; Venkatesan, K.; Bisht, D. Proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis. Proteome Sci. 2010, 8, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bespyatykh, J.; Shitikov, E.; Zgoda, V.; Smolyakov, A.; Zamachaev, M.; Dogonadze, M.; Zhuravlev, V.; Ilina, E. Changes in metabolism of Mycobacterium tuberculosis Beijing B0/W148 cluster against the background of anti-tuberculosis therapy. FEBS J. 2017, 284 (Suppl. 1), 347–347. [Google Scholar]
- Dasgupta, N.; Kapur, V.; Singh, K.K.; Das, T.K.; Sachdeva, S.; Jyothisri, K.; Tyagi, J.S. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 2000, 80, 141–159. [Google Scholar] [CrossRef] [PubMed]
- Converse, P.J.; Karakousis, P.C.; Klinkenberg, L.G.; Kesavan, A.K.; Ly, L.H.; Allen, S.S.; Grosset, J.H.; Jain, S.K.; Lamichhane, G.; Manabe, Y.C.; et al. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 2009, 77, 1230–1237. [Google Scholar] [CrossRef] [Green Version]
- Cao, G.; Howard, S.T.; Zhang, P.; Hou, G.; Pang, X. Functional analysis of the EspR binding sites upstream of espR in Mycobacterium tuberculosis. Curr. Microbiol. 2013, 67, 572–579. [Google Scholar] [CrossRef]
- Koul, A.; Choidas, A.; Treder, M.; Tyagi, A.K.; Drlica, K.; Singh, Y.; Ullrich, A. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 2000, 182, 5425–5432. [Google Scholar] [CrossRef] [Green Version]
- Kraeva, N.; Leštinová, T.; Ishemgulova, A.; Majerová, K.; Butenko, A.; Vaselek, S.; Bespyatykh, J.; Charyyeva, A.; Spitzová, T.; Kostygov, A.Y.; et al. LmxM.22.0250-Encoded Dual Specificity Protein/Lipid Phosphatase Impairs Leishmania mexicana Virulence In Vitro. Pathogens 2019, 8, 241. [Google Scholar] [CrossRef] [Green Version]
- Benjak, A.; Sala, C.; Hartkoorn, R.C. Whole-transcriptome sequencing for high-resolution transcriptomic analysis in Mycobacterium tuberculosis. Methods Mol. Biol. 2015, 1285, 17–30. [Google Scholar]
- Van Embden, J.D.; Cave, M.D.; Crawford, J.T.; Dale, J.W.; Eisenach, K.D.; Gicquel, B.; Hermans, P.; Martin, C.; McAdam, R.; Shinnick, T.M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J. Clin. Microbiol. 1993, 31, 406–409. [Google Scholar] [CrossRef] [Green Version]
- Mokrousov, I.; Vyazovaya, A.; Zhuravlev, V.; Otten, T.; Millet, J.; Jiao, W.W.; Shen, A.D.; Rastogi, N.; Vishnevsky, B.; Narvskaya, O. Real-time PCR assay for rapid detection of epidemiologically and clinically significant Mycobacterium tuberculosis Beijing genotype isolates. J. Clin. Microbiol. 2014, 52, 1691–1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bespyatykh, J.A.; Zimenkov, D.V.; Shitikov, E.A.; Kulagina, E.V.; Lapa, S.A.; Gryadunov, D.A.; Ilina, E.N.; Govorun, V.M. Spoligotyping of Mycobacterium tuberculosis complex isolates using hydrogel oligonucleotide microarrays. Infect. Genet. Evol. 2014, 26, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Supply, P.; Allix, C.; Lesjean, S.; Cardoso-Oelemann, M.; Rüsch-Gerdes, S.; Willery, E.; Savine, E.; de Haas, P.; van Deutekom, H.; Roring, S.; et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006, 44, 4498–4510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bespyatykh, J.; Shitikov, E.; Andrei, G.; Smolyakov, A.; Klimina, K.; Veselovsky, V.; Malakhova, M.; Georgij, A.; Dogonadze, M.; Manicheva, O.; et al. System OMICs analysis of Mycobacterium tuberculosis Beijing B0/W148 cluster. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Gonzalez, R.H.; Bonnal, R.; Caccamo, M.; Maclean, D. Bio-samtools: Ruby bindings for SAMtools, a library for accessing BAM files containing high-throughput sequence alignments. Source Code Biol. Med. 2012, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Seemann, T. GitHub-Tseemann/Snippy: Rapid Haploid Variant Calling and Core Genome Alignment. 2015. Available online: https://github.com/tseemann/snippy (accessed on 23 January 2020).
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-Metagenomes from Highly Chimeric Reads; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar]
- Schleusener, V.; Köser, C.U.; Beckert, P.; Niemann, S.; Feuerriegel, S. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: Comparison of automated analysis tools. Sci. Rep. 2017, 7, 46327. [Google Scholar] [CrossRef] [Green Version]
- Hawkey, J.; Hamidian, M.; Wick, R.R.; Edwards, D.J.; Billman-Jacobe, H.; Hall, R.M.; Holt, K.E. ISMapper: Identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genom. 2015, 16, 667. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinform. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 January 2020).
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, D.R. Degust: Interactive RNA-seq Analysis. Available online: http://victorian-bioinformatics-consortium.github.io/degust/ (accessed on 20 January 2020).
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bespyatykh, J.; Smolyakov, A.; Guliaev, A.; Shitikov, E.; Arapidi, G.; Butenko, I.; Dogonadze, M.; Manicheva, O.; Ilina, E.; Zgoda, V.; et al. Proteogenomic analysis of Mycobacterium tuberculosis Beijing B0/W148 cluster strains. J. Proteom. 2019, 192, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. 1995, 57, 289–300. [Google Scholar] [CrossRef]
| Drug* (Resistance Associated Gene/Protein) | Clinical Isolates | ||
| RUS_B0 | 3955 | 2093 | |
| STR (Rpsl) | R (K43R) | R (K43R) | R (K43R) |
| INH (KatG) (inhA) | R (S315T) | R (S315T) | R (S315T) R+** (t-8a) |
| RIF (RpoB) | R (S450L) | R (S450L) | R (S450L) |
| EMB (EmbB) | R (Q497R) | R (Q497R) | R (Q497R) |
| ETH (ethA) | R (110delA) | R (110delA) | R (110delA) |
| FQ (GyrA) | S | R (D94A) | R (D94A) |
| KAN, CAP, AM (rrs) | R (a1401g) | R (a1401g) | R (a1401g) |
| PZA (pncA) | R (t-11c) | R (t-11c) | R (t-11c) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bespyatykh, J.; Shitikov, E.; Bespiatykh, D.; Guliaev, A.; Klimina, K.; Veselovsky, V.; Arapidi, G.; Dogonadze, M.; Zhuravlev, V.; Ilina, E.; et al. Metabolic Changes of Mycobacterium tuberculosis during the Anti-Tuberculosis Therapy. Pathogens 2020, 9, 131. https://doi.org/10.3390/pathogens9020131
Bespyatykh J, Shitikov E, Bespiatykh D, Guliaev A, Klimina K, Veselovsky V, Arapidi G, Dogonadze M, Zhuravlev V, Ilina E, et al. Metabolic Changes of Mycobacterium tuberculosis during the Anti-Tuberculosis Therapy. Pathogens. 2020; 9(2):131. https://doi.org/10.3390/pathogens9020131
Chicago/Turabian StyleBespyatykh, Julia, Egor Shitikov, Dmitry Bespiatykh, Andrei Guliaev, Ksenia Klimina, Vladimir Veselovsky, Georgij Arapidi, Marine Dogonadze, Viacheslav Zhuravlev, Elena Ilina, and et al. 2020. "Metabolic Changes of Mycobacterium tuberculosis during the Anti-Tuberculosis Therapy" Pathogens 9, no. 2: 131. https://doi.org/10.3390/pathogens9020131
APA StyleBespyatykh, J., Shitikov, E., Bespiatykh, D., Guliaev, A., Klimina, K., Veselovsky, V., Arapidi, G., Dogonadze, M., Zhuravlev, V., Ilina, E., & Govorun, V. (2020). Metabolic Changes of Mycobacterium tuberculosis during the Anti-Tuberculosis Therapy. Pathogens, 9(2), 131. https://doi.org/10.3390/pathogens9020131






